Abstract
Autophagy is an evolutionarily conserved lysosomal pathway for degrading cytoplasmic proteins, macromolecules, and organelles. While autophagy has become one of the most attractive topics in cancer research, the current autophagy literature is often viewed as confusing, because of its association with apparently contradictory roles, such as survival and cell death. Autophagy can serve as a tumor suppressor, as a partial reduction in autophagic capacity or defective autophagy (e.g., heterozygous knockdown BECN1 (+/−) in mice) provides an oncogenic stimulus, causing malignant transformation and spontaneous tumors. In addition, autophagy seems to function as a protective cell survival mechanism against environmental and cellular stress (e.g., nutrient deprivation, hypoxia and therapeutic stress) and causes resistance to antineoplastic therapies. Recent studies have demonstrated that the inhibition of autophagy in cancer cells may be therapeutically beneficial in some circumstances, as it can sensitize cancer cells to different therapies, including DNA-damaging agents, antihormone therapies (e.g., tamoxifen), and radiation therapy. This supports the hypothesis that inhibiting autophagy can negatively influence cancer cell survival and increase cell death when combined with anticancer agents, providing a therapeutic advantage against cancer. On the other hand, the induction of autophagy by the inhibition of anti-autophagic proteins, such as Bcl-2, PKCδ, and tissue transglutaminase 2 (TG2), may lead to autophagic cell death in some apoptosis-resistant cancers (i.e., breast and pancreatic cancers), indicating that the induction of autophagy alone may also be used as a potential therapy. Overall, the data suggest that, depending on the cellular features, either the induction or the inhibition of autophagy can provide therapeutic benefits to patients and that the design and synthesis of the first-generation modulators of autophagy may provide the tools for proof of concept experiments and the impetus for translational studies that may ultimately lead to new therapeutic strategies in cancer.
Keywords: autophagy, programmed cell death, apoptosis, Bcl-2, Beclin 1, siRNA, small-molecule inhibitors, cancer
Introduction
Autophagy is an evolutionarily conserved and highly regulated process of large-scale lysosomal degradation of long-lived proteins, macromolecules, ribosomes, and organelles, such as the endoplasmic reticulum, Golgi apparatus, and mitochondria. The term autophagy is derived from the Greek roots “auto” (self) and “phagy” (to eat) and means self-digestion. It is considered to be a physiological mechanism that may serve as a means of temporary survival, and is triggered by starvation (amino acid and nutrient deprivation), hypoxia, and metabolic stress. Self-digestion provides a means of recycling macromolecules as an alternative energy source; however, if the cellular stress leads to continuous or excessively induced autophagy, cell death may ensue. Autophagy requires the sequestration of cytoplasmic content through the formation of double-membrane vesicles, mediated by a highly organized and hierarchical team of ATG proteins.1 The initial phagophores are formed from the endoplasmic reticulum and surround and pack organelles to form autophagosomes.1,2 Subsequently, autophagosomes merge with lysosomes and digest the contents (i.e., misfolded proteins and organelles), generating building blocks for the synthesis of macromolecules and metabolites for use as an energy source, eventually leading to either cell survival or cell death depending on the duration and severity of the process.1,2
The current autophagy literature, however, is often viewed as confusing because, depending on cell type and context, macroautophagy (autophagy from here on) seems to have different roles. For example, in fully transformed cancer cells it appears to function as a tumor suppressor as defective autophagy is associated with malignant transformation and carcinogenesis. However, in normal cells and in some cancer cells, it appears to function as a protective mechanism against cellular stress and yet the induction of autophagy is associated with cell death in some type of cancers.
Autophagy as a Tumor Suppressor Mechanism
Cancer cells often display a reduced autophagic capacity compared to their normal counterparts. Studies have shown that cancer cells express lower levels of the autophagy-related proteins LC3-II and Beclin 1 than normal epithelial cells,3,4 and that while heterozygous disruption of BECN1 promotes tumorigenesis4 the overexpression inhibits tumorigenesis,5 supporting the contention that defective autophagy or the inhibition of autophagy plays a role in malignant transformation. The BECN1 gene is deleted in about 40% of prostate, 50% of breast, and 75% of ovarian cancers.5–7 In addition, reduced expression of BECN1 has been reported in other types of cancers, including human colon cancer,8 brain tumors,9 hepatocellular carcinomas,10 and cervical cancers.11 Overall, the data suggest that a defective autophagic process is clearly linked to cancer development.
The most important evidence linking dysfunctional autophagy and cancer comes from studies demonstrating that the inhibition of autophagy in mice, by disruption of BECN1, increases cellular proliferation, increases the frequency of spontaneous malignancies (i.e., lung cancer, liver cancer, and lymphomas) as well as mammary hyperlasia and accelerates the development of carcinogen (i.e., hepatitis B virus)-induced premalignant lesions. In addition, transfection of MCF-7 breast cancer cells, that express low levels of Beclin 1, with the BECN1 gene, inhibits growth and tumor formation,5 further suggesting that Beclin 1 is a haploinsufficient tumor suppressor and that the defective autophagy may be critical for the malignant transformation of cells.4,12,13 Furthermore, defects in autophagy (e.g., reduced BECN1 expression) have not only been shown to be associated with a malignant phenotype, but also poor prognosis of cancer patients with hepatocellular carcinoma.14
Overall, the available evidence suggests that the expression of autophagic genes and their corresponding autophagic activities are frequently suppressed in cancer cells and that autophagy plays a critical role in oncogenesis as a tumor suppressor, which has an impact on patient response to therapy. Therefore, the induction of autophagy may help to reverse the malignant phenotype.
Autophagy as a Prosurvival and Resistance Mechanism against Cellular Stress and Cancer Therapeutics
The physiological function of autophagy is related to the maintenance of cellular homeostasis under cellular stress. Utilizing autophagy as a survival mechanism in the harsh tumor microenvironment, which is highly hypoxic and acidic, may work in favor of cancer cells. Indeed, some types of cancer cells may exploit autophagy as a means to adapt to the hypoxic, nutrient-limiting, and metabolically stressful tumor microenvironment and therapeutically induced cell stress or damage.15 A number of antineoplastic therapies, including radiation therapy, chemotherapy (e.g., doxorubicin, temozolomide, etoposide), histone deaceltylase inhibitors, arsenic trioxide, TNFα, IFNγ, imatinib, rapamycin, and anti-estrogen hormonal therapy (e.g., tamoxifen), have been observed to induce autophagy as a protective and prosurvival mechanism in human cancer cell lines (reviewed in refs. 3 and 16). In fact, the therapeutic efficacy of these agents can be increased if autophagy is inhibited.17–26 In addition, hypoxia induces autophagy that is independent of HIF1α, suggesting that autophagy is involved in cancer cell survival by increasing resistance to hypoxic stress in the tumor microenvironment. These findings support the hypothesis that these therapies induce a type of protective, prosurvival autophagy, which increases the cancer cells’ resistance to therapies, and that inhibition of autophagy may lead to increased cell death and inhibition of tumor growth.
Autophagy as a Prodeath and Type II Programmed Cell Death Mechanism
Apoptosis (programmed cell death-type I, PCD-type I) and necrosis are a well-known mechanism of cell death induced by anticancer therapies. Until recently, apoptosis was a synonym for programmed cell death and thought to be the major mechanism of cell death in response to chemo- and radiation therapy. However, emerging studies have demonstrated the existence of a nonapoptotic form of programmed death called autophagic cell death, which is now considered as a PCD-type II. In contrast to apoptosis, autophagic cell death, in general, is caspase-independent, does not involve classic DNA laddering and is believed to be a result of an extensive autophagic degradation of intracellular content.27
Studies show that cytotoxic signals can induce autophagy in cells that are resistant to apoptosis (apoptosis defective), such as those expressing high Bcl-2 or Bcl-XL, those lacking Bax and Bak, or those being exposed to pan-caspase inhibitors, such as zVAD-fmk.31 The proapoptotic Bcl-2 family member proteins Bak and Bax regulate the intrinsic apoptotic pathway by causing mitochondrial outer membrane permeabilization and cyctochrome c release. Bax−/− and Bak−/− knockout fibroblast cells are resistant to apoptosis and undergo autophagic cell death after the induction of death, following starvation, growth factor withdrawal, chemotherapy (etoposide) or radiation.28,31 The evidence suggests that autophagy leads to cell death in response to several compounds, including rottlerin29 (see also the next section for detailed mechanism) cytosine arabinoside,30 etoposide and staurosporine31 as well as growth-factor deprivation.30 A link between autophagy and related autophagic cell death has been demonstrated using pharmacological (e.g., 3-MA) and genetic (silencing of ATG5, ATG7 and BECN1) approaches for suppression of autophagy. For example, the knockdown of ATG5 or BECN1 in cancer cells containing defects in apoptosis leads to a marked reduction in autophagic cell death (and autophagic response) in response to cell death stimuli with no sign of apoptosis.29,32 Studies also suggest that apoptosis and autophagy are linked by effector proteins (e.g., Bcl-2, Bcl-XL, Mcl-1, ATG5, p53) and common pathways (e.g., PI3K/Akt/mTOR, NFκB, ERK).31–35 Overall, there is evidence that autophagy may function as a PCD-type II in cancer cells in which apoptosis is defective or hard to induce. Therefore it is reasonable to propose the notion that the induction of autophagic cell death may be used as a therapeutic strategy to treat cancer.
Targeting Autophagy as a Novel Cancer Therapy
Current data suggest that depending on the context, either the induction or the inhibition of autophagy can provide therapeutic benefits to patients and that the design and synthesis of modulators of autophagy may provide novel therapeutic tools and may ultimately lead to new therapeutic strategies in cancer.
Defects in apoptosis lead to increased resistance to chemotherapy, radiotherapy, some anticancer agents, and targeted therapies. Therefore, induction of autophagic cell death may be an ideal approach in those cancers that are resistant to apoptosis by anticancer therapies (e.g., chemotherapy, radiation). As explained in the previous section, cancer cells can undergo autophagic cell death when their apoptosis is inhibited, or they are resistant to therapy-induced apoptosis (e.g., in response to DNA-damaging agents such as etoposide), suggesting that autophagic cell death can be induced as an alternative cell-death mechanism when cells fail to undergo apoptosis. Therefore, induction of autophagic cell death may serve as a novel therapeutic tool to eliminate cancer cells with defective apoptosis, which is the case in many advanced, drug-resistant and metastatic cancers. We have recently demonstrated that the inhibition of some protein kinases (e.g., PKCδ in pancreatic cancer) or the targeting of key proteins that are involved in the suppression of autophagy (e.g., Bcl-2, TG2) can trigger autophagic cell death without any other treatment.
On the other hand, because a number of cancer therapies, such as radiation therapy, chemotherapy and targeted therapies (e.g., imatinib) induce autophagy as a protective resistance mechanism against anticancer therapies for cancer cell survival, the inhibition of autophagy can be used to enhance the efficacy of anticancer therapies.
1) Induction of autophagic cell death as a therapeutic strategy
a. Pancreatic cancer
Pancreatic adenocarcinoma, which is one of the most aggressive malignant diseases with one of the poorest prognoses (median survival time of 6 to 12 months), is virtually incurable. The major reason for the poor prognosis is that pancreatic cancer patients fail to undergo apoptosis due to intrinsic resistance to death receptor- and mitochondria-initiated apoptosis and do not respond to most conventional therapies, such as chemotherapy and radiotherapy. We hypothesize that induction of autophagic cell death may be a useful alternative approach to killing tumor cells, such as these that are notoriously resistant to virtually all chemotherapy- and radiation-induced apoptosis.
Pancreatic adenocarcinomas usually have dysregulation of the signaling pathways involved in apoptosis mediated by death ligands and cytotoxic drugs, which render this malignancy highly resistant to chemotherapy or radiotherapy, and consequently contribute to its rapid fatal course (please see ref. 49 for further details). Dysregulation of the expression of several antiapoptotic Bcl-2 family protein members, including Bcl-2, Bcl-XL, or Mcl-1, has been demonstrated, and shown to protect the pancreatic cancer cells from apoptosis induced by death ligands or chemotherapeutic drugs.49 Tyrosine kinase growth factor receptors and their ligands play different roles in cell proliferation, differentiation, invasion, metastasis, and angiogenesis. In pancreatic cancer a variety of these growth factor receptors are overexpressed, influencing the prognosis of the disease. For example, the concomitant presence of the epidermal growth factor receptor (EGFR) and its ligands EGF and amphiregulin is associated with enhanced tumor aggressiveness and shorter survival in patients.49 K-ras proto-oncogene and mutations of the p53 tumor-suppressor gene and the Smad4 gene are frequently dysregulated in the tumors. Overall, dysregulation of growth-promoting factors, growth inhibitory pathways, and gene mutations, combine to give pancreatic cancer cells a growth advantage, resistance to cell death by therapies and eventually faster progression and poor survival.
We recently reported that protein kinase C delta (PKCδ) positively regulates the expression of tissue transglutaminase 2 (TG2) which in turn constitutively suppresses autophagic cell death, suggesting a novel mechanism for the regulation of autophagic cell death.29,36 These findings provided the first evidence that the inhibition or knockdown of either PKCδ or TG2 induces massive autophagic cell death without induction of apoptosis and support the contention that the inhibition of molecules that suppress autophagy leads to autophagic cell death. We and others found that TG2 is overexpressed in the majority (> 80%) of pancreatic cancer cell lines and in tumors from patients with pancreatic cancer.29,37 Most importantly, increased levels of TG2 have been implicated in drug resistance and metastatic phenotypes in pancreatic, breast, and ovarian cancers as well as melanoma.37–39 TG2 binds to important tumor-suppressor proteins, including Rb and p53, which is involved in the regulation of autophagy, and can translocate to the nucleus where it associates with histones to regulate certain cellular functions.40 It is of interest to note that we found that the PKCδ/TG2 axis regulates downstream targets, including Bcl-2, NFκB, and phosphatidyl inositole-3-kinase (PI3K)/Akt/mTOR/P70S6K, signaling pathways that are known to be involved in the suppression of autophagy.36 Taken together, data from our studies and others suggest that targeting PKCδ and/or TG2 to induce autophagic cell death may be a promising therapeutic strategy for the treatment of pancreatic cancer. In support of this idea, we have also shown that the in vivo down-regulation of TG2 by liposomal small interfering RNA (siRNA), as a single agent or in combination with gemcitabine, inhibited tumor growth in nude mice bearing orthotopically implanted TG2-expressing human pancreatic cancer cells.41 Silencing of TG2 in these xenografts significantly inhibited cancer cell proliferation and growth of TG2-positive tumors.41 As expected, we have not found induction of apoptosis in tumors by tunnel assay analysis following knockdown of TG2 by nanoliposomal siRNA in vivo, suggesting that the treatment in fact induced autophagic death rather than apoptosis as demonstrated in in vitro studies using the same cell line.36 Overall, these studies suggest that induction of autophagic death may be a viable therapeutic strategy in apoptosis-resistant tumors.
b. Breast cancer
Breast cancer is the most common cancer and the second leading cause of death in the United States. The Bcl-2 protooncogene is overexpressed in half of all human malignancies and more than 60% of breast cancers and is thought to exert its oncogenic role by preventing cells from undergoing apoptosis. 42–44 Bcl-2 overexpression leads not only to the resistance of cancer cells towards chemotherapy, radiation, and hormone therapy but also to an aggressive tumor phenotype in patients with a variety of cancers.43,44
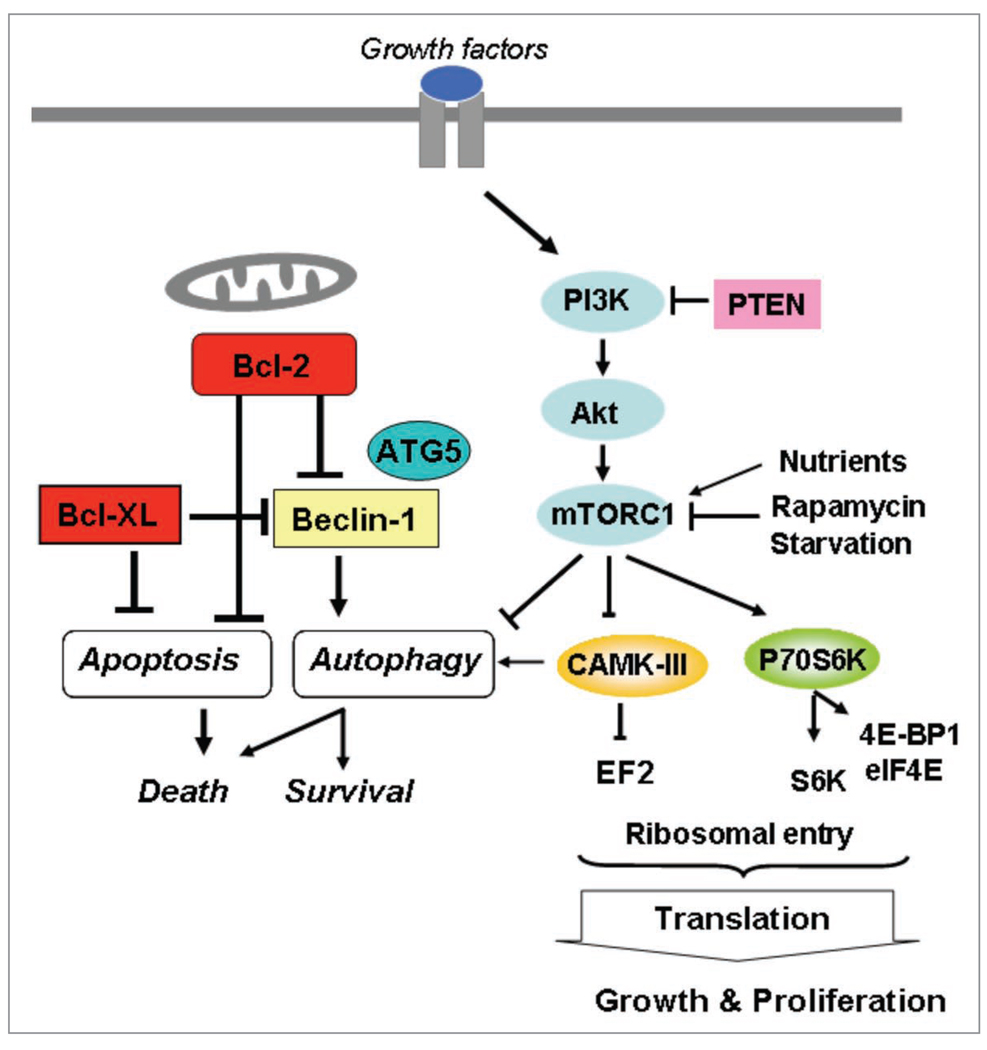
Our recent findings suggest that silencing Bcl-2 expression using siRNA in MCF-7 breast cancer cells led to significant autophagic, but not apoptotic cell death.32 We demonstrated that the knockdown of autophagy genes (e.g., ATG5 and BCN1) significantly inhibited both autophagy and cell death induced by Bcl-2 siRNA after a long-term treatment of up to seven days,32 suggesting that Bcl-2 knockdown causes autophagic cell death. MCF-7 cells are known to be caspase 3-deficient, providing a higher threshold for the induction of apoptosis, potentially rendering an autophagic cell death pathway more important. Furthermore, about 45% to 75% of tumor tissues from breast cancer patients do not have detectable caspase 3 expression,45,46 consistent with the notion that the inhibition of Bcl-2 in patient tumors causes autophagic cell death, and further highlighting the significance of our finding in MCF-7 cells. In addition, we reported that doxorubicin predominantly induces autophagy at low doses and apoptosis at high doses. Furthermore, the combination of Bcl-2 siRNA treatment with a low dose of doxorubicin enhanced the autophagic response, tumor growth inhibition, and cell death.32 These results provided the first evidence that targeted silencing of Bcl-2 induces autophagic cell death in breast cancer cells and that targeting Bcl-2 may be used as a therapeutic strategy alone (Fig. 1) or in combination with chemotherapy in patients with breast cancer cells that have higher apoptotic thresholds due to caspase 3 deletion and that overexpress Bcl-2.
Figure 1.
Regulation of autophagy and apoptosis is intertwined. The crosstalk between these through Bcl-2 and Bcl-XL may determine the predominant response to anticancer therapies. Downstream targets of mTor, EF2K (a.k.a.CAMK-III) and P70S6K, play a role in the regulation of translation and autophagy.
2) Inhibition of autophagy as a therapeutic approach for enhancing efficacy of anticancer therapies
Many studies have described the role of autophagy as a protective mechanism for cell survival that is initiated in response to metabolic or therapeutic stress. Recent studies suggest that the inhibition of autophagy may be a useful approach in terms of enhancing cell death caused by antineoplastic agents. Autophagy may delay apoptotic cell death caused by DNA-damaging agents and hormonal therapies such as tamoxifen.20–26,47,48 These studies have demonstrated that autophagy can be inhibited by knockdown of autophagy-related genes (ATG5, ATG6/BECN1, ATG10 and ATG12) using siRNA, or treatment of cells with pharmacological inhibitors e.g., 3-methyladenine, hydroxychloroquine, bafilomycin A1, or monensin), indicating that inhibition of autophagy may enhance the efficacy of current therapies. Therefore, we propose that inhibitors of autophagy may either enhance the efficacy of anti-tumor therapy or promote cell death, not only in primary cancer types but also in advanced-stage cancers and metastatic tumors that are considered drug resistant or apoptosis resistant, such as chemotherapy-resistant cancers. Both approaches, inhibition of either the prosurvival or induction of prodeath mechanism of autophagy, are supported by our own published and unpublished studies as well as several published papers, which will be discussed further (for further information please refer to a review by Rubinstein et al., 2007, ref. 87).
Pros and Cons of Therapeutically Targeting the Prodeath and Prosurvival Functions of Autophagy
Because autophagy may play a role as a cell survival pathway in response to therapeutic and cellular stresses in the tumor microenvironment (which is highly acidic and hypoxic) the induction of autophagy may work in favor of cancer cells.15 Therefore, inhibition of protective autophagy may break the resistance mechanism for survival of the harsh tumor microenvironment and lead to cell death. In addition, cancer therapies, such as radiation therapy, chemotherapy and targeted therapies (e.g., Imatinib), induce autophagy as a protective or survival mechanism against anticancer therapies by sequestering damaged mitochondria for cancer cell survival. The inhibition of autophagy may enhance efficacy of anticancer therapies. This approach may also lead to some side effects, however, since the inhibition of autophagy by pharmacological inhibitors or by genetically targeting autophagy genes may lead to unexpected consequences, such as decreased genomic stability. Such concerns may be alleviated if therapies are administered at a certain therapeutic window to minimize side effects to normal tissues or such therapies are specifically targeted to tumor cells using nanotechnology or tumor-targeting carriers.
On the other hand, those cancer cells that are resistant to apoptosis-inducing therapies, including radiation, chemotherapy, and growth factor antagonists can be killed by agents that induce massive autophagic response, which eventually kills cells by autophagic cell death (or type II-PCD) as an alternative death pathway without caspase activation. This approach may be highly useful for specifically advanced cancer and therapy-resistant cancers. In addition to persistent and massive autophagy, induced therapies may lead to autophagic cell death;3 autophagy may trigger cell death through the induction of apoptosis or necrosis in response to a selective targeting of a growth factor-inducing signal.84 Targeting cancer cells that often have dysregulated or reduced autophagic capacity by these type of therapies may not affect normal cells due to a tightly controlled autophagic response. In this regard, the ideal targets would be those overexpressed in cancer cells and those negatively regulate autophagy. Identifying targets whose manipulation may induce death-promoting activity of autophagy needs to be further studied as described below.
Methods for Developing Strategies for Targeting Autophagy
Several approaches have been used to target autophagy in the literature. These include (1) Silencing of genes involved in autophagy (e.g., BECN1/ATG6, ATG5, ATG7 and ATG8); (2) inhibition of signaling pathways regulating autophagy (PI3K/Akt/mTOR pathway); (3) Inhibition of autophagosome formation and function (bafilomycin A, hydrochloroquine). However, novel targets may be identified to induce or block autophagy in cancer cells.
Identification of Autophagy-Related Genes by siRNA Screening
The key components of autophagy signaling and genes involved in the formation of the autophagosome may be blocked by small interfering RNA. This technology holds promise as a therapeutic intervention and in functional studies involving the targeted silencing of genes in cancer cells.50 The function of these genes can be studied by transfecting cancer cells with a double-stranded siRNA targeting each gene and assessing a variety of end points, such as cell death, proliferation or certain phenotypes, following silencing of the gene. For example, the inhibition or induction of autophagy may be used to identify genes involved in autophagy using a well-established cell culture system that that has previously been transfected to express GFP-LC3. The accumulation or reduction of an autophagy-related punctate appearance (which indicates formation of autophagosomes containing the LC3-II marker protein) in cells after incubation with thousands of siRNA duplexes using screening technology may be used to identify genes mediating autophagic death and to screen for the compounds that enhance or block autophagy.51,52 For example, to identify compounds or treatments that inhibit autophagy, cells may be grown in 96-well plates transfected with GFP-LC3 and stimulated to undergo autophagy. Later individual wells may be analyzed for a reduction or induction of the punctate pattern formation following the treatment of each well with a specific siRNA duplex. Such a high-throughput approach may facilitate the rapid identification or monitoring of genes that are involved in the autophagic process. Similarly, one could easily identify compounds that modulate autophagy and thereby identify cellular pathways involved in this process. Recently, using siRNA to screen members of the kinome Chan et al., identified ULK1 as a multidomain modulator of autophagy.52 Once the genes mediating autophagy have been identified, cancer cells may be targeted using siRNA or small molecule inhibitors. Eventually, such agents are expected to find application in preclinical models and clinical settings.
Targeting Protein Kinases Using Small-Molecule Inhibitors
Inhibiting protective autophagy by targeting protein kinases that regulate autophagy using small-molecule inhibitors may be a useful approach. Currently, we are exploring the potential of using small molecules to inhibit selected protein kinases involved in the regulation of autophagy as a novel therapeutic strategy for cancer treatment. Despite enormous advances in our understanding of autophagy through studies in model organisms such as yeast, which have led to the characterization of many of the core components of the autophagy machinery, the signal transduction pathways that regulate autophagic processes are still unclear.53 While a number of protein kinases have been reported to regulate the induction of autophagy following nutrient deprivation or other cell stresses, to date, only the following protein kinases have been reported to induce ‘protective’ autophagy in cancer cells in response to cytotoxic agents: AMP-activated protein kinase (AMPK),54–57 jun N-terminal kinase (JNK),58,59 eukaryotic elongation factor-2 kinase (eEF2K),60 extracellular signal-regulated kinases 1 and 2 (ERK1/2),61,62 protein kinase C,59 PKR-like endoplasmic reticulum kinase (PERK),63,64 and glycogen synthase kinase 3 (GSK3) beta65 (Fig. 2).
Figure 2.
Protein kinase signaling pathways implicated in the regulation of autophagy. Inhibition of a particular kinase or kinases may help blocking induction of protective autophagy and enhance efficacy of anticancer therapies. On the other hand inhibition of some kinases, such as PKCδ, may lead to autophagic cell death in certain type of cancers.36
In some cases (e.g., AMPK and JNK), there is mechanistic evidence linking the kinases to known regulators of autophagy. For example, AMPK inhibits the phosphatidylinositol kinase homolog mammalian target of rapamycin (mTOR) under conditions of energy stress.55 mTOR is activated by nutrients and amino acids, as well as mitogenic signaling molecules such as insulin, and is an important negative regulator of autophagy.66 Inhibition of mTOR by AMPK is, therefore, believed to be the mechanism by which AMPK induces protective autophagy.67 JNK has been shown to promote the expression of the proautophagy protein BECN-1 in a c-Jun-dependent manner following ceramide treatment. 68 BECN-1 is an important autophagy protein, which is recruited to a multiprotein complex containing the lipid kinase Vps34, which induces nucleation of autophagosomes.69,70 JNK has also been shown to promote autophagy by inducing the release of BECN-1 from an inhibitory Bcl-2–BECN-1 complex.71,72 The protein serine/threonine kinase eEF2K is negatively regulated by mTOR and promotes autophagy in several tumor cells in response to cell stress,60,73,74 suggesting a possible link between the inhibition of global protein synthesis and the induction of autophagy. The mechanisms underlying the activity of ERK2, PKC, GSK3, and PERK are presently unknown.
Other protein kinases implicated in the promotion of autophagy, but not yet evaluated as therapeutic targets, include death-associated protein kinase (DAPK)75 and Unc-51-like kinase 1 (ULK-1)76,77. DAPK interacts with and is activated by the microtubule-associated protein MAP1B, which also binds the autophagy protein LC3. DAPK is reported to promote Vps34 activation by promoting the release of BECN1 from inhibitory complexes.78 ULK-1 appears to be a major downstream effector of mTOR and may be essential for the induction of autophagy.79 While further work is clearly necessary to evaluate the precise functions of these and other protein kinases, it is apparent that several protein kinases represent potentially promising targets for therapeutic intervention.
Identifying small-molecule inhibitors of protein kinases
Protein kinases comprise the largest enzyme family in the human genome, with just over 500 representatives. They share a conserved region of approximately 250 amino acids, known as the catalytic domain, which is required for their activity. The fold of this domain as well as a number of key catalytic residues is conserved, and the ATP binding site is formed in the cleft created by the two characteristic lobes of the catalytic domain. Most drug-development programs have focused on targeting inhibitors to this binding site, producing a host of novel molecules exhibiting favorable absorption, distribution, metabolism, and excretion properties, as well as high potency for the target protein kinase.
Protein kinases vary greatly in their complexity, often containing multiple domains of diverse function. However, for the purpose of screening for kinase inhibitors it is often possible to express and purify a truncated form of the kinase containing the catalytic domain. Most successful protein kinase inhibitors are designed to inhibit the ability of the protein kinase to bind ATP. A useful place to begin a search for a small-molecule ATP-competitive inhibitor is to screen a collection of potential lead compounds. A number of good-quality libraries of small molecules are commercially available in formats compatible with screening methods. These libraries may be ‘focused,’ exhibiting characteristics of known protein kinase inhibitors, or they may be collections of compounds that possess other favorable characteristics, such as high structural diversity, drug-like properties, and favorable Astex’s “Rule of Three” parameters.80
Protein kinases may be screened using a variety of assays to monitor their catalytic activity. An approach taken in our laboratory is to utilize the properties of the fluorescent moiety SOX.81,82 This moiety can be incorporated into a peptide substrate without greatly compromising the efficiency of the catalytic reaction.81,82 Monitoring the change in fluorescence of the SOX moiety conveniently follows the kinase reaction. Typically, a two- to six-fold change in fluorescence is observed upon full conversion of substrate to product, which is generally sufficient to robustly monitor reaction progress. Fluorescent SOX substrates are available commercially but are relatively easy to prepare in the laboratory.81,82 While this screening approach has only been used for a limited number of protein kinases, it is likely to be of wide applicability, as it can potentially be introduced into a number of protein kinase consensus sequences, without compromising the reaction. Once hits have been validated and their mechanism of inhibition established, traditional medicinal chemistry approaches are necessary to develop a lead compound into a potent bioactive inhibitor.
Concluding Remarks
Currently, the molecular mechanisms underlying the regulation of autophagy and the role of autophagy in cancer cells are not completely understood, but are beginning to be revealed. The reduced autophagic capacity seen in tumors, compared to their normal counterparts, may be linked to malignant transformation and cancer progression. The role of autophagy in established tumors may be related to the adaptation to hypoxic, nutrient-limiting and metabolically stressed environments as well as resistance to therapy-induced stress. Therefore, an understanding of the role of autophagy in cancer cell survival vs. prodeath may help in determining its therapeutic potential. While there is still much to learn about the molecular mechanisms underlying autophagy, it appears that modulators of autophagy may have a number of potential benefit. Inhibitors of autophagy may enhance the efficacy of currently used antineoplastic agents in cancer cells, while promoters of autophagy may induce autophagy-mediated cell death in cancers with high thresholds to apoptosis. Both strategies have significant potential to be translated into the clinic.
Acknowlegements
The projects mentioned here were supported by grants from Susan Komen for the Cure (KG080889) (BO) and by NCI grant (U54 RFA CA096300, GLB) by grants from the NIH (GM59802) (KND) and the Welch Foundation (F1390) (KND) and (H-F-0032) (KND).
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 3.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 4.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the Beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 6.Aita VM, Liang XH. Cloning and genomic organization of beclin1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 7.Shen Y, Li D, Wang L, Deng R, Zhu XF. Decreased expression of autophagy-related proteins in malignant epithelial ovarian cancer. Autophagy. 2008;4:1067–1068. doi: 10.4161/auto.6827. [DOI] [PubMed] [Google Scholar]
- 8.Koneri K, Goi T, Hirono Y. Beclin1 gene inhibits tumor growth in colon cancer cell lines. Anticancer Res. 2007;27:1453–1457. [PubMed] [Google Scholar]
- 9.Miracco C, Cosci E. Protein and mRNA expression of autophagy gene Beclin1 in human brain tumours. Int J Oncol. 2007;30:429–436. [PubMed] [Google Scholar]
- 10.Daniel F, Legrand A, Pessayre D. Beclin1 mRNA strongly correlates with Bcl-XLmRNA expression in human hepatocellular carcinoma. Cancer Invest. 2007;25:226–231. doi: 10.1080/07357900701206323. [DOI] [PubMed] [Google Scholar]
- 11.Wang ZH, Peng ZL, Duan ZL, et al. Expression and clinical significance of autophagy gene Beclin1 in cervical squamous cell carcinoma. Sichuan Da Xue Xue Bao Yi Xue Ban 200. 37:860–863. [PubMed] [Google Scholar]
- 12.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Becloin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edinger AL, Thompson CB. Defective autophagy leads to cancer. Cancer Cell. 2003;4:422–424. doi: 10.1016/s1535-6108(03)00306-4. [DOI] [PubMed] [Google Scholar]
- 14.Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z, Shi GM, Wang XY, Ke AW, Wu B, Fan J. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008;68:9167. doi: 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
- 15.Rouschop KM, Wouters BG. Regulation of autophagy through multiple independent hypoxic signaling pathways. Curr Mol Med. 2009;9:417–424. doi: 10.2174/156652409788167131. [DOI] [PubMed] [Google Scholar]
- 16.Djavaheri-Mergny M, Botti J, Codogno P. Autophagy and Autophagic Cell Death. In: Gewirtz A, editor. Apoptosis, Senescence, and Cancer. Vol. 93. 2007. p. 107. [Google Scholar]
- 17.Sotelo J, Briceno E, Lopez-Gonzalez MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144:337–343. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 18.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 19.Paglin S, Hollister T, Delohery T, Hackett N, McMahill M. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001;61:439–444. [PubMed] [Google Scholar]
- 20.Boya P, Gonzalez-Polo R-A, Casares N, Perfettini J-L, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Pierron G, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, Gorski SM. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112:389–403. doi: 10.1007/s10549-007-9873-4. [DOI] [PubMed] [Google Scholar]
- 22.Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 25.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 26.Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14:548–558. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- 27.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moretti K, Attia, Kim Lu. Crosstalk between Bak/Bax and mTOR signaling regulates radiation-induced autophagy. Autophagy. 2007;3:142–144. doi: 10.4161/auto.3607. [DOI] [PubMed] [Google Scholar]
- 29.Akar U, Ozpolat B, Mehta K, Fok F, Lopez Berestein G. Tissue transglutaminase (TG2) inhibits autophagy in pancreatic cancer cells. Mol Cancer Res. 2007;5:241–249. doi: 10.1158/1541-7786.MCR-06-0229. [DOI] [PubMed] [Google Scholar]
- 30.Xue L, Fletcher GC, Tolkovsky AM. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol Cell Neurosci. 1999;14:180–198. doi: 10.1006/mcne.1999.0780. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 32.Akar U, Chaves-Reyez A, Barria M, Tari A, Sanguino A, Kondo Y, Kondo S, Arun B, Lopez-Berestein G, Ozpolat B. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4:669–679. doi: 10.4161/auto.6083. [DOI] [PubMed] [Google Scholar]
- 33.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nature Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 35.Lockshin RA, Zakeri Z. Cell death in health and disease. J Cell Mol Med. 2007;11:1214–1224. doi: 10.1111/j.1582-4934.2007.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozpolat B, Akar U, Mehta K, Fok J, Kondo Y, Lopez-Berestein G. Mol Cancer Res 2007; 5:241-49. PKCδ and tissue transglutaminase (TG2) are novel inhibitors of autophagy. Autophagy. 2007;3:480–483. [Google Scholar]
- 37.Verma A, Wang H, Manavathi B, Fok JY, Mann AP, Kumar R, Mehta K. Increased expression of tissue transglutaminase in pancreatic ductal adenocarcinoma and its implications in drug resistance and metastasis. Cancer Res. 2006;66:10525–10533. doi: 10.1158/0008-5472.CAN-06-2387. [DOI] [PubMed] [Google Scholar]
- 38.Fesus L, Szondy Z. Transglutaminase 2 in the balance of cell death and survival. FEBS Lett. 2005;579:3297–3302. doi: 10.1016/j.febslet.2005.03.063. [DOI] [PubMed] [Google Scholar]
- 39.Mehta K, Fok J, Miller FR, Koul D, Sahin AA. Prognostic significance of tissue transglutaminase in drug resistant and metastatic breast cancer. Clin Cancer Res. 2004;10:8068–8076. doi: 10.1158/1078-0432.CCR-04-1107. [DOI] [PubMed] [Google Scholar]
- 40.Oliverio S, Amendola A, Di Sano F, Farrace MG, Fesus L, Nemes Z, et al. Tissue transglutaminase-dependent posttranslational modification of the retinoblastoma gene product in promonocytic cells undergoing apoptosis. Mol Cell Biol. 1997;17:6040–6048. doi: 10.1128/mcb.17.10.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma A, Guha S, Jansina F, et al. Therapeutic significance of elevated tissue transglutaminase expression in pancreatic cancer. Clin Cancer Res. 2008;14:2476. doi: 10.1158/1078-0432.CCR-07-4529. [DOI] [PubMed] [Google Scholar]
- 42.Buchholz T, Davis D, McConkey DJ, Symmans F, Valero V, Tucker SL, Pusztai L, Cristofanilli M, Esteva FJ, Hortobagyi G, Sahin A. Chemotherapy-induced apoptosis and Bcl-2 levels correlate with breast cancer response to chemotherapy. Cancer J. 2003;9:33–41. doi: 10.1097/00130404-200301000-00007. [DOI] [PubMed] [Google Scholar]
- 43.Bishop JM. Molecular themes in oncogenesis. Cell. 1991;64:235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- 44.Reed JC. Bcl-2: prevention of apoptosis as a mechanism of drug resistance. Hematol Oncol Clin North Am. 1995;9:451–473. [PubMed] [Google Scholar]
- 45.Devarajan E, Sahin AA, Chen JS, Krishnamurthy RR, Aggarwal N, Brun AM, et al. Down-regulation of caspase 3 in breast cancer: a possible mechanism for chemoresistance. Oncogene. 2002;21:8843–8851. doi: 10.1038/sj.onc.1206044. [DOI] [PubMed] [Google Scholar]
- 46.Devarajan E, Chen J, Multani AS, Pathak S, Sahin AA, Mehta K. Human breast cancer MCF-7 cell line contains inherently drug-resistant subclones with distinct genotypic and phenotypic features. Int J Oncol. 2002;20:913–920. [PubMed] [Google Scholar]
- 47.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 48.Sotelo J, Briceño E, López-González MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144:337–343. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 49.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 50.Melnikova I. RNA-based therapies. Nat Rev Drug Dis. 2007;6:863–864. doi: 10.1038/nrd2314. [DOI] [PubMed] [Google Scholar]
- 51.Ovcharenko D, Jarvis R, Hunicke-Smith S, Kelnar K, Brown D. High-throughput RNAi screening in vitro: from cell lines to primary cells. RNA. 2005;11:985–993. doi: 10.1261/rna.7288405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 53.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 54.Herrero-Martín G, Høyer-Hansen M, García-García C, Fumarola C, Farkas T, López-Rivas A, Jäättelä M. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–685. doi: 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meijer AJ, Codogno P. AMP-activated protein kinase and autophagy. Autophagy. 2007;3:238–240. doi: 10.4161/auto.3710. [DOI] [PubMed] [Google Scholar]
- 56.Harhaji-Trajkovic L, Vilimanovich U, Kravic-Stevovic T, Bumbasirevic V, Trajkovic V. AMPK-mediated autophagy inhibits apoptosis in cisplatin-treated tumor cells. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00663.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Wu Y, Cheng Y, Zhao Z, Tashiro S, Onodera S, Ikejima T. (2008) Fas-mediated autophagy requires JNK activation in HeLa cells. Biochem Biophys Res Commun. 2008;377:1205–1210. doi: 10.1016/j.bbrc.2008.10.151. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Wu Y, Cheng Y, Zhao Z, Tashiro S, Onodera S, Ikejima T. Involvement of PKC signal pathways in oridonin-induced autophagy in HeLa cells: a protective mechanism against apoptosis. Biochem Biophys Res Commun. 378:273–278. doi: 10.1016/j.bbrc.2008.11.038. [DOI] [PubMed] [Google Scholar]
- 60.Wu H, Zhu H, Liu DX, Niu TK, Ren X, Patel R, Hait WN, Yang JM. Silencing of elongation factor-2 kinase potentiates the effect of 2-deoxy-D-glucose against human glioma cells through blunting of autophagy. Cancer Res. 2009;69:2453–2460. doi: 10.1158/0008-5472.CAN-08-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shinojima N, Yokoyama T, Kondo Y, Kondo S. Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy. Autophagy. 2007;3:635–637. doi: 10.4161/auto.4916. [DOI] [PubMed] [Google Scholar]
- 62.Sivaprasad U, Basu A. Inhibition of ERK attenuates autophagy and potentiates tumour necrosis factor-alpha-induced cell death in MCF-7 cells. J Cell Mol Med. 2008;12:1265–1271. doi: 10.1111/j.1582-4934.2008.00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park MA, Zhang G, Norris J, Hylemon PB, Fisher PB, Grant S, Dent P. Regulation of autophagy by ceramide-CD95-PERK signaling. Autophagy. 2008;4:929–931. doi: 10.4161/auto.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park MA, Curiel DT, Koumenis C, Graf M, Chen CS, Fisher PB, Grant S, Dent P. PERK-dependent regulation of HSP70 expression and the regulation of autophagy. Autophagy. 2008:4364–4367. doi: 10.4161/auto.5593. [DOI] [PubMed] [Google Scholar]
- 65.Wang SH, Shih YL, Kuo TC, Ko WC, Shih CM. Cadmium toxicity toward autophagy through ROS-activated GSK-3beta in mesangial cells. Toxicol Sci. 2009;108:124–131. doi: 10.1093/toxsci/kfn266. [DOI] [PubMed] [Google Scholar]
- 66.Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36:2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 67.Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 2008;15:1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 68.Li DD, Wang LL, Deng R, Tang J, Shen Y, Guo JF, et al. The pivotal role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression during anticancer agents-induced autophagy in cancer cells. Oncogene. 2009;28:886–898. doi: 10.1038/onc.2008.441. [DOI] [PubMed] [Google Scholar]
- 69.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei Y, Sinha S, Levine B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy. 2008;4:949–951. doi: 10.4161/auto.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hait WN, Wu H, Jin S, Yang JM. Elongation factor-2 kinase: its role in protein synthesis and autophagy. Autophagy. 2006;2:294–296. doi: 10.4161/auto.2857. [DOI] [PubMed] [Google Scholar]
- 74.Wu H, Yang JM, Jin S, Zhang H, Hait WN. Elongation factor-2 kinase regulates autophagy in human glioblastoma cells. Cancer Res. 2006;66:3015–3023. doi: 10.1158/0008-5472.CAN-05-1554. [DOI] [PubMed] [Google Scholar]
- 75.Gozuacik D, Kimchi A. DAPk protein family and cancer. Autophagy. 2006;2:74–79. doi: 10.4161/auto.2.2.2459. [DOI] [PubMed] [Google Scholar]
- 76.Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5:85–87. doi: 10.4161/auto.5.1.7180. [DOI] [PubMed] [Google Scholar]
- 77.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1-ATG13-FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13- independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Congreve M, Carr R, Murray C, Jhoti H. A ‘rule of three’ for fragment-based lead discovery? Drug Discov Today. 2003;8:876–877. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 81.Lukovic E, Gonzalez-Vera JA, Imperiali B. Recognition-domain focused chemosensors: versatile and efficient reporters of protein kinase activity. J Am Chem Soc. 2008;130:12821–12827. doi: 10.1021/ja8046188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shults MD, Carrico-Moniz D, Imperiali B. Optimal Sox-based fluorescent chemosensor design for serine/threonine protein kinases. Anal Biochem. 2006;352:198–207. doi: 10.1016/j.ab.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 83.Rubinsztein D, Gestwicki JE, Klionsky D. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 84.Scott RC, Juhász G. Neufeld TP Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]