Summary
Interleukin-1 alpha (IL-1α) is a pro-inflammatory cytokine that is implicated in the initiation/maintenance of graft-versus-host disease (GVHD) and the immune response to infection. A cytosine (C) to thymine (T) transition at position −889 is believed to influence gene transcription. A previous single institution study showed that the presence of at least one IL1A T allele in the donor was associated with improved survival after unrelated donor haematopoietic stem cell transplant and lower transplant-related mortality if the donor and recipient each possessed the IL1A T allele. The present study sought to confirm these results in a larger homogeneous population. Thus the study population included 426 patients older than 18 years with chronic myeloid leukaemia (CML), transplanted in first chronic phase and receiving a total body irradiation and cyclophosphamide preparative regimen. Donor recipient pairs were categorised into four groups according to the presence or absence of an IL1A T allele in the donor and recipient. There were no significant differences in patient, donor and transplant characteristics between the groups. We did not observe an association with IL-1α genotype in donor and/or recipient and transplant-outcome. These data suggest that the outcome of unrelated donor transplant for CML is not influenced by IL-1α genotype.
Keywords: interleukin-1, polymorphisms, chronic myeloid leukaemia, unrelated donor, haematopoietic stem cell transplantation
Allogeneic haematopoietic stem cell transplantation (HSCT) offers a potential cure for a variety of malignant and non-malignant conditions. However, regimen-related toxicity, graft-versus-host disease (GVHD), and opportunistic infections (Kernan et al, 1993) still remain the major cause of morbidity and mortality after unrelated donor (URD) allogeneic HSCT. In addition to identifying patients and transplant-related prognostic factors, identifying favourable factors in the donor may aid in donor selection and may improve outcomes after HSCT.
Non-human leucocyte antigen (HLA) genetic polymorphism has been shown to influence risk of GVHD and HSCT mortality. Middleton et al (1998) showed that the homozygous d3 genotype of the tumour necrosis factor (TNF)-α microsatellite, and the presence of IL10 alleles with greater numbers of dinucleotide repeats were preferentially associated with grade III/IV GVHD in sibling transplant recipients. In studies of 993 recipients of matched sibling donor transplant, Lin et al (2003, 2005 showed that interleukin 10 (IL-10) genotype of the recipient and IL-10 receptor genotype of the donor modified the risk of GVHD. Associations between IL-6 and interferon-gamma genotype and susceptibility to GVHD have also been shown in sibling donor HSCT recipients (Cavet et al, 2001; Socie et al, 2001). Data addressing this issue in the unrelated donor HSCT setting are scarce, however Keen et al (2004) have reported association of TNF-α and IL-10 genotypes with toxicity after unrelated donor transplant. Taken together, these data suggest that non-HLA genetic variation influences risk of GVHD and HSCT mortality, and that it might be possible to predict HSCT outcome from a profile of donor or recipient risk factors, including cytokine polymorphisms.
Interleukin-1 is a pro-inflammatory cytokine implicated in the initiation and maintenance of GVHD and the immune response to infection (Ferrara & Deeg, 1991). The IL-1 gene family includes three members [IL1A (IL-1α), IL1B (IL-1β) and IL1N alias IL1RA (IL-1 receptor agonist; IL-1RA)] that mediate immune and inflammatory responses through two specific cell surface receptors. IL-1α and IL-1β are agonists and IL-1RA is a competitive receptor antagonist. A cytosine (C) to thymine (T) transition at position −889 in the IL1A promoter is believed to influence gene transcription. The T/T genotype creates the consensus site for a novel transcription factor (Skn-1) and is associated with a significant increase in promoter activity compared with the C/C genotype.
Three previous studies have examined IL-1 polymorphism and outcome of sibling donor bone marrow transplantation. Cullup et al (2001) studied IL1B and IL1N polymorphism and showed modest evidence for an association between donor IL-1RA genotype and incidence of acute GVHD. In a larger study, Lin et al (2000) reported little association between donor or recipient IL-1β and IL-1RA genotypes and frequency of GVHD. Cullup et al (2003) have also reported the association of IL1A −889 polymorphism in the donor genotype with the occurrence of chronic GVHD. Neither of these studies investigated IL1A polymorphism or examined URD HSCT recipients.
A previous single institution report of 90 recipients of unrelated donor HCT showed improved survival at 1 year if at least one IL1A T allele was present in the donor. Lower TRM was seen if both donor and recipient possessed the IL1A T allele (MacMillan et al, 2003). While these data showed a striking reduction in TRM associated with IL-1 genotype, numbers were small and these studies were performed in a heterogeneous patient population. In this study, we sought to replicate these findings in a larger and more uniform population.
Methods
Inclusion criteria
The study population included 426 recipients of unrelated donor HSCT, aged 18–60 years, transplanted in first chronic phase of chronic myeloid leukaemia (CML), between 1990 and 2002 in the US, using donors facilitated by the National Marrow Donor Program (NMDP). Patients had to have received a total body irradiation-containing conditioning regimen. Excluded were recipients of peripheral blood stem cells, disease status other than first chronic phase CML, and recipients of second transplant.
IL-1 genotyping of donor and recipient
Donor and recipient DNA were obtained from the NMDP repository and normalised to 10 ng/μl. The Institutional Review Board of the NMDP approved release of donor and recipient samples from the repository for genotyping of IL1A polymorphism and subsequent correlation with transplant-outcome. Donor-recipient pairs were genotyped for the IL1A polymorphism using a high throughput polymerase chain reaction (PCR) assay (5′nuclease allelic discrimination assay-TaqMan; Applied Biosystems, Foster City, CA, USA). We use gene-specific PCR primers (forward primer – 5′-ACT AGG CTG GCC ACA GGA ATT AT-3′; reverse primer – 5′-CCA GAA GCC AGT GGC TAA GTT T-3′) and fluorogenic probes [VIC (wild type)-CTT CAA TGG TGT TGC; FAM (variant)-CCT TCA ATG ATG TTG] for allelic discrimination. PCR cycling reactions were performed in 96-well microtitre plates in a GeneAmp PCR System 9600 (Perkin-Elmer, Norwalk, CT, USA). For each 25 μl reaction, a 10 ng DNA template was added to the reaction mixture containing wild-type VIC and variant FAM probe, PCR Mastermix (Applied Biosystems) along with forward and reverse primer (final concentration of each primer 7·5 μmol/l). Thermocycling was performed with initial 50°C incubation for 2 min followed by a 10-min incubation at 95°C. A two-step cycling reaction was performed for 40 cycles with denaturation at 95°C for 15 s, and annealing and extension at 60°C for 1 min. Results were analysed by the automated TaqMan allelic discrimination assay using sequence detection system software 2.1 (ABI TaqMan 7700; Applied Biosystems). Genotyping results were duplicated in 10% of samples; concordance between repeats was 100%. Further-more, 10% of the samples were also genotyped using direct sequencing; concordance with TaqMan genotyping was 100%.
Donor and recipient pairs were grouped into four mutually exclusive categories for analyses of genotypes (Hurme & Santtila, 1998). Similar to MacMillan et al (2003), the four groups were based on the presence or absence of a T-allele in donor and/or in recipient (only recipient has T-allele, only donor has T-allele, both donor and recipient have T-allele and neither donor or recipient have a T-allele).
Definition of clinical endpoints
Diagnosis of acute and chronic GVHD was based on local institutional criteria with overall grade of acute GVHD assigned retrospectively by the NMDP based on stage of involvement reported for each individual organ (Przepiorka et al, 1995). Transplant-related mortality was defined as death during a continuous remission. Relapse was defined as haematological leukaemia recurrence.
Statistical analysis
Baseline variables were compared between the four donor-recipient IL-1 genotype groups as described earlier using the chi-square statistic for categorical variables and the Kruskal–Wallis test for continuous variables. The probability of overall survival was calculated using the Kaplan–Meier estimator (Klein & Moeschberger, 2003). For overall survival, death from any cause was considered an event, and patients surviving at last follow-up were censored. Probabilities of acute and chronic GVHD, transplant-related mortality and relapse were calculated using the cumulative incidence function estimator (Klein & Moeschberger, 2003). For GVHD, death without the event was the competing risk; for transplant-related mortality, relapse was the competing event; and, for relapse, transplant-related mortality was the competing event.
Multivariate analysis was performed using the Cox proportional hazards regression model (Cox, 1972). All multivariate models were constructed using stepwise forward selection, with a P-value ≤ 0·05 considered significant. All variables met the proportional hazards assumption. The variable for donor-recipient IL-1 genotype was retained in all steps of model building. Other variables considered were recipient and donor age, sex of the recipient and donor, cytomegalovirus (CMV) serostatus of the recipient and donor, time from diagnosis to transplantation, recipient performance score at transplantation, GVHD prophylaxis, year of transplant and donor- recipient HLA compatibility. Disease status and conditioning regimen were not tested as all patients were in first chronic phase and received a total body irradiation based regimen. All reported P-values are two-sided. All analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC, USA).
Results
Patient and transplant characteristics
Patient, donor and transplant characteristics are shown in Table I. The median age at transplantation was 39 years (range 18–59 years) and 57% of patients were male. The median age of donors was 38 years (range 18–57 years). Sixty-four per cent of patients received grafts from donors matched at HLA-A, -B and -DRB1 (allele-level typing), 22% were mismatched at ≥1-loci (allele-level typing) but matched by antigen-level typing and 14% were mismatched at 1-locus by antigen-level typing. There was no difference in the genotype frequency between recipients and donors (c/c: 48% vs. 46%, c/t: 44% vs. 45%, t/t: 8% vs. 9%) and were consistent with those predicted under the conditions of the Hardy–Weinberg equilibrium. The median time from diagnosis to transplantation was 13 months. All patients received total body irradiation (myeloablative dose) and cyclophosphamide for conditioning. The median follow-up of surviving patients was over 7 years.
Table I.
Patient, donor and transplant characteristics by recipient and donor IL-1 genotype
| Variable | IL-1 genotype of the donor and recipient |
|||
|---|---|---|---|---|
| T-allele present in the recipient |
T-allele present in the donor |
T-allele present in recipient and donor |
T-allele absent in recipient and donor |
|
| Number of patients | 90 | 102 | 130 | 104 |
| Male | 48 (53) | 60 (59) | 76 (58) | 60 (58) |
| Performance score, ≥90 | 77 (86) | 87 (85) | 117 (90) | 90 (87) |
| Year of transplant | ||||
| 1990–1994 | 34 (38) | 42 (41) | 38 (29) | 32 (31) |
| 1995–1999 | 45 (50) | 46 (45) | 79 (61) | 55 (53) |
| 2000–2002 | 11 (12) | 14 (14) | 13 (10) | 17 (16) |
| GVHD prophylaxis | ||||
| T-cell depletion | 21 (23) | 19 (19) | 23 (18) | 19 (18) |
| Ciclosporin-based | 59 (65) | 70 (69) | 94 (72) | 71 (70) |
| Tacrolimus-based | 10 (11) | 13 (13) | 13 (10) | 14 (13) |
| D-R CMV serostatus | ||||
| D (−)/R (−) | 33 (37) | 37 (36) | 44 (34) | 27 (26) |
| D (+)/R (+) | 19 (21) | 18 (18) | 25 (19) | 19 (18) |
| D (+)/R (−) | 14 (16) | 16 (16) | 17 (13) | 24 (23) |
| D (−)/R (+) | 24 (27) | 26 (25) | 43 (33) | 32 (31) |
| Unknown | 5 (5) | 1 (1) | 2 (2) | |
| D-R HLA disparity | ||||
| Allele-matched | 63 (70) | 60 (59) | 82 (63) | 68 (65) |
| Allele-mismatch | 15 (17) | 26 (25) | 30 (23) | 22 (21) |
| Antigen-mismatch | 12 (13) | 16 (16) | 18 (14) | 14 (13) |
| Median follow-up of survivors (months) | 90 (12–144) | 98 (9–170) | 84 (22–168) | 91 (31–168) |
The numbers in brackets denote percentages.
Patient, donor and transplant characteristics were similar among the four cohorts using the chi square test for categorical values and the Kruskal–Wallis test for continuous variables.
D, donor; R, recipient; M, male; F, female; CMV, cytomegalovirus.
Graft-versus-host disease
Rates of grade 2–4 and grade 3–4 acute GVHD did not differ by donor-recipient IL-1 genotype (Table II). Similarly, rates of chronic GVHD did not differ by donor-recipient IL-1 genotype (Table II). The probabilities of grade 2–4 acute GVHD at day-100 were 62%, 60%, 56% and 55% when T-allele was present in only the recipient, present in only the donor, present in both the donor and recipient or absent in both donor and recipient, respectively. Corresponding probabilities of chronic GVHD at 5-years were 52%, 51%, 54% and 54%.
Table II.
Results of multivariate analysis of transplant outcome by donor and recipient IL-1 genotype
| Outcome | N1/N2 | Relative risk (95% confidence interval) |
P-value |
|---|---|---|---|
| Grade 2–4 acute GVHD | |||
| T-allele absent in recipient and donor | 56/94 | 1·00 | 0·682* |
| T-allele present in the recipient | 50/79 | 1·19 (0·81–1·75) | 0·369 |
| T-allele present in the donor | 50/83 | 1·01 (0·69–1·49) | 0·947 |
| T-allele present in recipient and donor | 63/112 | 0·95 (0·66–1·36) | 0·790 |
| Grade 3–4 acute GVHD | |||
| T-allele absent in recipient and donor | 28/94 | 1·00 | 0·095* |
| T-allele present in the recipient | 33/79 | 1·52 (0·92–2·53) | 0·104 |
| T-allele present in the donor | 20/83 | 0·78 (0·44–1·39) | 0·397 |
| T-allele present in recipient and donor | 33/112 | 0·98 (0·59–1·61) | 0·923 |
| Chronic GVHD | |||
| T-allele absent in recipient and donor | 53/101 | 1·00 | 0·739* |
| T-allele present in the recipient | 43/85 | 1·01 (0·67–1·52) | 0·958 |
| T-allele present in the donor | 53/94 | 1·22 (0·83–1·79) | 0·313 |
| T-allele present in recipient and donor | 64/122 | 1·10 (0·76–1·59) | 0·611 |
| Transplant-related mortality | |||
| T-allele absent in recipient and donor | 58/104 | 1·00 | 0·504* |
| T-allele present in the recipient | 49/90 | 1·14 (0·78–1·67) | 0·508 |
| T-allele present in the donor | 53/102 | 0·90 (0·62–1·31) | 0·592 |
| T-allele present in recipient and donor | 75/130 | 1·16 (0·82–1·64) | 0·397 |
| Relapse | |||
| T-allele absent in recipient and donor | 5/104 | 1·00 | 0·942* |
| T-allele present in the recipient | 6/90 | 1·41 (0·43–4·64) | 0·567 |
| T-allele present in the donor | 5/102 | 1·06 (0·31–3·68) | 0·923 |
| T-allele present in recipient and donor | 7/130 | 1·21 (0·38–3·82) | 0·742 |
| Overall mortality | |||
| T-allele absent in recipient and donor | 61/104 | 1·00 | 0·528* |
| T-allele present in the recipient | 54/90 | 1·16 (0·80–1·68) | 0·427 |
| T-allele present in the donor | 57/102 | 0·93 (0·65–1·33) | 0·691 |
| T-allele present in recipient and donor | 81/130 | 1·16 (0·83–1·62) | 0·385 |
N1, number of events; N2, number evaluable.
3-degree freedom test.
Transplant-related mortality
Transplant-related mortality rates did not differ by donor-recipient IL-1 genotype (Table II, Fig 1). In all groups, transplant-related mortality was higher in recipients of mismatched transplants [≥1-allele mismatch: relative risk (RR) 1.99, P < 0·001; 1-antigen mismatch: RR 2·05, P < 0·001] compared with matched transplants.
Fig 1.
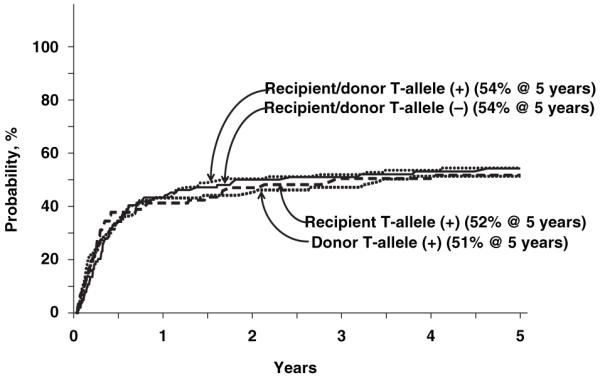
Probability of transplant-related mortality by donor and recipient IL-1 genotype.
Relapse
Relapse rates did not differ by donor-recipient IL-1 genotype (Table II). The probabilities of relapse at 5-years were 7%, 5%, 5% and 4% when T-allele was present in only the recipient, present in only the donor, present in both the donor and recipient or absent in both donor and recipient respectively. As expected, relapse rates were higher in recipients of T-cell depleted bone marrow grafts regardless of donor-recipient IL-1 genotype (RR 4·10, P < 0·001). Of the 23 patients with haematological relapse, three received donor leucocyte infu- sion (DLI) and three underwent a second transplant. Two patients who received DLI and one of the patients receiving a second transplant had received T-cell depleted bone marrow grafts.
Overall mortality
Overall mortality rates did not differ by donor-recipient IL-1 genotype (Table II, Fig 2). Mortality rates were higher after mismatched transplants (≥1-allele mismatch: RR 1·92, P < 0·001; 1-antigen mismatch: RR 1·94, P < 0·001) and in recipients of T-cell depleted grafts (RR 1·43, P = 0·017) regardless of donor-recipient IL-1 genotype. Frequent causes of death included infection (23%), GVHD (19%) and interstitial pneumonitis (17%). Others include: recurrent leukaemia (4%), graft failure (5%), haemorrhage (8%), organ failure (11%) and other causes (13%). Causes of death did not differ by donor-recipient IL-1 genotype.
Fig 2.
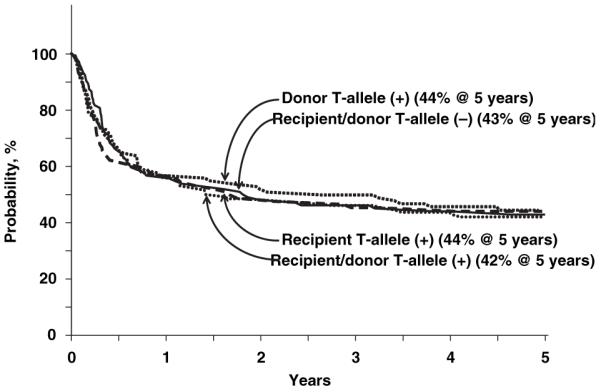
Probability of overall survival by donor and recipient IL-1 genotype.
Discussion
A number of studies have investigated the role of cytokine gene polymorphisms in susceptibility to post-HSCT complications, such as GVHD and transplant-related mortality. For example, possible roles for polymorphisms in TNFα, IL-10 and members of the IL-1 family in outcome of sibling donor HSCT have been addressed (Middleton et al, 1998; Cavet et al, 1999; Lin et al, 2000; Cullup et al, 2001, 2003, ).
The objective of the current study was to examine the association between the presence of the IL1A T allele and outcomes after URD transplantation. Previously MacMillan et al (2003) reported lower transplant-related mortality and improved overall survival after URD transplantation when an IL1A T allele was present in both the recipient and the donor. We sought to confirm these findings in a larger and relatively homogenous cohort by limiting the study population to patients with CML in first chronic phase who received a uniform conditioning regimen, and by focussing on the IL1A −889 polymorphism. Our findings differed from that reported by MacMillan et al (2003). We did not observe a significant association between the presence of an IL1A T allele in the recipient and/or donor and transplant-related mortality, GVHD or overall survival after URD transplant for CML in first chronic phase after adjusting for significant prognostic factors. We speculate that the differing results of these two studies reflected the relatively small (97 cases) and heterogeneous population in the first study. A limitation of this analysis is the heterogeneity of HLA matching. Although the number of cases with HLA mismatch did not differ by IL-1 categories, it remains possible that undetected HLA disparity outside of HLA-A, -B and -DRB1 could have confounded the ability to measure independent effects of IL1A polymorphism. These findings illustrate the need for replication of association studies in an independent dataset before clinical application.
Acknowledgements
This study was supported by Public Health Service Grant U24-CA76518–08 from the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, and the National heart, Lung and Blood Institute, ‘Cancer Free Kids’, Cincinnati, OH and the National Marrow Donor Program partly through funding from the Health Resources and Services administration (no. 204–97–0036) and the Office of Naval Research (N00014–99–2-0006 and N00014–5-1–0859). The views expressed in this article are those of the authors and do not reflect the official policy or position of the Department of Navy, the Department of Defense or the US government.
References
- Cavet J, Middleton PG, Segall M, Noreen H, Davies SM, Dickinson AM. Recipient tumor necrosis factor-alpha and interleukin-10 gene polymorphisms associate with early mortality and acute graft-versus host disease severity in HLA-matched sibling bone marrow transplants. Blood. 1999;94:3941–3946. [PubMed] [Google Scholar]
- Cavet J, Dickinson AM, Norden J, Taylor PR, Jackson GH, Middleton PG. Interferon-gamma and interleukin-6 gene polymorphisms associate with graft-versus-host disease in HLA matched sibling bone marrow transplantation. Blood. 2001;98:1594–1600. doi: 10.1182/blood.v98.5.1594. [DOI] [PubMed] [Google Scholar]
- Cox DR. Regression models and life tables. Journal of the Royal Statistical Society. 1972;34:187. [Google Scholar]
- Cullup H, Dickinson AM, Jackson GH, Taylor RP, Cavet J, Middleton PG. Donor interleukin 1 receptor antagonist genotype associated with acute graft-versus-host disease in human leucocyte antigen matched sibling allogeneic transplants. British Journal of Haematology. 2001;113:807–813. doi: 10.1046/j.1365-2141.2001.02811.x. [DOI] [PubMed] [Google Scholar]
- Cullup H, Dickinson AM, Cavet J, Jackson GH, Middleton PG. Polymorphisms of interleukin-1alpha constitute independent risk factors for chronic graft-versus-host disease after allogeneic bone marrow transplantation. British Journal of Haematology. 2003;122:778–787. doi: 10.1046/j.1365-2141.2003.04510.x. [DOI] [PubMed] [Google Scholar]
- Ferrara JL, Deeg HJ. Graft-versus-host disease. The New England Journal of Medicine. 1991;324:667–674. doi: 10.1056/NEJM199103073241005. [DOI] [PubMed] [Google Scholar]
- Hurme M, Santtila S. IL-1 receptor antagonist (IL-1Ra) plasma levels are co-ordinately regulated by both IL-1Ra and IL-1beta genes. European Journal of Immunology. 1998;28:2598–2602. doi: 10.1002/(SICI)1521-4141(199808)28:08<2598::AID-IMMU2598>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Keen LJ, DeFor TE, Bidwell JL, Davies SM, Bradley BA, Hows JM. Interleukin-10 and tumor necrosis factor alpha region haplotypes predict transplant-related mortality after unrelated donor stem cell transplantation. Blood. 2004;103:3599–3602. doi: 10.1182/blood-2002-11-3568. [DOI] [PubMed] [Google Scholar]
- Kernan NA, Bartsch G, Ash RC, Beatty PG, Champlin R, Filipovich A, Gajewski J, Hansen JA, Hanslee-Downey J, McCullough J, McGlave P, Perkins HA, Phillips GL, Sanders J, Stroncek D, Thomas ED, Blume KG. Analysis of 462 transplantations from unrelated donors facilitated by the National Marrow Donor Program. The New England Journal of Medicine. 1993;328:593–602. doi: 10.1056/NEJM199303043280901. [DOI] [PubMed] [Google Scholar]
- Klein J, Moeschberger M. Survival Analysis: Techniques of Censored and Truncated Data. Springer-Verlag; New York, NY: 2003. [Google Scholar]
- Lin M-T, Martin PJ, Tseng L-H, Singleton K, Martin E, Petersodrf E, Hansen J. Correlation between polymorphisms of the IL-1 gene complex and development of acute graft-versus-host disease after hematopoietic cell transplantation. Blood. 2000;96:396a. [Google Scholar]
- Lin MT, Storer B, Martin PJ, Tseng LH, Gooley T, Chen PJ, Hansen JA. Relation of an interleukin-10 promoter polymorphism to graft-versus-host disease and survival after hematopoietic-cell transplantation. The New England Journal of Medicine. 2003;349:2201–2210. doi: 10.1056/NEJMoa022060. [DOI] [PubMed] [Google Scholar]
- Lin MT, Storer B, Martin PJ, Tseng LH, Grogan B, Chen PJ, Zhao LP, Hansen JA. Genetic variation in the IL-10 pathway modulates severity of acute graft-versus-host disease following hematopoietic cell transplantation: synergism between IL-10 genotype of patient and IL-10 receptor beta genotype of donor. Blood. 2005;106:3995–4001. doi: 10.1182/blood-2004-11-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan ML, Radloff GA, DeFor TE, Weisorf DJ, Davies SM. Interleukin-1 genotype and outcome of unrelated donor bone marrow transplantation. British Journal of Haematology. 2003;121:597–604. doi: 10.1046/j.1365-2141.2003.04314.x. [DOI] [PubMed] [Google Scholar]
- Middleton PG, Taylor PR, Jackson G, Proctor SJ, Dickinson AM. Cytokine gene polymorphisms associating with severe acute graft-versus-host disease in HLA-identical sibling transplants. Blood. 1998;92:3943–3948. [PubMed] [Google Scholar]
- Przepiorka D, Weisdorf D, Martin P, Klingemann HG, Beatty P, Hows J, Thomas ED. 1994 Consensus Conference on acute GVHD grading. Bone Marrow Transplantation. 1995;15:825–828. [PubMed] [Google Scholar]
- Socie G, Loiseau P, Tamouza R, Janin A, Busson M, Gluckman E, Charron D. Both genetic and clinical factors predict the development of graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Transplantation. 2001;72:699–706. doi: 10.1097/00007890-200108270-00024. [DOI] [PubMed] [Google Scholar]


