Summary
The translational apparatus of the bacterial cell remains one of the principal targets of antibiotics for the clinical treatment of infection worldwide. Since the introduction of specific translation inhibitors into clinical practise in the late 1940’s, intense efforts have been made to understand their precise mechanisms of action. Such research has often revealed significant and sometimes unexpected insights into many fundamental aspects of the translation mechanism. Central to progress in this area, high-resolution crystal structures of the bacterial ribosome identifying the sites of antibiotic binding are now available, which, together with recent developments in single-molecule and fast-kinetic approaches, provide an integrated view of the dynamic translation process. Assays employing these approaches and focusing on specific steps of the overall translation process are amenable for drug-screening. Such assays, coupled with structural studies, have the potential not only to accelerate the discovery of novel and effective antimicrobial agents, but also to refine our understanding of the translation mechanism, since antibiotics often stabilize specific functional states of the ribosome and allow distinct translation steps to be dissected in molecular detail.
Keywords: antibiotics, drug development, fast kinetics, protein synthesis, ribosome structure, single molecule FRET, translation inhibition
The essentials of protein synthesis
The translation machinery is responsible for the accurate conversion of the genetic information within messenger RNA (mRNA) into a corresponding polypeptide sequence. The ribosome provides the platform upon which the mRNA is recognized and n“decoded” by transfer RNAs (tRNAs). Evolutionarily, tRNAs connect the RNA and protein worlds: at one end is an anticodon sequence that is complementary to a specific mRNA codon, whereas on the other is an amino acid linked to the 3′ CCA terminus by an ester linkage. tRNAs thus serve as adaptor molecules in protein synthesis by specifying the incorporation of a specific amino acid for each mRNA codon. The process of translation occurs in four principal stages: initiation, elongation, termination and recycling. Polymerization of the polypeptide chain occurs during the elongation phase, as the ribosome traverses the open reading frame (ORF) of the mRNA selecting a specific aminoacyl-tRNA (aa-tRNA) at each codon triplet step.
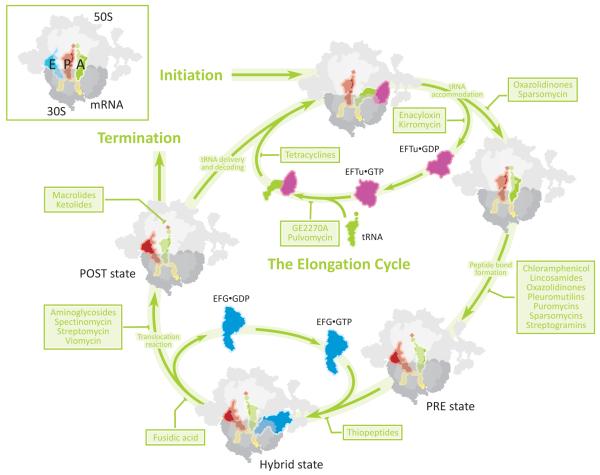
The ribosome (70S in bacteria) possesses three main tRNA binding sites: the Aminoacyl (A)-, Peptidyl (P)- and Exit (E)-sites, located at the interface of the small (30S) and large (50S) ribosomal subunits (inset to Figure 1). The L-shaped tRNA molecules are oriented such that the tRNA anticodon-mRNA codon interactions take place on the 30S subunit, while the 3′-CCA termini interact with the 50S subunit. The process of initiation, facilitated by initiation factors, places the unique, initiator fMet-tRNAfMet at the P site of the ribosome where it interacts with the start codon of the mRNA. Following initiation, ribosomes enter into the elongation phase of protein synthesis (Figure 1). Elongation lies at the heart of protein synthesis and involves the entry and movement of tRNAs through the three tRNA binding sites (A →P →E) of the ribosome in a cyclic fashion. The number of elongation cycles is dictated by the length of the ORF and the polypeptide being synthesized. During elongation, aa-tRNAs are selected by the ribosome according to the mRNA codon presented at the A site of the 30S subunit, in a process referred to as decoding (Figure 1). The delivery of the aa-tRNAs is a multistep, induced fit process that is facilitated by elongation factor Tu (EF-Tu) and utilizes GTP hydrolysis. Complementary base-pairing interactions between the tRNA anticodon and mRNA codon stimulates EF-Tu to hydrolyse GTP and dissociate from the ribosome, allowing aa-tRNA to fully accommodate into the peptidyltransferase center (PTC) of the large subunit. Peptide-bond formation subsequently occurs between adjacently bound peptidyl- and aminoacyl-tRNAs and transfers the growing polypeptide chain from P-site tRNA to A-site tRNA, leaving deacylated-tRNA in the P site. This ribosomal state, referred to as the pre-translocation complex (PRE state) (Figure 1), is highly dynamic in nature. In this complex, A- and P-site tRNAs reversibly oscillate between classically-defined A/A and P/P configurations and so-called hybrid states (A/P, P/E) wherein the 3′-CCA ends of both tRNAs move with respect to the large subunit while remaining relatively fixed with respect to the small subunit. Hybrid tRNA configurations are facilitated after peptide bond formation by the E-site’s capacity to stably bind deacylated or uncharged tRNA (Figure 1). The elongation cycle progresses by the translocation of A- and P-site tRNAs with respect to the small subunit. This complex multistep process, mediated by elongation factor-G (EF-G) catalyzed GTP hydrolysis, moves the mRNA-tRNA2 complex into the P- and E-sites. In so doing, a post-translocation complex (POST state) is formed, and the next downstream mRNA codon enters the A site (Figure 1). As repetitive elongation cycles continue, with the ribosome alternating between globally distinct PRE and POST states, the nascent polypeptide chain passes through a tunnel in the large ribosomal subunit and emerges into the cytoplasm where protein folding takes place. When a stop codon of the mRNA ORF enters into the A-site, the ribosome diverts into the termination phase. Here protein release factors specifically recognize the stop codon and mediate water-catalyzed hydrolysis of the nascent peptide from the P-site tRNA. The ribosomal subunits are then separated from deacylated tRNA and mRNA through a multistep, factor-mediated process termed recycling.
Figure 1. Sites of antibiotic action during the elongation cycle of protein synthesis.
Schematic representation of the different steps of the elongation cycle with the sites of inhibition of the major classes of antibiotics.
Antibiotics and ribosome function
As seen in Figure 1, protein synthesis, and in particular the elongation cycle, is a key target for antibiotic-mediated regulation. In fact, antibiotics have been identified that inhibit almost every step of the elongation cycle. Thus, studies into antibiotic action provide not only insight into the mechanism of inhibition of the antibiotic, but also a means of exploring the fundamental mechanism of protein synthesis. Antibiotics are chemical substances that are produced by one organism to kill another. The first antibiotics brought into clinical use were the sulfonamides (1932), which target folate metabolism, and β-lactams, such as penicillin (1940), which prevent bacterial cell wall synthesis (Fischbach and Walsh, 2009). However, the four major classes of antibiotics to subsequently enter into clinical practise all target the translational apparatus. These include the phenylpropanoids, such as chloramphenicol and tetracyclines in 1949, followed closely by the aminoglycosides, such as streptomycin (1950), and the macrolides, for example erythromycin (1952). A wealth of literature addressing the chemistry, selectivity and inhibitory properties of antibiotics was available soon after their discovery and clinical implementation (reviewed by Gale et al., 1972). As highlighted in the book chapter of Gale and coworkers, the sites of interaction of these important inhibitors required the development of cell-free in vitro translation systems using crude extracts that were competent for mRNA-directed polypeptide synthesis. These systems were first utilized in the 1960s to decipher the genetic code (reviewed by Rheinberger, 2004). Together with the subsequent development of model systems to monitor individual steps of protein synthesis, a more detailed understanding of the steps of protein synthesis inhibited by each antibiotic emerged.
Correlating structural and biochemical studies of antibiotic action
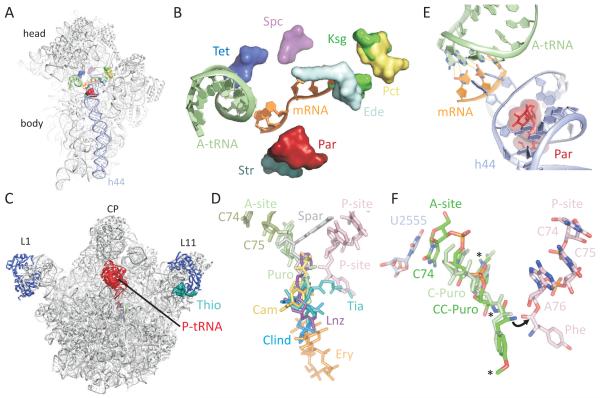
The turn of the century saw the first crystal structures of antibiotics bound to the 30S and 50S subunits (reviewed by Wilson, 2009). In 2009, work in this area carried out in the Steitz, Ramakrishnan and Yonath laboratories was recognized for its importance and impact with the Nobel Prize in Chemistry. Today, all major classes of ribosome-targeting antibiotics have been visualized in complex with a ribosomal particle (see Figure 2A-D). As foreshadowed by earlier biochemical studies, such structures definitively revealed that antibiotics predominantly target highly-conserved, functional centres of the bacterial ribosome including the pathway of tRNA2-mRNA movement through the small subunit (Figure 2A, B) as well as the PTC and adjacent nascent peptide exit tunnel on the large subunit (Figure 2C, D). Atomic-resolution structures of the ribosome, interpreted in the context of the prior decades of biochemical research, provided unparalleled insights into the core activities of the ribosome and ultimately culminated in precise articulations of the mechanisms of tRNA decoding and peptide-bond formation, as well as the antibiotic inhibition of these processes by, for example, the aminoglycoside family of antibiotics and puromycin, respectively (reviewed by Ogle and Ramakrishnan, 2005; Schmeing and Ramakrishnan, 2009; Simonovic and Steitz, 2009).
Figure 2. Binding sites of antibiotics on the ribosome.
Superimposition of binding sites of antibiotics (colored) that target the (A, B) small 30S (yellow) and (C, D) large 50S (blue) subunits, with the position of the mRNA (orange), A-tRNA (green) and helix 44 (blue) shown in (B) and P-tRNA (red) shown in (C). (E) Binding of paromomycin (Par) induces an extruded conformation for A1492 and A1493 of the 16S rRNA, which monitor the codon-anticodon interaction of the mRNA (orange)-tRNA (green) duplex. (F) Peptide bond formation results from nucleophilic attack of the α-amino group of the A-tRNA onto the carbonyl carbon of the P-tRNA (arrowed). The C74 of the A-tRNA stacks on U2555 of the 23S rRNA to re-position the A-tRNA (compare C-Puro and CC-Puro) for the nucleophilic attack.
The aminoglycoside family of antibiotics has long been known for its ability to stimulate translational misreading. In the mid-1960’s, the Gorini lab demonstrated that, while streptomycin inhibited incorporation of phenylalanine (Phe) during translation of a synthetic poly(U) mRNA, the misincorporation of other amino acids was stimulated, in particular isoleucine and tyrosine (Davies et al., 1964). Streptomycin was shown to do so by stabilizing the binding of “incorrect” Ile-tRNA to ribosomes programmed with UUU (Phe) triplet codons (Kaji and Kaji, 1965; Pestka et al., 1965). Similar misreading effects were also observed in the presence of aminoglycosides, such as kanamycin, neomycin, hygromycin B and gentamicin (Davies et al., 1965). In the presence of aminoglycosides, the estimated level of translational accuracy decreased from a normal frequency of 1 × 10−3 −10−4 misincorporation events per codon, to as much 1 in 10−1 to 10−2 (see Zaher and Green (2009) and references therein). Chemical probing experiments performed in the Noller lab indicated that aminoglycosides bind at the top of helix 44 (h44) of the 30S subunit, in close proximity to the decoding site (Moazed and Noller, 1987). Subsequent NMR structures of the aminoglycoside paromomycin bound to small RNA fragments mimicking h44 indicated that drug binding stabilizes distinct, extruded conformations of universally-conserved nucleotides A1492 and A1493 within the h44 internal asymmetric loop, which Puglisi and coworkers suggested were important for monitoring the codon-anticodon duplex (Fourmy et al., 1996). This model was borne-out by the structures of the 30S subunit with mRNA and tRNA in the A site, in the presence and absence of paromomycin (reviewed by Ogle and Ramakrishnan, 2005). These structures revealed that the ribosome utilizes nucleotides A1492 and A1493 to monitor the geometry of the tRNA-mRNA decoding interaction. Cognate codon-anticodon interactions stabilize the extruded conformation of residues A1492 and A1493 promoting aa-tRNA accommodation at the A site (Figure 2E). Similarly, by stabilizing A1492 and A1493 in extruded conformations, aminoglycosides, such as paromomycin, stabilize the binding of near-cognate tRNAs as well as promote their accommodation on the ribosome, whereas in contrast, streptomycins reduce the rate at which cognate tRNAs are selected and slightly enhance near-cognate tRNA binding (Gromadski and Rodnina, 2004; Karimi and Ehrenberg, 1994). However, it should be noted that misreading contributes only part of the inhibitory effect of aminoglycosides, since the stabilization of tRNA in the A-site by aminoglycosides inhibits translocation (Peske et al., 2004) and promotes back-translocation (Shoji et al., 2006).
Puromycin inhibits growth across all three kingdoms, and thus is not used clinically, but nevertheless has been an important tool for studying the peptidyltransferase reaction. Puromycin structurally mimics the terminal aminoacyl-adenosine moiety of aminoacyl-tRNA (Yarmolinsky and de la Haba, 1959) (Figure 2F). Puromycin acts as an acceptor in the peptide bond forming reaction (Gilbert, 1963) to release the nascent peptide from peptidyl-tRNA and the ribosome (Allen and Zamecnik, 1962; Morris and Schweet, 1961). Historically, the classical definitions of A- and P-site are derived from the inability or ability, respectively, of aa- or peptidyl-tRNAs to react with puromycin (Bretscher and Marcker, 1966). In line with mounting experimental evidence that the ribosome was a ribozyme, photo-crosslinking and chemical probing of aa-tRNA and puromycin derivatives bound to ribosomes identified specific 23S rRNA nucleotides as comprising the site of peptide-bond formation (reviewed by Polacek and Mankin, 2005). Supporting this notion, resistance mutations and chemical probing studies of PTC inhibitors identified nucleotides in the same region of the 23S rRNA (see Polacek and Mankin, 2005; Wilson, 2009).
The visualization of puromycin analogues mimicking (i) the substrates (Figure 2F), (ii) transition state and (iii) post-peptide bond formation products through crystallographic analyses led to identification of the exact site of puromycin action and provided an atomic understanding of the mechanism of peptide bond formation (reviewed by Simonovic and Steitz, 2009). Trapping these A- and P-site substrates in a pre-peptide bond formation state was possible because the CC-puromycin analogues used had a stable amide linkage between the ribose and the amino acid (Figure 2F), rather than a labile ester linkage as in aa-tRNA. The structures reveal that the active site of the ribosome is composed exclusively of rRNA, although there are extensions of some ribosomal proteins, such as L27 in bacteria and L10e in eukaryotes, which contact the CCA-end of P-tRNA and thus may contribute to positioning of the substrates (Voorhees et al., 2009). Comparison of the ribosome structures of peptidyl-tRNA at the P-site with C74C75-puromycin, or C75-puromycin at the A-site, revealed an induced-fit mechanism in which the substrates and active-site residues reposition to allow the peptidyl-transferase reaction to proceed (Schmeing et al., 2005). Specifically, C74 stacks with U2555, shifting the A-site substrate down to position the α-amino group for nucleophilic attack on the ester carbon of the P-tRNA (Figure 2F). Thus, the antibiotic puromycin has enabled detailed insight into how binding and positioning of the substrates contributes to the 2 × 107-fold enhancement in rate of peptide bond formation (Sievers et al., 2004).
The “fastlane” to antibiotic inhibition of the translational apparatus
The study of the rates of enzyme reactions, and the factors that modulate such rates, has often proved valuable as an approach to understanding enzyme mechanisms. Ribosome-catalysis of translation is a multi-step process, and a goal of rate studies on the ribosome has been the elucidation of the detailed kinetic mechanisms by which ribosomes carry-out specific reactions of the catalytic cycle, and how antibiotics affect such mechanisms. The most informative of such studies have been single-turnover transient kinetics ensemble experiments (stopped flow and quenched flow; reviewed by for example Beringer and Rodnina, 2007; Wintermeyer et al., 2004) and single molecule dynamics studies (see below). Stopped-flow experiments typically employ fluorescently-labelled ribosomal ligands, such as elongation factors, mRNAs, and tRNAs, as well as fluorescently labelled ribosomes, and monitor changes in fluorescence that characterize a particular reaction within the overall process of protein synthesis. In addition, the change in fluorescence when Pi (inorganic phosphate) binds to a fluorophore-labelled derivative of phosphate binding protein (PBP) permits determination of the rate of Pi release into solution following ribosome-dependent GTP hydrolysis. This process occurs following the binding of IF2·GTP during initiation, of both ternary complex (aa-tRNA·EF-Tu·GTP) and EF-G·GTP during elongation, and of RF3·GTP during termination. Changes in light-scattering have also been employed in studies of 70S initiation complex (70SIC) formation from 30S and 50S subunits (Antoun et al., 2006; Grigoriadou et al., 2007; Grunberg-Manago et al., 1975; Milon et al., 2008), and 70S dissociation into 30S and 50S subunits during ribosome recycling (Hirokawa et al., 2008; Pavlov et al., 2008). Quenched-flow kinetics are used to determine the rates of the two chemical steps catalysed by the ribosome, namely peptidyl transferase and GTP hydrolysis.
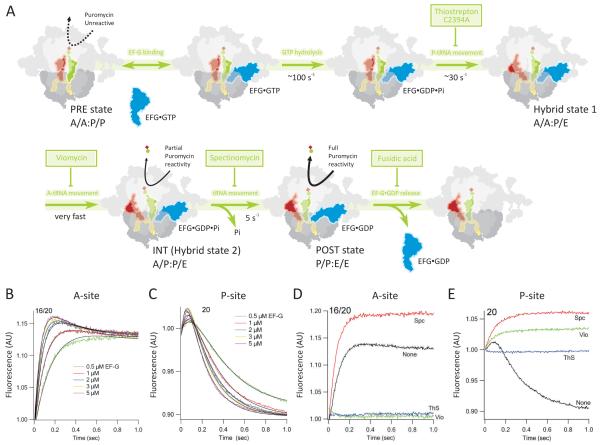
Application of such probes for measuring particular reactions of protein synthesis, either simultaneously or in parallel, allows formulation of detailed multi-step kinetic schemes that demonstrate the order of specific steps (e.g. ligand-binding, GTP-hydrolysis, conformational change, Pi release) within these overall reactions. Moreover, when two fluorescently-labeled components that form a good FRET pair are used together in a stopped-flow experiment, changes in FRET efficiency can be monitored as a function of time to allow structural changes to be attributed to a given step within the overall reaction (Milon et al., 2008; Qin et al., 2009; Seo et al., 2006; Wintermeyer et al., 2004). The availability of detailed multi-step kinetic schemes provides an excellent framework for understanding the mechanism of action of a given antibiotic, allowing identification of the precise step or steps that are targeted by that antibiotic. Below we present the results of studies illustrating how such identifications were achieved for antibiotics inhibiting four distinct steps during the translocation reaction (see also Peske et al., 2004). Related studies have been carried out for antibiotic effects on 70SIC formation (Grigoriadou et al., 2007; Milon et al., 2008; Qin et al., 2009), and PRE complex formation (Gromadski and Rodnina, 2004). Single turnover studies from several laboratories (Pan et al., 2007; Savelsbergh et al., 2003; Studer et al., 2003; Walker et al., 2008) have led to a comprehensive kinetic model for EF-G·GTP-dependent translocation, as shown in Figure 3A. In this model, EF-G·GTP binding to the PRE complex is followed by a rapid GTP hydrolysis step that triggers a conformational change in the ribosome leading to tRNA and mRNA movement and formation of the POST complex. The use of PRE complexes containing either proflavin-labelled fMetPhe-tRNAPhe (prf16/20) in the A-site or proflavin-labeled tRNAfMet (prf20) in the P-site provides two measures of the kinetics of tRNA movement during translocation (Figure 3B, C) (Pan et al., 2007). The biphasic spectral changes seen with each labelled tRNA allowed identification of a kinetically competent intermediate, denoted the INT complex, during translocation, which is rapidly formed following EF-G·GTP binding to the PRE complex and GTP hydrolysis, and is then more slowly converted to a POST complex. The rate of the latter reaction correlates closely with two other measures of translocation, the increases in reactivity of peptidyl tRNA toward a peptidyl acceptor molecule, such as the antibiotic puromycin (Pan et al., 2007), and the rate of mRNA movement (Liu et al., 2010; Savelsbergh et al., 2003). Movements of A- and P-tRNAs are strongly coupled during both INT formation and INT conversion to POST. Peptidyl-tRNA within the INT complex occupies a hybrid site, having puromycin reactivity intermediate between those of the PRE (almost zero) and POST complexes. The similarities in the rate constants for INT formation and for conformational change in the ribosome following EF-G·GTP binding (Seo et al., 2006) strongly suggest that the conformational change is rapidly propagated to the tRNA binding sites, triggering INT formation. Related experiments demonstrate that mRNA movement occurs during the slower, second phase of the reaction (Liu et al., 2010; Savelsbergh et al., 2003).
Figure 3. Fast kinetic analysis of translocation inhibitors.
(A) A scheme for EF-G mediated catalysis of translocation and sites of antibiotic interference. For the PRE complex only the classic state is shown, for simplicity (the hybrid state is omitted). Note that spectinomycin does not inhibit P release.
(B-E) EF-G-dependent translocation measured by change in the fluorescence prf-labeled tRNAs as a function of EF-G concentration, using E. coli PRE complexes containing (B) fMetPhe-tRNAPhe(prf16/20), or (C) prf-tRNAfMet(prf20). (D, E) as in (B, C), respectively, but with 1 μM EF-G and the absence (none, black) or presence of 1 mM spectinomycin (Spc, red), 5 μM viomycin (Vio, green) or 5 μM thiostrepton (ThS, blue).
The effects of three antibiotics on translocation, as measured by changes in the fluorescence of labeled tRNAs, are shown in Figure 3D, E (Pan et al. 2007; see also Peske et al., 2004). Although thiostrepton allows EF-G·GTP binding and GTP hydrolysis, it inhibits Pi release and the conformational changes following GTP hydrolysis (Rodnina et al., 1999; Seo et al., 2006), and thus prevents movement of either tRNA (Figure 3A). Viomycin breaks the strong coupling of tRNA movement during INT formation, permitting some P-tRNA movement (Figure 3E), while totally preventing A-tRNA movement (Figure 3D), resulting in the accumulation of the P/E complex that is an intermediate between the PRE and INT complexes (Figure 3A). This result is fully consistent with functional (Feldman et al., 2010) and structural (Stanley et al., 2010) results showing that viomycin stabilizes peptidyl-tRNA binding to the A-site decoding center within a PRE complex. Spectinomycin does not interfere with INT formation, but rather selectively stabilizes the INT complex, inhibiting its conversion to POST (Figure 3A). This is consistent with structural studies showing that binding of spectinomycin traps a distinct swiveling state of the head domain of the 30S subunit (Borovinskaya et al., 2007) and suggests that completion of the translocation process requires the full swiveling movement of the head. Interestingly, spectinomycin also stabilizes an intermediate with properties similar to those of the INT complex during LepA-facilitated back-translocation (Liu et al., 2010).
A fourth important antibiotic inhibitor of EF-G·GTP dependent translocation is fusidic acid (FA), which stabilizes EF-G·GDP on the ribosome, thus inhibiting EF-G turnover in translocation or in GTP hydrolysis. Fusidic acid has little if any effect on any of the other steps shown in Figure 3A (Savelsbergh et al., 2009; Seo et al., 2006). FA binds tightly to the EF-G·GDP·ribosome complex (Kd 0.4 μM; Willie et al., 1975), and acts as a “slow” inhibitor of EF-G·GTPase on the ribosome, decreasing the apparent initial turnover rate and eventually halting GTPase turnover altogether. Consistent with this model are both ensemble (Seo et al., 2006) and single molecule (Wang et al., 2007) FRET studies showing time-dependent FA stabilization of a specific conformation of the EF-G ribosome complex. Structural studies show that, within this complex, fusidic acid binds to EF-G in a pocket surrounded by the switch II region of the G domain (Gao et al., 2009). Such binding prevents the conformational change of switch II that would normally occur after GTP hydrolysis and that is required for EF-G·GDP release from the ribosome. Ribosome recycling following termination of protein synthesis is also strongly inhibited by fusidic acid mediated stabilization of EF-G·GDP binding to the ribosome (Savelsbergh et al., 2009).
Antibiotics modulate the energy landscape of ribosome dynamics
The first systems for investigating ribosome function and antibiotic action at the single-molecule scale emerged within the past 8-10 years and are thus in relative infancy. Nevertheless, early studies have already revealed important insights into the mechanism of translation (reviewed by Aitken et al., 2010; Marshall et al., 2008), often derived from the use of antibiotics which are highlighted here. The nature of single-molecule studies entails tracking individual ribosomes during the process of translation. Such efforts necessitate immobilizing one or more components of the translation apparatus to detect translation reactions over extended periods. The first single-molecule studies of ribosome function focused on detecting PTC activity using puromycin (Sytnik et al., 1999). Subsequent advances in CCD technologies, microfluidic devices and fluorophore photophysics have since allowed the tracking of complete protein synthesis reactions (see, for example, Uemura et al., 2010; Wen et al., 2008).
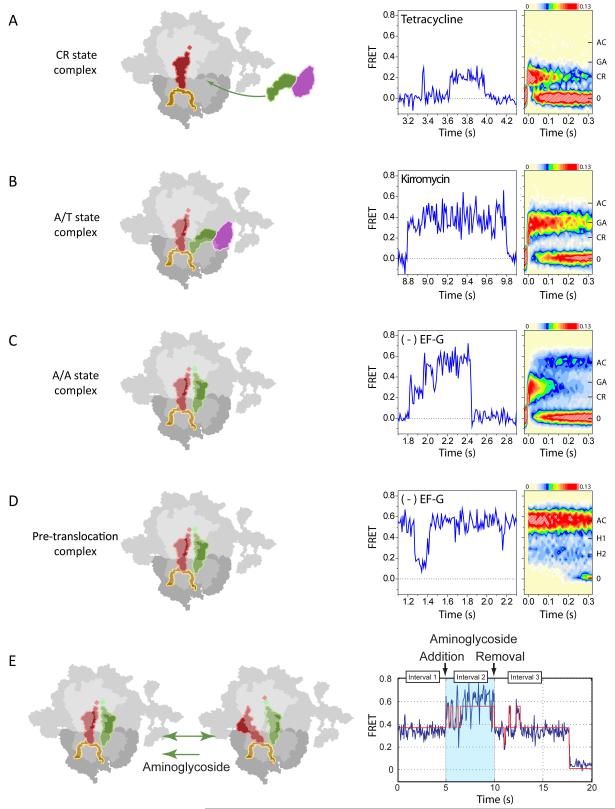
Puromycin reactivity assays were first used to demonstrate robust tRNA selection and translocation activities of surface-immobilized ribosome complexes taking advantage of the distinct reactivities of PRE and POST state ribosome complexes (Blanchard et al., 2004b). The interaction of ternary complex with the ribosome was investigated under pre-steady state conditions, probing the codon-dependence of the interaction and the sensitivity of the system to the known antibiotic inhibitors of tRNA selection: tetracycline and kirromycin (Blanchard et al., 2004a). This was achieved by taking advantage of established methods to site-specifically label P-site tRNA and aa-tRNA in the ternary complex with fluorescent probes via naturally-occurring post-transcriptional tRNA modifications (Plumbridge et al., 1980; Wintermeyer and Zachau, 1979). In this way, entry of ternary complex into the A site and the selection process could be monitored from the perspective of fluorescence resonance energy transfer (FRET) (Figure 4). Here, structural data suggested that complete aa-tRNA accommodation and formation of the PRE complex should result in a high-FRET state corresponding to classically-bound A- and P-site tRNA configurations.
Figure 4. Antibiotic action on ribosomal tRNA dynamics using smFRET.
Schematic representations of ribosomal states (left panels), representative smFRET data (center panels) and iteratively post-synchronized population FRET histograms (right panels) reveal structural and kinetic features of the ternary complex-ribosome interaction in the presence of the inhibitors (A) tetracycline, (B) kirromycin or (C) uninhibited, where aa-tRNA can enter the ribosome via transient CR and GA states on path to the fully-accommodated, AC, state, followed by peptide-bond formation. (D) In the absence of EF-G, A- and P-site tRNAs fluctuate between classical and hybrid configurations within the PRE complex stemming from thermally-accessible conformational processes intrinsic to the system. (E) Steady-state measurements demonstrate that tRNA dynamics in the PRE complex are strongly influenced by aminoglycoside-class antibiotics that reversibly bind the small subunit decoding region (Feldman et al., 2010). Iterative iterative, post-synchronized population FRET histogram is achieved by synchronizing all FRET observations for each individual FRET trajectory above the noise threshold (0.12 FRET) to t=0. Left and right data panels have the same Y-axis (FRET); accommodated (AC), GTPase activated (GA), and codon recognition (CR) states are indicated for reference. The color map applied to population histograms (right data panels) is encoded from light blue (lowly populated) to red (highly populated).
Textbook knowledge of the selection process suggested that tetracycline should efficiently block tRNA binding at the A site (Figure 1), as the primary tetracycline binding site overlaps with the position of the anticodon-stem-loop of an A-tRNA (Figure 2A, B). However, while single-molecule observations generally supported this notion, in the presence of tetracycline, ternary complex was shown to transiently interact (~50-100 ms lifetime) with the A-site (Figure 4A, unpublished data) in a manner consistent with the established bimolecular rate constant of the ternary complex-ribosome interaction dependent on the nature of the codon-anticodon interaction (Blanchard et al., 2004a). These results revealed that tetracycline principally blocks the selection process at a step after initial binding and codon recognition (Figure 4A).
In line with earlier observations that slow GTP hydrolysis occurs in tetracycline’s presence, higher FRET states were observed to be transiently sampled. This insight predicted that GTP hydrolysis occurred within higher-FRET states. This model was tested by performing experiments in the presence of non-hydrolyzable GTP analogues and kirromycin, an antibiotic that binds directly to EF-Tu to block aa-tRNA selection immediately subsequent to GTP hydrolysis (reviewed by Parmeggiani and Nissen, 2006). Like GDPNP, kirromycin efficiently blocked the selection process within a stabilized intermediate FRET state (Figure 4B). These data suggested that the cognate ternary complex moves on the ribosome subsequent to codon recognition to adopt stabilizing interactions that are required for GTP hydrolysis to occur. This is consistent with structures of the kirromycin-stalled EF-Tu·ribosome complex where EF-Tu is trapped in a high-affinity state on the ribosome with extensive interaction observed between the G-domain of EF-Tu and the large ribosomal subunit (Stark et al., 1997). A key feature of smFRET observations of the selection process was evidence of transient ternary complex dynamics, visualized by the existence of short-lived FRET excursions during the uninhibited selection process (Figure 4C). In the case of tetracycline, transient dynamics occurred to FRET states consistent with those observed in the presence of GDPNP and kirromycin (Figure 4C), whereas with kirromycin, transient dynamics occurred to even higher FRET states, consistent with those of the completely accommodated aa-tRNA configuration.
While the precise nature and role of conformational events during the selection process remain elusive, persistent dynamics within the PRE complex, following peptide bond formation have been more extensively explored (Munro et al., 2007). Detailed investigations into the molecular basis of transient dynamics in the PRE translocation showed that A- and P-site tRNAs reversibly exchange between classical and hybrid states through thermally-driven, conformational processes within the ribosome (Figure 4D). Such observations led to formulation of the metastable energy landscape hypothesis (Munro et al., 2009), which drew upon well-established observations that small proteins tend to adopt an ensemble of native state conformations that exist in dynamic, ligand-dependent exchange. This postulate predicts that intrinsic dynamics, which were previously shown to be a fundamental feature of smaller protein enzymes, also applied to ribosome function. In the native state ensemble view, dynamics in the PRE complex reflect the sampling of functionally-relevant ribosome conformations required for the downstream process of translocation. To test this model, the nature of steady-state fluctuations in the PRE complex were carried out in the absence and presence of specific aminoglycoside members (Feldman et al., 2010). As shown for paromomycin (Peske et al., 2004), aminoglycosides alter the PRE complex energy landscape leading to stabilization of peptidyl-tRNA in the classical A site (Figure 4E). For the specific aminoglycosides investigated, the extent of this stabilization was found to directly correlate with translocation inhibition. However, evidence of a second titratable binding site was observed for the antibiotic neomycin whose saturation ultimately led to the stabilization of hybrid tRNA configurations and almost complete inhibition of translocation (Feldman et al., 2010). Interestingly, bimodal titration behaviour was also observed for the chemically distinct peptide antibiotic, viomycin. Such observations, along with those showing that the potent translocation inhibitors spectinomycin and hygromycin B operate through mechanisms distinct from the aminoglycoside class (Peske et al., 2004), consistent with the notion that the translocation process is both complex and multistep in nature.
Antibiotics and translational-regulation
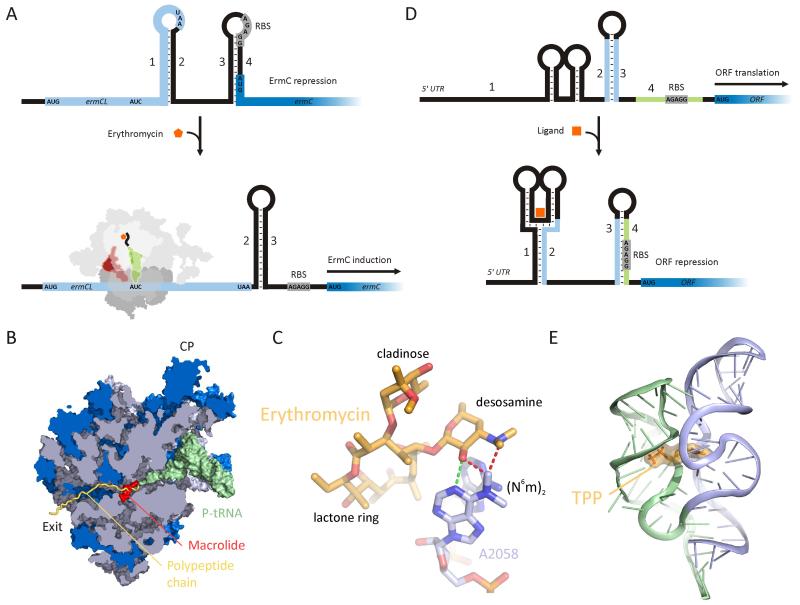
The cell makes use of numerous molecular sensors and switches that respond to small molecule effectors to control the production and function of proteins. Indeed, a wide variety of regulated bacterial operons have been discovered where ribosome-stalling during translation of a short upstream open reading frame (uORF), or so-called leader peptide, occurs in response to a small molecule, which in turn regulates expression of a downstream cistron (Lovett and Rogers, 1996; Ramu et al., 2009; Tenson and Ehrenberg, 2002). A well-characterized example of such a translational attenuation system is present in the E. coli tryptophanase operon where the presence of free tryptophan causes ribosome-stalling during translation of the TnaC leader peptide, allowing transcription and translation of the downstream tryptophan-catabolising enzymes (Gong and Yanofsky, 2002). Similarly, translation of leader peptides is utilized by bacteria to control the inducible expression of genes conferring resistance to antibiotics, such as chloramphenicol and the macrolide antibiotic erythromycin (Lovett and Rogers, 1996; Ramu et al., 2009). For example, in the absence of erythromycin, the expression of the erythromycin resistance gene, ermC, is repressed because the ribosome-binding site (RBS) and start codon are sequestered within a stem-loop secondary structure in the 5′ UTR of the mRNA (Figure 5A). In contrast, sub-inhibitory concentrations of erythromycin leads to ribosome stalling during translation of the ermCL leader peptide, which promotes the formation of an alternative stem-loop structure, exposing the RBS of ermC and thus allowing ErmC induction (Figure 5A). Mutations in the leader peptide abolish induction, indicating that the ErmCL sequence itself is an essential component of the stalling mechanism. Indeed, cryo-EM reconstruction of a TnaC-stalled ribosome reveals that the TnaC leader peptide makes distinct interactions with components of the ribosomal tunnel (Seidelt et al., 2009). Moreover, macrolide antibiotics, such as erythromycin, bind within the ribosomal tunnel adjacent to the PTC (Figure 5B) and inhibit translation by preventing the egression of the polypeptide chain. Erythromycin allows synthesis of oligopeptides of 8-11 amino acids before peptidyl-tRNA drop-off is induced (Tenson et al., 2003). Therefore it is likely that the ErmC leader peptide interacts directly with drug, consistent with the observation that the nature of the macrolide antibiotic, in particular, the presence of a C3-cladinose, has also been shown to be critical for stalling (Vazquez-Laslop et al., 2008). The ermC gene encodes a methyltransferase that modifies nucleotide A2058 of the 23S rRNA (Figure 5C). A2058 comprises part of the erythromycin binding site, forming a hydrogen bond to the C5-desosamine sugar of erythromycin (Figure 5C). Presumably, dimethylation of A2058 prevents stable binding of erythromycin because the N6 methyl group encroaches on the binding position of the drug (Figure 5C). The ermCL operon is thus auto-regulatory since N6-m2A2058 resistant ribosomes will not be subject to erythromycin-mediated translational stalling and therefore will promote ermC repression.
Figure 5. Antibiotic-mediated translation regulation.
(A) Schematic showing erythromycin induction of ErmC expression. In the absence of erythromycin, stem combinations 1 + 2 and 3 + 4 are favored leading to repression of ErmC due to the sequestering of the SD and AUG start codon of ErmC within the 3+ 4 stem-loop. However, in the presence of erythromycin, erythromycin-induced stalling of the ribosome at the AUC (Ile) codon during translation of the ermC leader, leads to an alternative 2 + 3 stem-loop, allowing ribosomes to access the SD and AUG start codon of the ermC gene.
(B) Transverse section of the ribosomal tunnel to reveal the binding site of the macrolides, such as erythromycin (orange), relative to P-tRNA (green) and putative path of nascent chain (blue).
(C) The methyltransferase ErmC dimethylates 23S rRNA nucleotide A2058, which prevents stable binding of erythromycin because of the close distance between the methyl groups of A2058 and the desosamine sugar of erythromycin (red dashed lines). In the absence of methylation, A2058 forms a hydrogen bond with the desosamine sugar (green dashed line).
(D) Schematic showing translational repression mediated by ligand binding to a riboswitch. Ligand binding leads to an alternative 1 + 2 and 3 + 4 stem-loop combination that sequesters the SD sequence of the downstream ORF leading to translational repression.
(E) Structure of the ligand TPP (thiamine pyrophosphate) bound to the E. coli TPP riboswitch.
Alternative small molecule-dependent regulatory systems have been identified that utilize metabolite-binding RNAs: These riboswitches usually reside within the 5′ UTRs of mRNAs and can adopt alternative conformations in response to ligand-binding to modulate expression of a downstream gene (reviewed by Roth and Breaker, 2009; Serganov and Patel, 2009). A large variety of riboswitches have been discovered that monitor the level of distinct metabolites within the cell, ranging from glycine, lysine, and S-adenosylmethionine to adenine, guanine and thiamine pyrophosphate (TTP). In the latter example, high concentrations of TPP lead to repression of downstream ORFs involved in thiamine biosynthesis (Figure 5D) (Winkler et al., 2002). The TPP riboswitch is a three-way helical junction (Figure 5E) (Serganov and Patel, 2009), which in the presence of TPP adopts a conformation that sequesters the RBS of the downstream ORF within an alternate stem-loop structure (Figure 5D) (Winkler et al., 2002). Because riboswitches bind small ligands and often control the expression of essential metabolic genes, they have been proposed as potential drug discovery targets (reviewed by Blount and Breaker, 2006). Indeed, a number of antibacterial metabolic analogues have already been reported: Pyrithiamine, a thiamine analogue, which probably targets the TPP riboswitch; lysine analogues, such as L-aminoethylcysteine and DL-4-oxalysine, that bind to the lysC riboswitch and inhibit Bacillus subtilis growth, as well as the riboflavin analogue, roseoflavin, which binds and regulates the FMN riboswitch. Furthermore, mutations within lysine and FMN riboswitches have been identified that confer resistance to the relevant metabolic analogues by disrupting binding to the riboswitch. Before the discovery of naturally occurring riboswitches, RNA aptamers were evolved in vitro with high affinity for small molecules, such as dyes, proteins, and antibiotics, including tetracyclines and aminoglycosides. In some cases, these artificial aptamers have been developed successfully to function as riboswitches for regulation of translation (reviewed by Suess and Weigand, 2008).
Development of novel translational inhibitors
The prevalence of antibiotic-resistant strains of pathogenic bacteria within the clinical setting is ever-increasing, prompting the need for the development of novel and more potent antibiotics (reviewed by Fischbach and Walsh, 2009). Following the introduction of the quinolones (ciprofloxacin) and streptogramins (Synercid) into clinical practise the early 1960’s, only three truly new classes of antibiotics have followed in the subsequent ~50 years; the oxazolidinones (linezolid, 2000), lipopeptides (daptomycin, 2003) and more recently the pleuromutilins (retapamulin, 2007). Nevertheless, in the intervening years a number of new semi-synthetic derivatives have been developed based on the original natural parent compound, for example, telithromycin from erythromycin and tigecycline from tetracycline (Fischbach and Walsh, 2009). These second and third generation antibiotics display improved activity against some multi-drug resistant pathogenic strains, while still utilizing the same core scaffold and binding site as the original parent compound and are thus ultimately vulnerable to some level of cross-resistance. While linezolid is a truly synthetic compound and therefore has not had prolonged exposure to bacterial populations, the binding site at the PTC of the ribosome (Figure 6A) (Ippolito et al., 2008; Wilson et al., 2008), overlaps with many other natural antibiotics, such as sparsomycin, chloramphenicols and pleuromutilins (e.g. tiamulin) (Figure 2D). Although resistance to linezolid has so far been very infrequent, cross-resistance to other antibiotics, such as pleuromutilins, has been documented in laboratories as well as in clinical isolates (see Wilson, 2009). The next generation oxazolidinones are already in clinical trials, such as radezolid and TR-701, both of which display improved antimicrobial activity against linezolid-resistant strains (Shaw et al., 2008; Wimberly, 2009). Nevertheless, discovery and development of new antimicrobial inhibitors continues and the ribosome as a target remains one of the major players (reviewed by Sutcliffe, 2005).
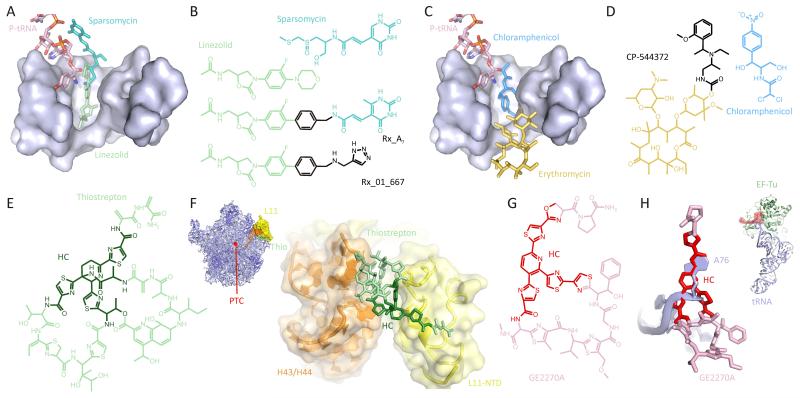
Figure 6. Development of novel antimicrobial inhibitors.
(A) Binding site of CCA-end of P-tRNA (pink), sparsomycin (teal) and linezolid (pale green) at the PTC of the bacterial 50S subunit.
(B) Chemical structures of sparsomycin, linezolid and the hybrid Rx_A1 compound.
(C) Binding site of CCA-end of P-tRNA (pink), chloramphenicol (blue) and erythromycin (yellow) at the PTC of the 50S subunit.
(D) Chemical structures of the hybrid macrolide CP-544372 and chloramphenicol.
(E) Chemical structure and (F) binding site of thiostrepton on the 50S subunit. The heterocyclic core (HC) of thiostrepton is highlighted in dark green. Inset shows 50S subunit with thiostrepton (green), L11 (yellow) and helix 43/44 (orange).
(G) Chemical structure and (H) binding site of GE2270A on EF-Tu. The heterocyclic core (HC) of GE2270A is highlighted in red and A76 of tRNA (pale blue) is shown for comparison. Inset shows structure of ternary complex EF-Tu-GDPNP-tRNA with relative position of GE2270A (red).
Indeed, the availability of atomic structures for the ribosome as well as many ribosome-antibiotic complexes (Figure 2) provides an excellent basis for the rational design of novel inhibitors (reviewed by Franceschi and Duffy, 2006). One approach to derive novel inhibitors takes advantage of the close proximity, or overlap, in binding site of antibiotics to generate so-called hybrid antibiotics. For example, Rib-X pharmaceuticals have designed a series of compounds based initially on the relative juxtaposition of the binding sites of linezolid and sparsomycin (Hansen et al., 2003) on the large subunit (Figure 6A), for example Rx_A7 (Figure 6B) (reviewed by Franceschi and Duffy, 2006; Wimberly, 2009). Different bridging elements were employed and the terminal aromatic group was optimized, producing biaryloxazolidinones, such as Rx_01_667 (Figure 6B), that are vastly superior to linezolid and active against linezolid-resistant strains (Skripkin et al., 2008). Similarly, bridged aromatic rings were attached to the cladinose ring of erythromycin to generate 4″-O-heteroarylcarbomyl derivatives of macrolides, such as CP-544372, with the intention to direct the sidechain into the binding site of chloramphenicol at the PTC (Figure 6C, D) (Xu et al., 2008). Many of these compounds have improved antimicrobrial activity against a variety of different erythromycin-resistant strains (Xu et al., 2008). Semi-synthetic aminoglycoside derivatives and mimetics have also been developed where the 2-DOS ring is replaced by aminoazepane or other synthetic scaffolds, as well as hybrids that fuse hygromycin B and paromomycin, or neomycin B with chloramphenicol or linezolid (reviewed by Hermann, 2007).
To date, all the clinical classes of antibiotics that target the ribosome bind either to the decoding center on the small subunit (tetracyclines and aminoglycosides) or to the PTC on the large subunit (macrolides, streptogramins, oxazolidinones and pleuromutilins) (Figure 2). Thus, there is an interest in developing antibiotics that target other regions of the ribosome to reduce the likelihood of cross-resistance. A number of biochemical approaches are being taken to identify functionally important hotspots on the ribosome that do not overlap with known antibiotic binding sites (Laios et al., 2004; Yassin et al., 2005; Yassin and Mankin, 2007). In fact, indications exist that a number of compounds, such as TAN-1057, the orthosomycins, GE82832 and the NRI compounds do interact with unique sites on the ribosome (see Wilson, 2009), however no structural characterization for these compounds exists so far. In contrast, the thiopeptide class of antibiotics include two distinct subfamilies that have been well-characterized both biochemically and structurally: Thiopeptides, such as thiostrepton (Figure 6E), bind the large ribosomal subunit where they interact with ribosomal protein L11 and rRNA helices 43 and 44 - far from the PTC (Figure 6F) (Harms et al., 2008). Thiostrepton is a well-known translocation inhibitor that prevents stable binding of EF-G to the ribosome (Figure 1) (reviewed by (Wilson, 2009). In contrast, the GE2270A-like thiopeptides (Figure 6G) have a binding site on elongation factor EF-Tu that overlaps with the position of terminal A76 of tRNA (Figure 6H), and thus prevent formation of the ternary complex EF-Tu-GTP-tRNA (Figure 1) (reviewed by Parmeggiani and Nissen, 2006). Although thiopeptides are already in veterinary usage, their low water solubility and poor bioavailability has so far precluded use in human medicine. However, recent successes in the total synthesis of a number of thiopeptides, including thiostrepton and GE2270A, opens the way to identification of new derivatives and lead compounds (reviewed by Nicolaou et al., 2009). Indeed, screening a thiopeptide fragment library using a series of translation machinery assays led to the identification of a series of novel thiopeptide precursor compounds that were either themselves inhibitory or are able to relieve the inhibitory effects of their parent compounds, thiostrepton or GE2270T (Starosta et al., 2009). Two of the families contained six-membered nitrogen heterocycle core (HC in Figure 6E-H), analogous to the thiopeptide antibiotics thiostrepton and GE2270A. Interestingly, the HC precursors were able to compete with both GE2770A for binding to EF-Tu and with thiostrepton for ribosome binding, suggesting the potential of these precursors as lead compounds for development of dual inhibitors (Starosta et al., 2009).
Acknowledgments
Research in the laboratories of the authors is financed by the Deutsche Forschungsgemeinschaft (WI3285/1-1 to D.N.W.), the Human Frontiers of Science Organization (RGY0088/2008 to S.C.B. and D.N.W.) and the National Institutes of Health (5R01GM079238-03 and GM07739 to S.C.B and GM071014 to B.S.C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aitken CE, Petrov A, Puglisi JD. Single Ribosome Dynamics and the Mechanism of Translation. Annu Rev Biophys. 2010;39:491–513. doi: 10.1146/annurev.biophys.093008.131427. [DOI] [PubMed] [Google Scholar]
- Allen DW, Zamecnik PC. The effect of puromycin on rabbit reticulocyte ribosomes. Biochim. Biophys. Acta. 1962;55:865–874. doi: 10.1016/0006-3002(62)90899-5. [DOI] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 2006;25:2539–2550. doi: 10.1038/sj.emboj.7601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer M, Rodnina MV. The ribosomal peptidyl transferase. Mol Cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004a;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. USA. 2004b;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JH. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem Biol. 2007;2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher MS, Marcker KA. Polypeptidyl-sRibonucleic acid and amino-acyl-sRibonucleic acid binding sites on ribosomes. Nature. 1966;211:380–384. doi: 10.1038/211380a0. [DOI] [PubMed] [Google Scholar]
- Davies J, Anderson P, Davis BD. Inhibition of protein synthesis by spectinomycin. Science. 1965;149:1096–1098. doi: 10.1126/science.149.3688.1096. [DOI] [PubMed] [Google Scholar]
- Davies J, Gilbert W, Gorini L. Streptomycin, suppression, and the code. Proc. Natl Acad. Sci. USA. 1964;51:883–890. doi: 10.1073/pnas.51.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat Chem Biol. 2010;6:54–62. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Structure of the A-site of E. coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- Franceschi F, Duffy EM. Structure-based drug design meets the ribosome. Biochem. Pharmacol. 2006;71:1016–1025. doi: 10.1016/j.bcp.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Gale EF, Cundliffe E, Reynolds PE, Richmond MH, Waring MJ. Antibiotic inhibitors of ribosome function. In The molecular basis of antibiotic action. John Wiley and sons; Bristol, UK: 1972. pp. 278–379. [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Polypeptide synthesis in Escherichia coli. II. The polypeptide chain and S-RNA. J Mol Biol. 1963;6:389–403. doi: 10.1016/s0022-2836(63)80051-0. [DOI] [PubMed] [Google Scholar]
- Gong F, Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- Grigoriadou C, Marzi S, Kirillov S, Gualerzi CO, Cooperman BS. A quantitative kinetic scheme for 70 S translation initiation complex formation. J Mol Biol. 2007;373:562–572. doi: 10.1016/j.jmb.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat Struct Mol Biol. 2004;11:316–322. doi: 10.1038/nsmb742. [DOI] [PubMed] [Google Scholar]
- Grunberg-Manago M, Dessen P, Pantaloni D, Godefroy-Colburn T, Wolfe AD, Dondon J. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J. Mol. Biol. 1975;94:461–478. doi: 10.1016/0022-2836(75)90215-6. [DOI] [PubMed] [Google Scholar]
- Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Hermann T. Aminoglycoside antibiotics: old drugs and new therapeutic approaches. Cell Mol Life Sci. 2007;64:1841–1852. doi: 10.1007/s00018-007-7034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa G, Iwakura N, Kaji A, Kaji H. The role of GTP in transient splitting of 70S ribosomes by RRF (ribosome recycling factor) and EF-G (elongation factor G) Nucleic Acids Res. 2008;36:6676–6687. doi: 10.1093/nar/gkn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito JA, Kanyo ZF, Wang D, Franceschi FJ, Moore PB, Steitz TA, Duffy EM. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem. 2008;51:3353–3356. doi: 10.1021/jm800379d. [DOI] [PubMed] [Google Scholar]
- Kaji H, Kaji A. Specific binding of sRNA to ribosomes: effect of streptomycin. Proc Natl Acad Sci USA. 1965;54:213–219. doi: 10.1073/pnas.54.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R, Ehrenberg M. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur J Biochem. 1994;226:355–360. doi: 10.1111/j.1432-1033.1994.tb20059.x. [DOI] [PubMed] [Google Scholar]
- Laios E, Waddington M, Saraiya AA, Baker KA, O’Connor E, Pamarathy D, Cunningham PR. Combinatorial genetic technology for the development of new anti-infectives. Arch Pathol Lab Med. 2004;128:1351–1359. doi: 10.5858/2004-128-1351-CGTFTD. [DOI] [PubMed] [Google Scholar]
- Liu H, Pan D, Pech M, Cooperman BS. Interrupted catalysis: the EF4 (LepA) effect on back-translocation. J. Mol. Biol. 2010;396:1043–1052. doi: 10.1016/j.jmb.2009.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett PS, Rogers EJ. Ribosome regulation by the nascent peptide. Microbiol. Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annu Rev Biochem. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- Milon P, Konevega AL, Gualerzi CO, Rodnina MV. Kinetic checkpoint at a late step in translation initiation. Mol. Cell. 2008;30:712–720. doi: 10.1016/j.molcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Morris AJ, Schweet RS. Release of soluble protein from reticulocyte ribosomes. Biochim Biophys Acta. 1961;47:415–416. doi: 10.1016/0006-3002(61)90310-9. [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Navigating the ribosome’s metastable energy landscape. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou KC, Chen JS, Edmonds DJ, Estrada AA. Recent advances in the chemistry and biology of naturally occurring antibiotics. Angew Chem Int Ed Engl. 2009;48:660–719. doi: 10.1002/anie.200801695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmeggiani A, Nissen P. Elongation factor Tu-targeted antibiotics: four different structures, two mechanisms of action. FEBS Lett. 2006;580:4576–4581. doi: 10.1016/j.febslet.2006.07.039. [DOI] [PubMed] [Google Scholar]
- Pavlov MY, Antoun A, Lovmar M, Ehrenberg M. Complementary roles of initiation factor 1 and ribosome recycling factor in 70S ribosome splitting. EMBO J. 2008;27:1706–1017. doi: 10.1038/emboj.2008.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- Pestka S, Marshall R, Nirenberg M. RNA codewords and protein synthesis. V. Effect of streptomycin on the formation of ribosome-sRNA complexes. Proc Natl Acad Sci USA. 1965;53:639–646. doi: 10.1073/pnas.53.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge JA, Baumert HG, Ehrenberg M, Rigler R. Characterisation of a new, fully active fluorescent derivative of E. coli tRNAPhe. Nucleic Acids Res. 1980;8:827–843. [PMC free article] [PubMed] [Google Scholar]
- Polacek N, Mankin AS. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit Rev Biochem Mol Biol. 2005;40:285–311. doi: 10.1080/10409230500326334. [DOI] [PubMed] [Google Scholar]
- Qin H, Grigoriadou C, Cooperman BS. Interaction of IF2 with the ribosomal GTPase-associated center during 70S initiation complex formation. Biochemistry. 2009;48:4699–4706. doi: 10.1021/bi900222e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol Microbiol. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- Rheinberger H-J. A history of protein biosynthesis and ribosome research. 2004. [Google Scholar]
- Rodnina MV, Savelsbergh A, Matassova NB, Katunin VI, Semenkov YP, Wintermeyer W. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl Acad. Sci. USA. 1999;96:9586–9590. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol. Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. Rna. 2009;15:772–780. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache JP, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H, Abedin S, Kamp D, Wilson DN, Nierhaus KH, Cooperman BS. EF-G-dependent GTPase on the ribosome. Conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Amino acid recognition and gene regulation by riboswitches. Biochim Biophys Acta. 2009;1789:592–611. doi: 10.1016/j.bbagrm.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KJ, Poppe S, Schaadt R, Brown-Driver V, Finn J, Pillar CM, Shinabarger D, Zurenko G. In vitro activity of TR-700, the antibacterial moiety of the prodrug TR-701, against linezolid-resistant strains. Antimicrob Agents Chemother. 2008;52:4442–4447. doi: 10.1128/AAC.00859-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers A, Beringer M, Rodnina MV, Wolfenden R. The ribosome as an entropy trap. Proc. Natl. Acad. Sci. U S A. 2004;101:7897–7901. doi: 10.1073/pnas.0402488101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovic M, Steitz TA. A structural view on the mechanism of the ribosome-catalyzed peptide bond formation. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbagrm.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skripkin E, McConnell TS, DeVito J, Lawrence L, Ippolito JA, Duffy EM, Sutcliffe J, Franceschi F. R chi-01, a new family of oxazolidinones that overcome ribosome-based linezolid resistance. Antimicrob Agents Chemother. 2008;52:3550–3557. doi: 10.1128/AAC.01193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat. Struct. Mol. Biol. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Rinkeappel J, Brimacombe R, Wintermeyer W, Vanheel M. Visualization of Elongation Factor Tu On the Escherichia Coli Ribosome. Nature. 1997;389:403–406. doi: 10.1038/38770. [DOI] [PubMed] [Google Scholar]
- Starosta AL, Qin H, Mikolajka A, Leung GY, Schwinghammer K, Nicolaou KC, Chen DY, Cooperman BS, Wilson DN. Identification of distinct thiopeptide-antibiotic precursor lead compounds using translation machinery assays. Chem Biol. 2009;16:1087–1096. doi: 10.1016/j.chembiol.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- Suess B, Weigand JE. Engineered riboswitches: overview, problems and trends. RNA Biol. 2008;5:24–29. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JA. Improving on nature: antibiotics that target the ribosome. Curr Opin Microbiol. 2005;8:534–542. doi: 10.1016/j.mib.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Sytnik A, Vladimirov S, Jia Y, Li L, Cooperman BS, Hochstrasser RM. Peptidyl transferase center activity observed in single ribosomes. J. Mol. Biol. 1999;285:49–54. doi: 10.1006/jmbi.1998.2312. [DOI] [PubMed] [Google Scholar]
- Tenson T, Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 2003;330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc Natl Acad Sci U S A. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qin H, Kudaravalli RD, Kirillov SV, Dempsey GT, Pan D, Cooperman BS, Goldman YE. Single-molecule structural dynamics of EF-G--ribosome interaction during translocation. Biochemistry. 2007;46:10767–10775. doi: 10.1021/bi700657d. [DOI] [PubMed] [Google Scholar]
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willie GR, Richman N, Godtfredsen WP, Bodley JW. Some characteristics of and structural requirements for the interaction of 24,25-dihydrofusidic acid with ribosome - elongation factor G complexes. Biochemistry. 1975;14:1713–1718. doi: 10.1021/bi00679a025. [DOI] [PubMed] [Google Scholar]
- Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci U S A. 2008;105:13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT. The use of ribosomal crystal structures in antibiotic drug design. Curr Opin Investig Drugs. 2009;10:750–765. [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem. Soc. Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Zachau HG. Fluorescent derivatives of yeast tRNAPhe. Eur. J. Biochem. 1979;98:465–475. doi: 10.1111/j.1432-1033.1979.tb13207.x. [DOI] [PubMed] [Google Scholar]
- Xu P, Liu L, Jin ZP, Wang GQ, Liu J, Li Y, Lei PS. Synthesis and antibacterial activity of 4″-O-heteroarylcarbamoyl derivatives of macrolide. Bioorg Med Chem Lett. 2008;18:5507–5511. doi: 10.1016/j.bmcl.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Yarmolinsky MB, de la Haba G. Inhibition by puromycin of amino acid incorporation into protein. Proc Natl Acad Sci USA. 1959;45:1721–1729. doi: 10.1073/pnas.45.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin A, Fredrick K, Mankin AS. Deleterious mutations in small subunit ribosomal RNA identify functional sites and potential targets for antibiotics. Proc Natl Acad Sci U S A. 2005;102:16620–16625. doi: 10.1073/pnas.0508444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin A, Mankin AS. Potential new antibiotic sites in the ribosome revealed by deleterious mutations in RNA of the large ribosomal subunit. J Biol Chem. 2007;282:24329–24342. doi: 10.1074/jbc.M703106200. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]