Abstract
There has been growing debate over the nature of the genetic contribution to individual susceptibility to common complex diseases such as diabetes, osteoporosis, and cancer. The ‘Common Disease, Common Variant (CDCV)’ hypothesis argues that genetic variations with appreciable frequency in the population at large, but relatively low ‘penetrance’ (or the probability that a carrier of the relevant variants will express the disease), are the major contributors to genetic susceptibility to common diseases. The ‘Common Disease, Rare Variant (CDRV)’ hypothesis, on the other hand, argues that multiple rare DNA sequence variations, each with relatively high penetrance, are the major contributors to genetic susceptibility to common diseases. Both hypotheses have their place in current research efforts.
A Brief History of the Debate
Debates concerning precisely how genetic variations contribute to phenotypic expression have been at the heart of a great deal of biomedical research for more than a century. In fact, one of the most contentious yet insightful of these debates occurred at the turn of the 20th century and was rooted in positions championed by two opposing intellectual camps. The ‘Mendelians,’ in the form of William Bateson, Hugo de Vries, and others, focused on discrete gene-based units of inheritance and Mendel’s laws as the fundamental factors responsible for phenotypic expression and phenotypic similarities and differences across generations. On the other hand, the ‘Biometricians,’ as represented primarily by Karl Pearson, focused on the measurement and statistical analysis of continuous phenotypes such as height as well as the variation exhibited by such phenotypes within a population. The Biometricians rejected aspects of what is known today as Mendelian genetics as espoused by the ‘Mendelian’ camp at the time due to the fact that discrete units of heredity, such as Mendelian-segregating genes, could not, it seemed to them, explain the continuous range of phenotypic variation seen in real populations.
The debate between the Mendelians and Biometricians was resolved, to a high degree, by RA Fisher among others. Fisher essentially argued that multiple genes, in the form in which the Mendelians believed them to exist, each following Mendel’s laws yet working collectively (primarily additively), could influence phenotypic expression and hence the continuous variation that a phenotype might exhibit in the population at large [1]. The historical vagaries surrounding the Mendelians’ and Biometricians’ opposition of each other, as well as Fisher’s and others’ contribution to the resolution of this opposition, have been very elegantly and richly detailed by William Provine in his book “The Origins of Theoretical Population Genetics” [2].
The distinction between overt, single gene-based, Mendelian forms of the inheritance and the more polygenic or multifactorial forms of inheritance of the type envisioned by the Biometricians and later refined by Fisher, provides context for contemporary debates concerning the genetic basis of complex disease entities such as hypertension, cancer and diabetes – especially the ‘Common Disease, Common Variant (CDCV) vs. Common Disease Rare Variant (CDRV) debate – in at least two ways. First, there is still debate over the actual number of genes or genetic variations that might influence any particular trait. For example, in the early-1960’s, a vigorous debate, very much analogous to the Mendelian/Biometrician debate, over the nature of essential or primary hypertension and its frequency in the population at large occurred. Basically, Sir Robert Platt argued that hypertension was due, in large part, to common genetic variations with relatively high ‘penetrance,’ whereas George Pickering argued that hypertension was due to the existence of a number of genetic variations, each with reduced penetrance (or ‘polygenes’), as comprehensively summarized by Swales [3]. Although most contemporary geneticists consider Pickering’s hypothesis to be more consistent with the empirical studies of the epidemiology of hypertension, there is clearly room for peaceful co-existence for the two perspectives since some overtly Mendelian forms of hypertension exist which are largely attributable to genetic variations with high penetrance (albeit with low frequency [4]). This is true of virtually all other common chronic diseases as well, since Mendelian, primarily single genetic defect-related, forms of most diseases have been identified.
Second – and more to the point of this review – although most geneticists would argue that the genetic basis of most common chronic diseases is more likely to be consistent with the Biometrician/Fisherian/Pickering view entailing multiple genetic factors working in aggregate, there is considerable debate over the actual frequency of the multiple genes and genetic variations that may be at play, and this issue is at the heart of the CDCV vs. CDRV debate.
The CDCV hypothesis has it roots in a number of publications, but one of the most prominent, by Reich and Lander [5], considered what they termed the ‘allelic spectrum of disease’ and used empirical data to qualify this spectrum. This spectrum is simply the totality of variations that contribute to a disease, including low penetrance, high prenetrance, common (i.e. variations having a frequency of greater than 1% in the population), and rare variations (i.e., variations with a frequency less than 1%). Essentially, Reich and Lander provided a theoretically perspective from which they attempted to “…weave together strands from the human mutation and population genetics literature to provide a framework for understanding and predicting the allelic spectra of disease genes. The theory does a reasonable job for diseases where the genetic etiology is well understood… [but] also has bearing on the Common Disease/Common Variants (CD/CV) hypothesis, predicting that at loci where the total frequency of disease alleles is not too small, disease loci will have relatively simple spectra.”[5] Although not complete advocates of the CDCV hypothesis, Lander and Reich concluded that, on the basis of the available data, the CDCV is not incompatible with many diseases.
A number of investigators have challenged the CDCV hypothesis and offered the alternative CDRV hypothesis in its place. For example, Pritchard [6], argued that population processes operative in the human lineage would be more likely to favor the existence of multiple rare variations contributing to disease rather than common variations. Essentially, Pritchard [6] posited that population-level processes influence the frequency of ‘deleterious’ (i.e., disease susceptibility) variations, such as mutation, random genetic drift, and purifying selection against susceptibility mutations. These processes, he argued, have acted on the human population during its expansion in the last few centuries or so, and have led to a situation in which the genomic positions or loci harboring variations underlying disease susceptibility are likely to be mildly deleterious (and hence not subject to overt selection), have a high overall mutation rate, have a total frequency that is quite high. and exhibit extensive allelic heterogeneity. Thus, Pritchard [6] argued that the notion that multiple, very recent rare variations contributing to disease arising in the last two centuries is more consistent with human population pathobiology than the notion that older, common variations are contributing to disease. For example, common variations are likely to be older and hence have been subjected to potential selective forces over time. By reaching an appreciable frequency, they therefore are not as likely to have been subjected to negative selection. Rare variations, on the other hand, are either likely to be new (i.e., only a few generations old) and hence not have been subjected to negative selection for a long time, or are rare because they are being selected against due to their deleterious nature. In this light, it is of interest that recent reports on the frequency of human alleles and their likely ‘functional’ or phenotypic effects suggest that less frequent variations are more likely to be functional than common variations [7].
The CDCV vs. CDRV debate is more than just an academic debate, as each position in the debate entails, or is consistent with, different strategies for the identification of variations contributing to disease susceptibility [8]. We take the view that both positions are more or less defensible and correct in that multiple common variations with low penetrance and multiple rare variations with moderate to high penetrance contribute to the expression and frequency of common human diseases in the population at large.
The Evidence for Each Hypothesis
Background Evidence
The evidence that multiple rare variations might be contributing to human phenotypic variation is consistent with some early in-depth sequencing and re-sequencing studies of human genic variation. For example, studies by Nickerson and colleagues in the late 1990’s on the lipoprotein lipase (LpL) gene suggested that a number of naturally occurring variations, both common and rare, are likely to influence LpL function. LpL is a gene known to be a contributor to cholesterol levels and ultimately, when dysregulated, to heart disease (e.g.,[9]). Follow-up survey sequencing studies by Nickerson and colleagues, among others, has consistently shown that rare, likely functionally significant, variations occur naturally in human genes of relevance to a number of human phenotypes and diseases (e.g., [10]). The identification of a number of rare variations from survey sequencing studies of physiologic important human genes is also consistent with studies of genes known to lead to rare diseases, such as cystic fibrosis and BRCA1 and BRCA2 forms of breast cancer, in which hundreds of rare, yet disease-causing, variations have been identified over the years [11, 12].
GWA Studies and Their Results
The availability of high-throughput genotyping technologies, coupled with the results of major polymorphism characterization efforts such as the International Hapmap Initiative, have made it possible to conduct genome wide association (GWA) studies seeking to identify common variations that are statistically linked with particular diseases [13]. To date, hundreds of GWA studies have been performed, with many having identified unequivocal, statistically compelling, associations between particular genetic variations and diseases of all sorts [14]. However, as successful as these studies have been in identifying such associations, the genetic variations identified to date from such studies collectively explain only a small fraction of the burden of any disease in the population at large. Importantly, intensive GWA studies investigating many traits and diseases have led to associations involving genes or genomic regions that typically have 30 to over 50 associated sequence variants, each of relatively low penetrance, with typical risk ratio of approximately 1.2. With rare exceptions, more than 90–95% of the heritable component of a disease has been left unexplained after extensive GWAS interrogation. This suggests that individual common inherited variations are not likely to explain the majority of common chronic disease prevalence and ultimately raises the question as to the nature of the remaining genetic factors contributing to disease, or what has been termed the ‘missing heritability’ of disease phenotypes [15]. The fact that GWA studies have seemingly reached their limits in the identification of common variations contributing to common diseases obviously leaves the door open for the discovery of multiple rare variations that contribute to common diseases (or possibly other forms of genetic and epigenetic variation).
Sequencing Studies
Many investigators have gone beyond survey sequencing of human genes to catalog rare sequence variations, to actually contrasting and comparing the frequency of rare variations in individuals with and without disease. Table 1 lists relevant studies. Virtually all of these studies have observed frequency differences between individuals with and without a particular disease phenotype for multiple rare single nucleotide polymorphisms (SNPs) in functionally-relevant (e.g., coding) regions of the genes studied. Although published studies of this sort may reflect publication bias (i.e., many other studies may have attempted to do this for different diseases and genes and simply did not find anything interesting), they do indicate that multiple rare variations are likely to be associated with some human diseases and disease-related phenotypes.
Table X.
Recent Sequencing Studies Linking Multiple Rare Variations to a Phenotype or Disease
| Reference | Gene | Phenotype | Results |
|---|---|---|---|
| 37 Nejentsev et al. (2009) | IFIH1 | Type 1 Diabetes | Multiple rare cSNPs are more frequent in T1D |
| 38 Marini et al. (2008) | MTHFR | Folate response | Multiple coding SNP effects are folate remedial |
| 39 Ji et al. (2008) | Salt handling genes | Blood Pressure | Multiple coding SNPs for individuals with low BP |
| 40 Azzopardi et al. (2008) | APC | Colorectal cancer | Multiple variations among colorectal cancer |
| 41 Masson et al. (2008) | CTRC | Pancreatitis | Multiple variations among pancreatitis patients |
| 42 Ma et al. (2007) | Toll-like receptors | Tuberculosis (TB) | Multiple coding variations influence TB |
| 43 Ahituv et al. (2007) | 58 different genes | Obesity | Multiple variations among obese patients |
| 44 Romeo et al. (2007) | ANGPTL4 | Elevated HDL | Multiple variations among high HDL patients |
| 45 Kotowski et al. (2006) | PCSK9 | Low LDL | Frequent nonsense mutations among low LDL |
| 46 Cohen et al. (2005) | PCSK9 | Heart disease | Multiple sequence variations among HD patients |
| 47 Cohen et al. (2006) | NPC1L1 | Low LDL | Multiple rare variants among low LDL patients |
| 48 Cohen et al. (2005) | PCSK9 | Low LDL | Frequent nonsense mutations among low LDL |
| 49 Cohen et al. (2004) | ABCA1,APOA1, LCAT | Low plasma HDL | Coding SNPs differences for low HDL patients |
One important aspect of these studies is that they focused on the identification of multiple rare variations, any one of which might often be possessed (in isolation of others) by the individuals with the disease phenotype of interest. This suggests that all the variations contributing to the pool of variations that were (collectively) greater in frequency among the individuals with the disease phenotype perturb the gene of interest in roughly the same way to induce that disease phenotype. In fact, such ‘allelic heterogeneity’ is an important feature in the formulation of the CDRV hypothesis, in that it is argued that although there may be many genes or genomic regions that might influence a disease, whereby each of these genes or genomic regions may harbor many different rare variations that affect these genes or regions (note that this ‘allelic heterogeneity’ argument has also been made in the context of somatic mutations that influence tumorigenesis, as discussed below). Many genes that have been implicated in common disease pathogenesis have been shown to harbor multiple functionally-significant, naturally occurring, rare (and common) variations (see, e.g., Table 9.1 [16]).
In addition to the identification of rare SNPs contributing to common diseases and disease-related phenotypes, there have been a number of studies investigating the contribution of rare structural variations to human phenotypic variation [17]. However, such studies are in their infancy and have often raised more questions than they have answered, as discussed below in the case of neuropsychiatric disease.
The Case of Multiple Rare Singleton Deletions and Neuropsychiatric Disease
A number of recent studies have considered the contribution of copy number variations (CNVs) to neuropsychiatric diseases, including autism [18], and schizophrenia [19]. Each of these studies provided compelling statistical evidence suggesting that, e.g., autistic, schizophrenic, or bipolar individuals are more likely to possess CNVs in their genome – and in particular deletions of genomic regions. The main theme of these studies is that they find evidence of not one particular CNV being present in the genomes of individuals with these conditions, but rather any of a number of rare CNVs [20]. The finding that there are more rare CNVs, as a whole, among individuals with neuropsychiatric conditions suggests that one of two sorts of phenomena must be a play: either the genomes of individuals with these conditions are unusually ‘fragile’ in the sense that deletions arise at arbitrary places in the genome and this reflects some fundamental genomic ‘lesion’ associated with the etiologies of these disorders, or the actual locations of the deletions is of crucial importance in that these locations, when perturbed, cause brain dysfunction. Both of these phenomena are problematic. It is quite unlikely that individuals with ‘fragile’ genomes would only manifest the unique features of autism, schizophrenia, and bipolar disorder and not other conditions such as mental retardation, metabolic problems, developmental anomalies, etc. In other words, the unique phenotypes of autism, schizophrenia, and bipolar seem too specific for a gross molecular lesion such as global genomic fragility. In addition, if the genomic locations affected by the slight increase in number of rare CNVs in cases versus controls actually do harbor genes that are specific to, e.g., schizophrenia, then it is important for the scientific community to demonstrate that this is the case. This would rule out the ‘fragile genome’ hypothesis as well as any belief that the ‘multiple rare CNVs and neuropsychiatric disease’ findings reflect false positive results.
Are Diseases Influenced by Rare Variations Familial?
One very interesting question that has been raised by researchers contemplating the role of rare variations in complex diseases is whether or not diseases that are actually influenced by rare variations are likely to exhibit familial clustering. For example, Bodmer and Bonilla [8] have argued that such diseases are not likely to be familial and provide some theoretical calculations to show why this is the case. The suggestion that diseases influenced by rare variations are not familial has important consequences in that it suggests that family-based studies of the type pursued via classical genetics techniques (such as linkage analysis) are not likely to be useful for discovering causative genetic variations [8]. However, the arguments by Bodmer and Bonilla [8] are problematic for at least two reasons.
First, if someone possesses a disease that is influenced by multiple rare inherited variations that work additively, then that individual’s parents obviously possessed the right constellation of variations to at least lead to a non-zero probability that they would produce an offspring that could ultimately manifest the disease. This fact would clearly lead to a higher probability of those parents producing another offspring with the phenotype than parents without the appropriate constellation of genetic variations. In this sense, one could say that the parents are ‘enriched’ for predisposing variations simply because they produced an offspring with the disease, and this enrichment could lead them to produce another offspring with the disease with a higher than average probablity. Note that this probability would clearly be a function of the number of variations that could contribute to the phenotype: if there was only single rare variant with low penetrance that could induce the phenotype, then the probability that an additional offspring would be produced by the parents is small. If, however, there are many variations that work additively, then the probability would be higher. The calculations by Bodmer and Bonilla [8] are consistent with the assumption of the existence of only a few predisposing variations with low penetrance, as opposed to any number of rare variations that work additively.
Second, the empirical evidence is consistent with the suggestion that diseases influenced by rare variations are indeed familial. Consider the simple fact that most common, chronic conditions such as diabetes, hypertension, and cancer have been subjected to GWA studies, as noted above, and the results of these studies suggest that common variations explain only a small fraction of these disease’s frequencies in the population at large. This leaves the door open to other genomic explanations for their frequency, such as the implication of multiple low frequency (MAF 1–5%) or rare (< 1%) variations. In fact, the lack of a major contribution to these diseases by common variations via GWA studies is motivating studies seeking to identify rare variations, including CNVs, that may contribute to them (see below). Virtually all the diseases for which GWA studies have been pursued – and for which the alternative CDRV hypothesis to the CDCV hypothesis is being explored – exhibit familial clustering.
The ‘1000 Genomes’ Project and Related individual Sequencing Projects
In order to adequately test the CDRV hypothesis against the CDCV hypothesis for any disease, rare variations have to first be identified among individuals with the disease. This requires DNA sequencing protocols. Although Table 1 documents studies that have exploited DNA sequencing technologies.to identify rare variations associated with different disease phenotypes, these studies were performed with a singular focus on a particular gene or genomic region. In order to facilitate the search for rare variations in different genes, if not the entire genome, using contemporary DNA sequencing technologies, the ‘1000 Genomes’ project was initiated (www.1000genomes.org/). This project seeks to characterize sequence variation in 1000 individuals in order to provide a baseline for further disease-oriented DNA sequencing studies as well as develop appropriate protocols and bioinformatics tools.
Insight into rare variations and their potential impact on phenotypic expression has also benefited from a number of large-scale sequencing projects that attempted to sequence and assemble the entire genomes of individual humans [21,22,23]. These studies identified hundreds of thousands of novel genomic variations across the individuals studied that are very likely to be rare in the population or have arisen de novo in the genomes of the individuals sequenced. Studies investigating the potential functional impact of these rare or de novo variations suggest that many of them are likely to be functional and phenotypically-relevant [24].
Cancer ‘Driver’ vs. ‘Passenger’ Mutations
A debate analogous to the debate about the role of common and rare variations in inherited or congenital diseases involves cancer genomics. This debate concerns the identification and differentiation of ‘driver’ mutations from ‘passenger’ mutations in tumoriigenesis. Driver mutations are those mutations that essentially cause or lead to tumoriigenesis. Passenger mutations, on the other hand, are simply those somatic mutations that build up over the unchecked cell replication that is the hallmark of cancer [25]. A number of very recent papers describing tumor sequencing and resequencing studies – many sparked by The Cancer Genome Atlas (TCGA) initiative (http://cancergenome.nih.gov/) – have identified a number of mutations in cancers [26, 27, 28, 29], some subsets of which are likely to be causative or driver mutations. However, attempts to identify causal or driver mutations among the discovered mutations on the basis of their frequency across different samples has been criticized as highly problematic [30, 31, 32]. The current or prevailing belief is that it is unlikely that a single, or even a few, commonly observed mutations are responsible for any one tumor type. Rather, the evidence to date is consistent with the notion that a number of perturbations in particular genes and genetic pathways induced more than likely by singular or rare mutations – all of which have similar tumorigenic effects – are responsible for tumorigenesis [25].
Verifying Findings Associated with Each Hypothesis
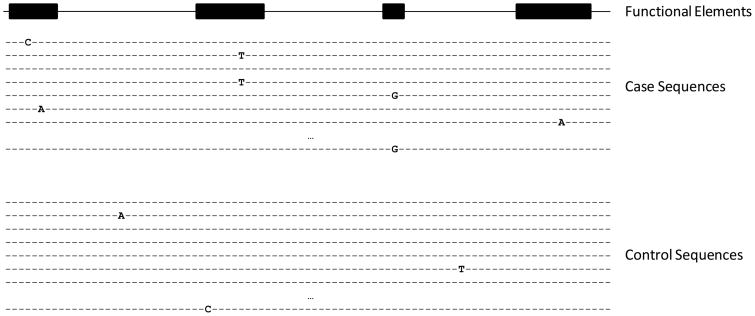
In order to substantiate claims about the role of either specific common or rare variations in disease, some form of validation of an initial finding implicating those variations is in order. For common variations implicated in GWA and candidate gene association studies, the sine qua non of validation is replication of the association in an independent population or sample of individuals than that used in the initial study [33]. However, replication studies of associations involving rare variations that exploit follow-up or ancillary populations is problematic given the infrequency of the variations of interest. This fact can be overcome to some degree by testing the hypothesis that the genes or genomic regions of interest have a collection of variations that, as a group, are more frequent among individuals with a disease phenotype than individuals without that phenotype (Figure 1). Statistical methods for carrying out such hypothesis tests are in their infancy, but will be crucial for advancing the CDRV hypothesis [16, 34, 35]. In addition to statistical evidence for associations between variations and a disease phenotype (whether implicating common or rare variations, or whether identified in an initial or replication study), it is important to assess the biological significance of the variation(s) in question via computational methods, laboratory assays or model systems.
Figure 1.
Hypothetical DNA sequences obtained from cases and controls for a genomic region of interest. The presence of an actual base (as opposed to a simple dot) indicates the presence of a non-reference or alternative allele. Dots indicate that the sequences are consistent with a reference. The shaded rectangles above the sequences reflect known functional elements at the positions indicated in the sequence below. Note that many of the variations possessed by the cases are rare and fall into the functional genomic regions. Relevant hypothesis tests would investigate the collective frequency difference of these variations between the case and control sequences. The definition and justification for grouping rare variations to be tested in this manner is crucial.
Conclusion
The contemporary CDCV vs. CDRV debate is, as noted, not only rooted in historical debates about the nature of phenotypic variation, but also implicates different strategies for identifying genetic variations that predispose individuals to a disease. It is safe to say, however, that strategies for uncovering common and rare variations should be pursued for any disease phenotype, and that the CDCV/CDRV debate should be seen as not an ‘either/or’ debate, but rather as a debate about the degree to which common and rare variations contribute to a particular disease phenotype. As noted, it is known that rare, Mendelian forms of most common chronic diseases for which the CDCV vs. CDRV debate has been invoked, exist. For example, Liddle’s syndrome is a very rare form of hypertension influenced by rare genetic variations [36] and familial breast cancer induced by BRCA1 and BRCA2 mutations implicates multiple highly penetrant, yet very rare, variations and yet both hypertension and breast cancer have more common forms for which GWA studies and related strategies have been, and should be, pursued.
The many discoveries resulting from GWA studies themselves suggest that there must be genetic factors contributing to common complex diseases that are simply not amenable to detection via the GWA study strategy, as emphasized throughout this review, since the variations identified via GWA only explain a small fraction of the prevalence of the diseases studied. Although it could be that the ‘missing heritability’ associated with these diseases that is not accounted for by common variations is accounted for by subtle gene x environment interactions, common CNVs with low penetrance, complicated epistatistic interactions involving many common variations, and/or epigenomic phenomena, to the exclusion of rare variations, this seems unlikely. In fact, evidence of the type provided in Table 1 suggests otherwise. Ultimately, the question as to the veracity (or the degree of veracity) of the CDCV hypothesis vs. the CDRV hypothesis for any particular disease, like virtually all scientific questions, is an empirical one.
Acknowledgments
The authors benefited from the following research grants: The National Institute on Aging Longevity Consortium (U19 AG023122-01); The NIMH-funded Genetic Association Information Network Study of Bipolar Disorder (R01 MH078151-01A1); National Institutes of Health grants: N01 MH22005, U01 DA024417-01, and P50 MH081755-01; and the Scripps Translational Sciences Institute Clinical Translational Science Award (U54 RR0252204-01). Additional funding came from Scripps Genomic Medicine and the Price Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fisher RA. The correlation between relatives on the supposition of mendelian inheritance. Philosophical Transactions of the Royal Society of Edinburgh in 1918. 52:399–433. [Google Scholar]
- 2.Provine WB. The Origins of the Theoretical Population Genetics. 2. Chicago: The University of Chicago Press; 2001. [Google Scholar]
- 3.Swales JD. Platt versus Pickering. London: Keynes Press; 1985. [Google Scholar]
- 4.Lifton RP. Molecular genetics of human blood pressure variation. Science. 1996;272:676–680. doi: 10.1126/science.272.5262.676. [DOI] [PubMed] [Google Scholar]
- 5.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **8.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. Provides an overview of issues associated with the CDCV hypotheses as well as calculations that shed light on situations in which it is likely that rare variations are responsible for a phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nickerson DA, Taylor SL, Weiss KM, Clark AG, Hutchinson RG, Stengård J, Salomaa V, Vartiainen E, Boerwinkle E, Sing CF. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat Genet. 1998;19:233–240. doi: 10.1038/907. [DOI] [PubMed] [Google Scholar]
- 10.Crawford DC, Carlson CS, Rieder MJ, Carrington DP, Yi Q, Smith JD, Eberle MA, Kruglyak L, Nickerson DA. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am J Hum Genet. 2004;74:610–622. doi: 10.1086/382227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bobadilla JL, Macek M, Jr, Fine JP, Farrell PM. Cystic fibrosis: a worldwide analysis of CFTR mutations--correlation with incidence data and application to screening. Hum Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 12.Szabo C, Masiello A, Ryan JF, Brody LC. The breast cancer information core: database design, structure, and scope. Hum Mutat. 2000;16:123–131. doi: 10.1002/1098-1004(200008)16:2<123::AID-HUMU4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 13.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *14.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. An important review of the contemporary progress made in identifying common genetic variations that influence common diseases through genome wide association (GWA) studies. The review also discusses limitations of the GWA study approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 16.Schork NJ, Wessel J, Malo N. DNA sequence-based phenotypic association analysis. Adv Genet. 2008;60:195–217. doi: 10.1016/S0065-2660(07)00409-9. [DOI] [PubMed] [Google Scholar]
- 17.Eichler EE, Nickerson DA, Altshuler D, Bowcock AM, Brooks LD, Carter NP, Church DM, Felsenfeld A, Guyer M, Lee C, et al. Completing the map of human genetic variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **19.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. The first paper to show that multiple rare copy number variations (CNVs) are more frequent in individuals with Schizophrenia than without, raising questions about the mechanism of action of these CNVs. [DOI] [PubMed] [Google Scholar]
- 20.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 21.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng PC, Levy S, Huang J, Stockwell TB, Walenz BP, Li K, Axelrod N, Busam DA, Strausberg RL, Venter JC. Genetic variation in an individual human exome. PLoS Genet. 2008;4:e1000160. doi: 10.1371/journal.pgen.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torkamani A, Verkhivker G, Schork NJ. Cancer driver mutations in protein kinase genes. Cancer Lett. 2008 Dec 9; doi: 10.1016/j.canlet.2008.11.008. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- *26.Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. One of the first large-scale DNA sequencing studies of tumors to reveal the frequency of multiple variations, many rare, in different genes that are likely to be driving tumorigenesis. [DOI] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Getz G, Höfling H, Mesirov JP, Golub TR, Meyerson M, Tibshirani R, Lander ES. Comment on “The consensus coding sequences of human breast and colorectal cancers. Science. 2007;317:1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- 31.Forrest WF, Cavet G. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317:1500. doi: 10.1126/science.1138179. author reply 1500. [DOI] [PubMed] [Google Scholar]
- 32.Rubin AF, Green P. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317:1500. doi: 10.1126/science.1138956. [DOI] [PubMed] [Google Scholar]
- 33.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 34.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoggart CJ, Whittaker JC, De Iorio M, Balding DJ. Simultaneous analysis of all SNPs in genome-wide and re-sequencing association studies. PLoS Genet. 2008;4:1000130. doi: 10.1371/journal.pgen.1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansson JH, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton RP. Hypertension caused by a truncated epithelial sodium channel gamma subunit: genetic heterogeneity of Liddle syndrome. Nat Genet. 1995;11:76–82. doi: 10.1038/ng0995-76. [DOI] [PubMed] [Google Scholar]
- 37.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare Variants of IFIH1, a Gene Implicated in Antiviral Responses, Protect Against Type 1 Diabetes. Science. 2009 Mar 5; doi: 10.1126/science.1167728. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marini NJ, Gin J, Ziegle J, Keho KH, Ginzinger D, Gilbert DA, Rine J. The prevalence of folate-remedial MTHFR enzyme variants in humans. Proc Natl Acad Sci U S A. 2008;105:8055–8060. doi: 10.1073/pnas.0802813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **39.Ji W, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. A comprehensive study that identified multiple rare coding variations in particular genes that are collectively increased in frequency among individuals with higher than average blood pressures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azzopardi D, Dallosso AR, Eliason K, Hendrickson BC, Jones N, Rawstorne E, Colley J, Moskvina V, Frye C, Sampson JR, et al. Multiple rare nonsynonymous variants in the adenomatous polyposis coli gene predispose to colorectal adenomas. Cancer Res. 2008;68:358–63. doi: 10.1158/0008-5472.CAN-07-5733. [DOI] [PubMed] [Google Scholar]
- 41.Masson E, Chen JM, Scotet V, Le Maréchal C, Férec C. Association of rare chymotrypsinogen C (CTRC) gene variations in patients with idiopathic chronic pancreatitis. Hum Genet. 2008;123:83–91. doi: 10.1007/s00439-007-0459-3. [DOI] [PubMed] [Google Scholar]
- 42.Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, Musser JM. Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS ONE. 2007;2:1318. doi: 10.1371/journal.pone.0001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahituv N, Kavaslar N, Schackwitz W, Ustaszewska A, Martin J, Hebert S, Doelle H, Ersoy B, Kryukov G, Schmidt S, et al. Medical sequencing at the extremes of human body mass. Am J Hum Genet. 2007;80:779–791. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kotowski IK, Pertsemlidis A, Luke A, Cooper RS, Vega GL, Cohen JC, Hobbs HH. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78:410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 48.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. Erratum in: Nat Genet 2005, 37:328. [DOI] [PubMed] [Google Scholar]
- 49.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–72. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]