Abstract
The transcription factor Hypoxia-Inducible Factor 1 (HIF-1) has emerged as a major antitumor molecular target. Inhibition of HIF-1 activation has been shown to suppress the growth, survival, and metastatic spread of hypoxic tumors. The NCI Open Repository of marine invertebrates and algae lipid extracts was evaluated for HIF-1 inhibitory activity in a T47D human breast tumor cell-based reporter assay. Bioassay-guided chromatographic separation of the active extract from the sponge Dendrilla nigra produced four new lamellarin-like phenolic pyrroles, which most closely resemble the structure of the known D. cactos compound lamellarin O. However, unlike lamellarins, the structures of neolamellarin A (1), neolamellarin B (2), 5-hydroxyneolamellarin B (3), and 7-hydroxyneolamellarin A (4) lack the carboxyl moiety at position C-2 of the substituted pyrrole ring and have a significantly different pattern of oxidation. Compound 4 was found to inhibit hypoxia-induced HIF-1 activation (IC50 1.9 μM) in T47D cells. Hypoxic induction of vascular endothelial growth factor (VEGF), a potent angiogenic factor and HIF-1 target gene, was also inhibited by 4 at the secreted protein level.
When the consumption of oxygen outstrips the supply, hypoxic tissue areas emerge. One major character of advanced solid tumors is the prevalence of heterogeneously distributed hypoxic regions. Tumor hypoxia exerts two opposite effects on tumor cells – 1) reduction of cellular metabolism and cell proliferation that leads to subsequent cell death; and 2) promotion of aggressive tumor growth by selecting highly mutated and malignant tumor cells.1 Extensive tumor hypoxia correlates poor clinical outcome and treatment resistance.1 Despite intensive drug discovery efforts, there is no approved drug that specifically targets tumor hypoxia at the present time.2
One of the key molecular targets for anti-tumor hypoxia drug discovery is the transcription factor hypoxia-inducible factor-1 (HIF-1), a heterodimer composed of the oxygen-regulated HIF-1α and the constitutively expressed HIF-1β subunits.3 Since the discovery of HIF-1 in the early 1990s,4 extensive studies on HIF-1 and cancer strongly support HIF-1 as a valid molecular target for drug discovery that targets tumor hypoxia. Preclinical studies revealed that HIF-1 inhibition retards tumor growth and improves treatment outcome when combined with radiation and chemotherapeutic agents.5–11 Small molecule inhibitors of HIF-1 activation represent potential tumor-selective agents with reduced levels of detrimental effects on the well-oxygenated normal tissues. Intensive research efforts have yielded a number of chemically diverse small molecule HIF-1 inhibitors that target various cellular pathways that regulate hypoxic signaling.2,12,13 The majority of small molecule HIF-1 inhibitors discovered are either natural products or derived from natural products. One potential limitation of these small molecule inhibitors is that many are believed to inhibit HIF-1 through molecular mechanisms that would typically be associated with cytotoxic effects.2,12,13 The challenge lies in the discovery of potent HIF-1 inhibitors with high therapeutic index towards tumor cells while sparing normal cells.
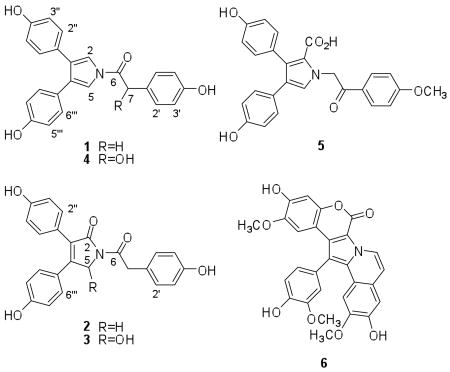
In search of potent and selective small molecule HIF-1 inhibitors, our research efforts take advantage of the vast biochemical diversity afforded by natural products to discover novel HIF-1 inhibitors that may function through mechanisms that have not previously been identified to regulate HIF-1 activation. Using a T47D human breast carcinoma cell-based reporter assay,14 the NCI Open Repository of marine invertebrates and algae lipid extracts was examined for HIF-1 inhibitory activity. The crude extract of the sponge Dendrilla nigra (Aplysillidae) inhibited hypoxia-induced HIF-1 activation (81% inhibition at 5 μg mL−1). Bioassay-guided chromatographic separation of the D. nigra extract yielded four new lamellarin-like phenolic pyrroles (1-4) that bear structural features similar to lamellarin O (5), originally isolated from the Australian sponge D. cactos.15
Over 30 lamellarins have been isolated from marine sponges (ie. Dendrilla cactos), ascidians (ie. Didemnum chartaceum), and opisthobranchs (Lamellaria sp.). With the exception of lamellarins O-R, lamellarins generally contain a pentacyclic (6H-[1]bezopyrano[4′,3′;4,5]pyrrolo[2,1-α]isoquinolin-one chromophore, as exemplified by lamellarin D (6), the most studied member of this class.16 Lamellarins have attracted a great deal of attention for their wide range of biological activities that include inhibition of HIV-1 intergrase, inhibition of topoisomerase I, and tumor cell cytotoxic effects that are insensitive to P-glycoprotein-mediated resistance and may even reverse MDR efflux.16
Results and Discussion
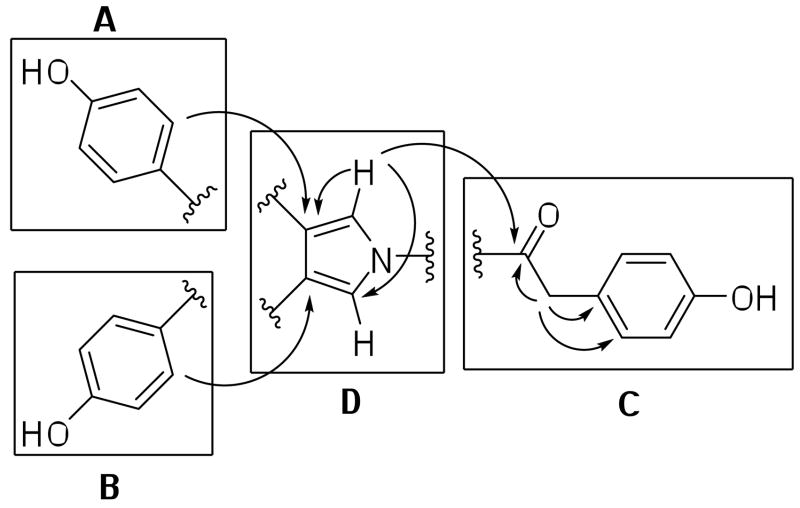
The crude lipid extract of the tropical marine sponge Dendrilla nigra, which was collected from shallow water in Saipan (NCI Open Repository collection #J002799), inhibited HIF-1 activation in T47D cells by 81% at 5 μg mL−1. The extract of D. nigra (5 g) was subjected to repeated Si gel CC to yield four new lamellarin-like alkaloids (1-4). Compound 1 was isolated as white needle-like crystals. Strong IR absorptions (3369 and 1697 cm−1) indicated the presence of phenolic and amide carbonyl functionalities, respectively. The 13C NMR and DEPT spectra showed twelve carbon resonances that included one carbonyl resonance at δ 168.8 (s, C-6), five quarternary carbon resonances, five sp2 aromatic carbon resonances and one sp3 methylene carbon resonance. The 1H NMR spectrum of 1 showed one methylene proton resonance at δ 4.20 (s, 2H-7), five aromatic proton resonances, and one downfield exchangeable phenolic proton resonance at δ 8.38 (s, 3H-OH). The HRESIMS, 1H and 13C NMR (Tables 1 and 2) supported the molecular formula C24H19NO4 and revealed that 1 possessed a symmetrical structure. The 1H-1H COSY correlations showed two 1H-1H spin systems: δ 7.23 (d, J = 8.4, 2H-2′,6′) was coupled to δ 6.83 (d, J = 8.4, 2H-3′,5′), and δ 7.09 (d, J = 8.4, 4H-2″,6″, 2‴,6‴) was coupled to δ 6.78 (d, J = 8.4, 4H-3″,5″, 3‴,5‴). These AA′XX′ patterns were indicative of three para-hydroxy-substituted aromatic rings. The two phenolic spin systems in partial structures A and B were found to be connected to partial structure C by the observation of long-range 1H-13C couplings between δ 4.20 (s, 4H-2″,6″,2‴,6‴) and 127.9 (2CH, C-3,4). The combination of long-range 1H-13C HMBC correlations from δ 7.09 (s, 2H-7) to 168.8 (C, C-6), 130.6 (CH, C-2′,6′), 124.8 (C, C-1′) and from δ7.23 (d, J = 8.4, 2H-2′,6′) to 39.8 (CH2, C-7), and 156.4 (CH, C-4′) (Figure 1) readily facilitated the establishment of three para-substituted phenolic partial structures A, B and C. Partial structures A and B exist in a similar magnetic environment and thus have identical chemical shifts. Taking into account partial structures A–C, only a C4H2N portion remains for the central connecting core structure of 1. Interpretation of the 1H NMR, 13C NMR, and 1H-13C HMBC correlations from δ 7.49 (s, 2H-2,5) to C-3,4 (127.9, C) and C-2,5 (117.2, CH), established a 1,3,4-trisubstituted 1H-pyrrole core partial structure D. The IR and the δ 168.8 chemical shift of C-6 support nitrogen substitution of the carbonyl moiety (i.e. amide). The final connectivity between each partial structure was determined from long-range 1H-13C HMBC correlations (as depicted in Figure 1). Thus the structure was assigned as a new lamellarin-like phenolic pyrroles metabolite neolamellarin A (1). Compound 1 can be thought of as a decarboxylated and alternatively oxidized form of lamellarin O.15
Table 1.
1H NMR data for 1-4 in acetone-d6 (400 MHz, unless otherwise noted)
| H | 1 | 2 | 3 | 4a |
|---|---|---|---|---|
| 2 | 7.49 (2H, s), H-2,5 | --- | --- | 7.48 (2H, s), H-2,5 |
| 5 | 4.65 (2H, s) | 6.54 (1H, s) | ||
| 7 | 4.20 (2H, s) | 4.24 (2H, s) | 4.26 (1Ha, d, 15.2) 4.16 (1Hb, d, 15.2) |
5.79 (1H, s) |
| 2′,6′ | 7.23 (2H, d, 8.4) | 7.17 (2H, d, 8.4) | 7.16 (2H, d, 8.4) | 7.42 (2H, d, 8.4) |
| 3′,5′ | 6.83 (2H, d, 8.4) | 6.75 (2H, d, 8.4) | 6.76 (2H, d, 8.4) | 6.83 (2H, d, 8.4) |
| 2″,6″ | 7.09 (4H, d, 8.4), H-2″,6″, 2‴,6‴ | 7.22 (2H, d, 8.4) | 7.21 (2H, d, 8.4) | 7.01 (4H, d, 8.4), H-2″,6″, 2‴,6‴ |
| 3″,5″ | 6.78 (4H, d, 8.4), H-3″,5″, 3‴,5‴ | 6.86 (2H, d, 8.4) | 6.85 (2H, d, 8.4) | 6.73 (4H, d, 8.4), H-3″,5″, 3‴,5‴ |
| 2‴,6‴ | 7.31 (2H, d, 8.4) | 7.39 (2H, d, 8.4) | ||
| 3‴,5‴ | 6.80 (2H, d, 8.4) | 6.78 (2H, d, 8.4) | ||
| OH | 8.38 (3H, s) | --- | 5.71 (1H, s), OH-5 | --- |
600 MHz
Table 2.
13C NMR data for 1-4 in acetone-d6 (100 MHz, unless otherwise noted)
| C | 1 | 2 | 3 | 4a |
|---|---|---|---|---|
| 2 | 117.2 CH | 170.6 C | 169.4 C | 118.5 CH |
| 3 | 127.9 C | 129.8 C | 129.7 C | 129.3 C |
| 4 | 127.9 C | 151.0 C | 152.5 C | 129.3 C |
| 5 | 117.2 CH | 50.7 CH2 | 81.5 CH | 118.5 CH |
| 6 | 168.8 C | 171.6 C | 172.0 C | 171.9 C |
| 7 | 39.8 CH2 | 42.1 CH2 | 42.6 CH2 | 73.9 CH |
| 1′ | 124.8 C | 126.6 C | 126.5 C | 131.5 C |
| 2′,6′ | 130.6 CH | 131.7 CH | 132.1 CH | 129.9 CH |
| 3′,5′ | 115.4 CH | 115.9 CH | 116.1 CH | 116.8 CH |
| 4′ | 156.4 C | 157.1 C | 157.4 C | 159.1 C |
| 1″ | 125.6 C | 123.8 C | 123.9 C | 126.5 C |
| 2″,6″ | 129.6 CH | 131.9 CH | 131.8 CH | 130.8 CH |
| 3″,5″ | 115.1 CH | 116.2 CH | 116.4 CH | 116.3 CH |
| 4″ | 156.4 C | 158.4 C | 158.9 C | 157.9 C |
| 1‴ | 125.6 C | 124.6 C | 123.9 C | 126.5 C |
| 2‴,6‴ | 129.6 CH | 130.6 CH | 132.2 CH | 130.8 CH |
| 3‴,5‴ | 115.1 CH | 116.4 CH | 116.4 CH | 116.3 CH |
| 4‴ | 156.4 C | 160.2 C | 160.2 C | 157.9 C |
125 MHz
Figure 1.
Selected 1H-13C HMBC (arrows) correlations connecting partial structures A–D for 1.
Compound 2 was isolated as yellow oil. As observed for 1, the IR spectrum contained strong bands at 3384 (phenol) and 1695 cm−1 (amide carbonyl). The HRESIMS, 1H and 13C NMR (Tables 1 and 2) supported the molecular formula C24H19NO5. The 1H NMR spectrum revealed six aromatic proton resonances and two methylene resonances. Analysis of the 13C and 13C DEPT NMR spectra revealed 18 carbon resonances, including two carbonyl resonances at δ170.6 (C, C-2) and 171.6 (C, C-6), eight quarternary carbon resonances, six aromatic methine resonances, and two methylene resonances at δ 50.7 (CH2, C-5) and 42.1 (CH2, C-7). Unlike the 3,4-disubstituted 1H-pyrrole structure observed in 1, compound 2 contained a significantly different set of 1H and 13C resonances: [δH 4.65 (2H, s) and δC 50.7 (CH2), 151.0 (C), 129.8 (C), 170.6 (C)] and lacked the symmetrical structure observed in 1, thus the chemical shifts of partial structures A and B were distinct. Comparison of the 1H and 13C NMR spectra of 2 with those of 1 indicated that the structure of 2 possessed the same p-substituted phenolic aromatic partial structures A, B and C, which were further confirmed by the 1H-1H COSY and 1H-13C HMBC correlations. According to the molecular formula, partial structure D was assigned the formula C4H2ON. The 1H-13C HMBC correlations from δ 4.65 (s, 2H-5) to 151.0 (C, C-4), 129.8 (C, C-3), 170.6 (C, C-2), from δ 7.22 (d, J = 8.4, 2H-2″,6″) to 129.8 (C, C-3), and from δ 7.31 (d, J = 8.4, 2H-2‴,6‴) to 151.0 (C, C-4) established that the structure of 2 (neolamellarin B) was similar to that of 1, except that 2 possessed a 1,3,5-trisubstitutedpyrrolidin-2-one core, rather than the 1,3,5-substituted-1H-pyrrole core observed in 1.
Compound 3 was isolated as yellow powder. Infrared absorbances, similar to those observed for 1 and 2, were observed at 3336 (phenol) and 1696 cm−1 (amide carbonyl). The molecular formula of 3 was determined to be C24H19NO6 by HRESIMS. The 1H and 13C NMR data indicated that the structure of 3 possessed the same A–C partial structures as ascribed to 1 and 2. Comparison of the 13C NMR spectrum of 3 with that of 2 suggested that the only difference is that the methylene carbon resonance at δ 50.7 (CH2, C-5) of 2 was replaced by a hydroxylated methine carbon resonance at δ 81.5 (CH, C-5) in 3. The 1H NMR resonance at δ 4.65 (s, 2H-5) observed in 2 was missing in the spectrum of 3. The 1H NMR spectrum of 3 also contained one additional oxygenated methine resonance at δ 6.54 (s, 1H-5) and one exchangeable hydroxyl proton resonance at δ 5.71 (s, OH-5). These observations were consistent with the differences in the 13C NMR spectrum. Furthermore, the C-7 methylene proton signal appeared as two doublets [δ 4.26 (d, J = 15.2, 1H-7a), 4.16 (d, J = 15.2, 1H-7b)] in 3, rather than as a singlet [δ 4.24 (s, 2H-7)] as observed in 2. This phenomenon can be explained by the presence of a chiral C-5 hydroxyl substituent that presumably forms a hydrogen-bond to the C-6 carbonyl to form a 6-membered ring, thus strongly influencing the magnetic environment of the diastereotopic protons H-7a and H-7b. The 1H-1H COSY and 1H-13C HMBC correlations supported the structure of this new compound, trivially named 5-hydroxyneolamellarin B (3).
Compound 4 was isolated as yellow oil. The molecular formula of 4 was deduced to be C24H19NO5 by HRESIMS, thus indicating that it may be an isomer of 2. Analysis of the 1H and 13C NMR spectra indicated that 4 was structurally very similar to 1. The only difference in the 13C NMR spectrum (Table 2) was that 4 contains one more oxygenated methine carbon resonance at 73.9 ppm (CH, C-7) in place of a methylene carbon resonance observed at 39.8 ppm (CH, C-7) in the spectrum of 1. The presence of a C-7 hydroxyl substituent was further supported by 1H-13C HMBC correlations from H-7 (5.79 ppm) to both the C-6 carbonyl carbon resonance at 171.9 ppm and the aromatic carbon resonances at C-2′/C-6′ (129.9 ppm). The lack of optical activity suggested that 4 may be racemic. This was not surprising due to the unique environment of the C-7 hydroxyl moiety that is both benzylic and alpha to the amide carbonyl. Thus the structure of 4 was deduced to be a simple hydroxylated analog of 1, trivially named 7(R/S)-hydroxyneolamellarin A (4).
The effects of 1-4 on HIF-1 activity in a T47D cell-based reporter assay were determined in a concentration-response study.14 The most active compound 4 inhibited hypoxia (1% O2)-induced HIF-1 activation with an IC50 value of 1.9 μM. At the highest concentration tested (10 μM), the percentage inhibition exerted by 1-4 on hypoxia-induced HIF-1 activation ranged from 26% (1), 23% (2), 22% (3), to 98% (4). To control for potentially non-specific interference due to the inhibitory effects on tumor cell proliferation/viability, a Neutral Red assay was performed. Under hypoxic conditions (1% O2 for 16 and 48 h, respectively), 1-4 (at concentrations up to 10 μM) did not significantly inhibit T47D cell proliferation/viability (< 20%), as measured by the Neutral Red assay. To discern the specificity of 4 on HIF-1 activation induced by hypoxia versus iron chelation, the effects of 4 on 1,10-phenanthroline (10 μM)-induced HIF-1 activation (IC50 3.7 μM) were evaluated in the T47D cell-based reporter assay. As shown in Figure 2, 4 inhibited both hypoxia- and iron chelator-induced HIF-1 activation in T47D cells and a more pronounced inhibition was observed towards hypoxia-induced HIF-1 activation. In a transformed 3T3 cell-based reporter assay, 4 did not affect luciferase expression from a control construct (pGL3-control) at concentrations that range from 1 to 10 μM (data not shown). These results suggest that 4 specifically inhibited HIF-1 activation in the luciferase-based reporter assay. Compounds 1-3 (10 μM) partially inhibited 1,10-phenanthroline-induced HIF-1 activation in T47D cells: 58 ± 7% (1), 42 ± 6% (2), and 33 ± 10% (3), respectively.
Figure 2.
Effects of 4 on HIF-1 activation. A T47D cell-based reporter assay was used to determine the effects of 4 on hypoxia- (1% O2; solid bar) and iron chelator-induced (1,10-phenanthroline, 10 μM, open bar) HIF-1 activation. Compound 4 was tested at the concentrations of 1, 3, and 10 μM and the positive control emetine was tested at 1 μM. Data shown are averages from one experiment performed in triplicate and the bars represent standard deviation.
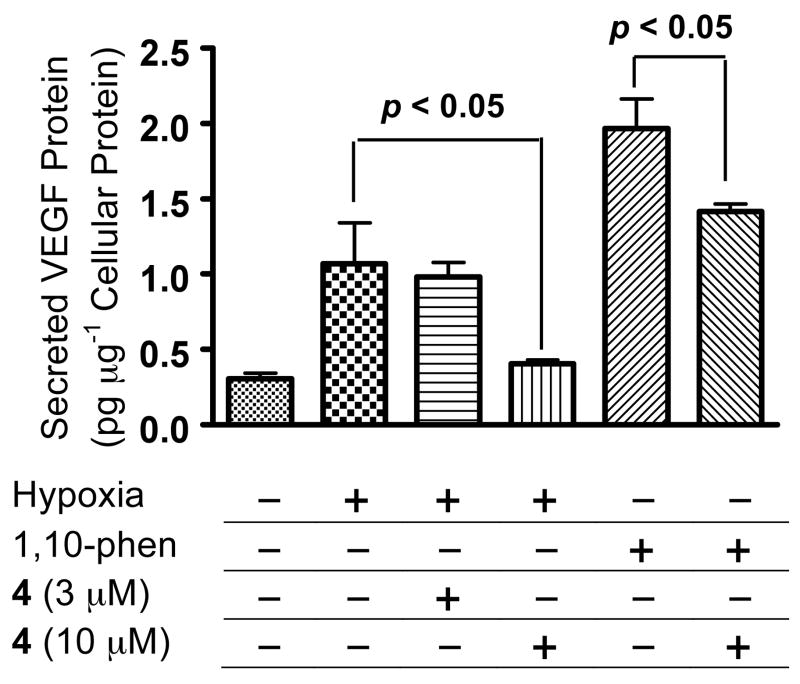
One of the most studied HIF-1 regulated target genes is vascular endothelial growth factor (VEGF), a key angiogenic factor that promotes tumor angiogenesis.17 The effects of 4 on the induction of secreted VEGF protein in T47D cells by hypoxia (1% O2) and 1,10-phenanthroline (10 μM) were examined and the data shown in Figure 3. Hypoxic exposure increased the level of secreted VEGF protein by 2.4-fold, and 4 (10 μM) blocked this induction (87%). No inhibitory effect was observed at a lower concentration (3 μM). Treatment with 1,10-phenanthroline increased secreted VEGF protein level by 5.3-fold, and 4 (10 μM) produced a 33% decrease in the induction. This result correlates with those obtained in the HIF-1 reporter assay. The inhibitory effect of 4 is more pronounced on hypoxia-induced HIF-1 and the downstream target gene (VEGF), relative to the effect on 1,10-phenanthroline-induced HIF-1 and VEGF.
Figure 3.
Effects of 4 on the induction of secreted VEGF protein. Exponentially grown T47D cells were exposed to 4 under hypoxic conditions (1% O2) or in the presence of 1,10-phenanthroline (10 μM) for 16 h. The level of secreted VEGF protein in the conditioned media was determined by ELISA and normalized to the level of protein in cellular lysates. Data shown are averages from one experiment performed in triplicate and the bars represent standard deviation. Data were compared by ANOVA followed by Dunnett’s multiple comparison test using GraphPad Prism 4 software. The p-values between the induced control and 4 treated samples are shown.
The effects of 1-4 were further evaluated on tumor cell proliferation/viability under normoxic conditions. Incubation (48 h) in the presence of 4 produced a marginal dose-dependent inhibition of T47D cell proliferation/viability [−8 ± 10% (1 μM), 34 ± 7% (10 μM)]. The 48 h duration of these assays appear to exert a low level of suppression of cell proliferation/viability at the highest concentration tested. This is in contrast to the lack of cytostatic/cytotoxic effects observed in the shorter incubation time (16 h). None of the other three compounds affected T47D cell proliferation/viability under normoxic conditions.
Lamellarin D (6) and related lamellarins that possess a planer pentacyclic chromophore are associated with DNA intercalation, metal-dependent cleavage, inhibition of topoisomerase I, and this planer system also appears to be linked to the tumor cell cytotoxic activity.16 The structure of 4 lacks the planer pentacylcic chromophore and appears unlikely to effect HIF-1 through the inhibition of topoisomerase I. The limited quantity of 4 prohibited mechanism of action studies and further investigation of its potential as a molecular-targeted antitumor agent. Efforts are required to synthetically produce 4 in order to further examine the relationship between lamellarins and the regulation of HIF-1 in tumors. In summary, compound 4 represents a bona fide HIF-1 inhibitor that suppresses the activation of HIF-1 and its downstream target VEGF in T47D cells.
Experimental Section
General Experimental Procedures
Melting points were measured using a MEL-TEMP® 3.0 micro-melting point apparatus and were uncorrected. Optical rotations were obtained on an AP IV/589-546 digital polarimeter. The IR spectrum was obtained using a Bruker Tensor 27 genesis Series FTIR. UV spectra were recorded on a Varian 50 Bio spectrophotometer. The NMR spectra were recorded in acetone-d6 on Bruker AMX-NMR spectrometers operating at either 400 MHz or 600 MHz for 1H and either 100 MHz or 150 MHz for 13C, respectively. The NMR spectra were recorded running gradients and solvent peaks (acetone-d6, δ 2.05 for 1H and δ 206.7 for 13C) were used as internal references. The HRESIMS spectra were measured using a Bruker Daltonic micro TOF with electrospray ionization. Silica gel (200–400 mesh) was used for column chromatography. The TLCs were run on Merck Si60F254 or Si60RP18F254 plates and visualized under UV at 254 nm and by heating after spraying with a 1% anisaldehyde solution in acetic acid:H2SO4 (50:1).
Sponge Material
The sponge extract was obtained from the National Cancer Institute’s Open Repository Program. Dendrilla cf. nigra (Aplysillidae) was collected at 8 m depth on February 18, 1993 (collection J002799) from Saipan, Commonwealth of the Northern Mariana Islands. The sample was identified by Dr. Michele Kelly (National Institute of Water and Atmospheric Research Limited, Auckland, New Zealand). It was frozen at −20 °C and ground in a meat grinder. A voucher specimen was placed on file with the Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution, Washington, D.C.
Extraction and Isolation
Ground sponge material was extracted with water. The residual sponge sample was lyophilized and extracted with CH2Cl2:MeOH (1:1), residual solvents were removed under vacuum, and the crude extract stored at −20 °C in the NCI repository at the Frederick Cancer Research and Development Center (Frederick, Maryland). The crude extract (5.0 g) (inhibited HIF-1 activation in T47D cells by 81% at 5 μg mL−1) was separated into 9 fractions by Si gel CC, using gradients of hexanes:EtOAc (100:0~0:100). Active fraction 7 that eluted with hexanes:EtOAc 1:1 (inhibited HIF-1 activation in T47D cells by 94% at 5 μg mL−1) was further separated by Sephadex LH-20 CC (eluted with 100% MeOH) and Si gel CC, eluted with hexanes:EtOAc (7:3), to produce 1 (120 mg, 2.4% yield). Active fraction 8, eluted with 100% EtOAc (inhibited HIF-1 activation in T47D cells by 96% at 5 μg mL−1), was further separated into 3 subfractions by Si gel CC, eluted with CH2Cl2:MeOH (93:7). Subfraction 8-2 (26 mg) was purified by Si gel CC, eluted with hexanes:EtOAc (65:35), to produce 2 (20 mg, 0.4% yield) and subfraction 8-3 (40 mg) was further purified by HPLC (Prodigy® 5μ, ODS-3 100 Å, 250 × 21.2 mm, isocratic 50% MeOH in H2O, 8.0 mL min−1), to produce 3 (4 mg, 0.08% yield, tR 58 min) and 4 (1 mg, 0.02% yield, tR 38 min).
Neolamellarin A (1), 3,4-bis(4-hydroxyphenyl)-1-[2-(4-hydroxyphenyl)-1-oxoethyl]-1H-pyrrole
White needles; mp 144–146 °C; UV (MeOH) λmax (log ε) 210 (4.53), 250 (4.41), 285 (4.27) nm; IR(film); νmax cm−1 3369, 1697, 1612, 1512, 1387, 1221, 1172; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 386.1385 [M+H]+ (calcd for C24H20NO4, 386.1392).
Neolamellarin B (2), 3,4-bis(4-hydroxyphenyl)-1-(2-(4-hydroxyphenyl)acetyl)-1H-pyrrol-2(5H)-one
Yellow oil; UV (MeOH) λmax (log ε) 224 (4.34), 330 (4.06) nm; IR(film); νmax cm−1 3384, 1695, 1607, 1513, 1362, 1236, 1103; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 424.1155 [M+Na]+ (calcd for C24H19NO5Na, 424.1161).
5-Hydroxyneolamellarin B (3), 5-hydroxy-3,4-bis(4-hydroxyphenyl)-1-(2-(4-hydroxyphenyl)acetyl)-1H-pyrrol-2(5H)-one
Yellow powder; [α]D25 +9.23 (c 0.065, acetone); UV (MeOH) λmax (log ε) 224 (4.47), 285 (4.14), 330 (4.18) nm; IR(film); νmax cm−1 3336, 1696, 1607, 1514, 1439, 1358, 1232, 1174, 1136; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 440.1121 [M+Na]+ (calcd for C24H19NO6Na, 440.1110).
7-(R/S)-Hydroxylamellarin A (4), 3,4-bis(4-hydroxyphenyl)-1-[2-(4-hydroxyphenyl)-2-(R/S)-hydroxy-1-oxoethyl]-1H-pyrrole
Yellow oil; [α]D25 0 (c 0.012, acetone); UV (MeOH) λmax (log ε) 285 (4.34) nm; IR(film); νmax cm−1 3354, 1695, 1609, 1512, 1440, 1365, 1221, 1176; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 402.1378 [M+H]+ (calcd for C24H20NO5, 402.1341).
T47D Cell-Based Reporter Assay for HIF-1 Activity
T47D cells (ATCC) were maintained in DMEM/F12 media with glutamine (Mediatech) supplemented with 10% FCS (v/v, Hyclone), and 50 U mL−1 penicillin G sodium and 50 μg mL−1 streptomycin (Biowhittaker) in a humidified environment under 5% CO2/95% air at 37 °C. Transfection, compound treatment, exposure to hypoxic conditions or a hypoxia mimetic (10 μM 1,10-phenanthroline), and measurement of luciferase activity were performed as described previously.14 The test compounds were prepared as 4 mM stock solutions in DMSO and stored at −20 °C.
ELISA for Secreted VEGF Protein
The plating of T47D cells, compound treatment, hypoxic exposure, and determination of secreted VEGF protein levels in the conditioned media by ELISA were as previously described.14,18 The only modification in the assay was that the cells were lysed with 250 μL M-PER mammalian protein extraction reagent (Pierce) after removal of the conditioned media, and protein concentrations in the cell lysates were determined using the Micro BCA Protein Assay Kit (Pierce). The amount of secreted VEGF protein was normalized to the amount of protein in the cellular lysates.
Neutral Red Assay for Cell Proliferation/Viability
Detailed description of the assay procedure was as previous published.19 The following formula was used to calculate % inhibition of cell proliferation/viability: % Inhibition = [1 − OD540(treated)/OD540(control)] × 100.
Acknowledgments
The authors thank the Natural Products Branch Repository Program at the National Cancer Institute for providing marine extracts from the NCI Open Repository used in these studies, D.J. Newman and E.C. Brown (NCI – Frederick, MD) for assistance with sample logistics and collection information, T. Smillie (UM) for coordinating sample acquisition from the NCI, H. Jia (UM) for performing luciferase assays in transformed 3T3 cells, S.W. Morris (St. Jude Children’s Research Hospital, Memphis, TN) for providing the transformed 3T3 cell line, S.L. McKnight (UT Southwestern Medical Center at Dallas) for providing the pTK-HRE3-luc construct, P.O. Sanduski (UM) for obtaining some NMR spectra, P. Carvalho (UM) for obtaining the HRESIMS data, and M.A. Avery for access to facilities and personnel within the UM Dept. of Medicinal Chemistry. This work was supported by the National Institutes of Health-NCI CA98787 (DGN/YDZ) and NOAA NURP/NIUST NA16RU1496. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Grant No. C06 RR-14503-01 from the National Institutes of Health.
References
- 1.Vaupel P, Mayer A. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Expert Opin Ther Targets. 2006;10:267–280. doi: 10.1517/14728222.10.2.267. [DOI] [PubMed] [Google Scholar]
- 3.Semenza GL. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 4.Semenza GL, Wang GL. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, Hankinson O, Pugh CW, Ratcliffe PJ. Proc Natl Acad Sci U S A. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan HE, Lo J, Johnson RS. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- 8.Kung AL, Wang S, Klco JM, Kaelin WG, Livingston DM. Nat Med. 2000;6:1335–1340. doi: 10.1038/82146. [DOI] [PubMed] [Google Scholar]
- 9.Kung AL, Zabludoff SD, France DS, Freedman SJ, Tanner EA, Vieira A, Cornell-Kennon S, Lee J, Wang B, Wang J, Memmert K, Naegeli HU, Petersen F, Eck MJ, Bair KW, Wood AW, Livingston DM. Cancer Cell. 2004;6:33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Unruh A, Ressel A, Mohamed HG, Johnson RS, Nadrowitz R, Richter E, Katschinski DM, Wenger RH. Oncogene. 2003;22:3213–3220. doi: 10.1038/sj.onc.1206385. [DOI] [PubMed] [Google Scholar]
- 11.Moeller BJ, Dreher MR, Rabbani ZN, Schroeder T, Cao Y, Li CY, Dewhirst MW. Cancer Cell. 2005;8:99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Nagle DG, Zhou YD. Curr Drug Target. 2006;7:355–369. doi: 10.2174/138945006776054979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melillo G. Cancer Metastasis Rev. 2007;26:341–352. doi: 10.1007/s10555-007-9059-x. [DOI] [PubMed] [Google Scholar]
- 14.Hodges TW, Hossain FC, Zhou YD, Nagle DG. J Nat Prod. 2004;67:767–771. doi: 10.1021/np030514m. [DOI] [PubMed] [Google Scholar]
- 15.Urban S, Butler MS, Capon RJ. Aust J Chem. 1994;47:1919–1924. [Google Scholar]
- 16.Bailly C. Curr Med Chem– Anti-Cancer Agents. 2004;4:363–378. doi: 10.2174/1568011043352939. [DOI] [PubMed] [Google Scholar]
- 17.Ferrara N, Gerber HP, LeCouter J. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 18.Mohammed KA, Hossain FC, Zhang L, Bruick RK, Zhou YD, Nagle DG. J Nat Prod. 2004;67:2002–2007. doi: 10.1021/np049753f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou YD, Kim YP, Mohammed KA, Jones DK, Muhammad I, Dunbar DC, Nagle DG. J Nat Prod. 2005;68:947–950. doi: 10.1021/np050029m. [DOI] [PMC free article] [PubMed] [Google Scholar]