Abstract
Variegated plants provide a valuable tool for studying chloroplast biogenesis by allowing direct comparison between green and white/yellow sectors within the same leaf. While variegated plants are abundant in nature, the mechanism of leaf variegation remains largely unknown. Current studies are limited to a few mutants in model plant species, and are complicated by the potential for cross-contamination during dissection of leaf tissue into contrasting sectors. To overcome these obstacles, an alternative approach was explored using tissue-culture techniques to regenerate plantlets from unique sectors. Stable green and pale yellow plants were developed from a naturally variegated Epipremnum aureum ‘Golden Pothos’. By comparing the gene expression between green and pale yellow plants using suppression subtractive hybridization in conjunction with homologous sequence search, nine down-regulated and 18 up-regulated genes were identified in pale yellow plants. Transcript abundance for EaZIP (Epipremnum aureum leucine zipper), a nuclear gene homologue of tobacco NTZIP and Arabidopsis CHL27, was reduced more than 4000-fold in qRT-PCR analysis. EaZIP encodes the Mg-protoporphyrin IX monomethyl ester cyclase, one of the key enzymes in the chlorophyll biosynthesis pathway. Examination of EaZIP expression in naturally variegated ‘Golden Pothos’ confirmed that EaZIP transcript levels were correlated with leaf chlorophyll contents, suggesting that this gene plays a major role in the loss of chlorophyll in the pale yellow sectors of E. aureum ‘Golden Pothos’. This study further suggests that tissue-culture regeneration of plantlets from different coloured sectors of variegated leaves can be used to investigate the underlying mechanisms of variegation.
Keywords: Golden Pothos, Mg-protoporphyrin IX monomethyl ester cyclase, tissue culture, transcript abundance, variegation formation
Introduction
Variegated plants are an ideal model for the study of chloroplast biogenesis because they have both green and white/yellow sectors on the same leaf, which can be used to compare differential gene expression directly and also for protein profiling in order to understand the co-ordinated expression of nuclear and organelle genes during chloroplast biogenesis (Sakamoto, 2003; Yu et al., 2007). Recent studies on variegated mutants of Arabidopsis such as immutans (im) (Aluru et al., 2001), yellow variegated 1 (var1) (Sakamoto et al., 2002) and yellow variegated 2 (var2) (Chen et al., 2000; Takechi et al., 2000), tobacco variegated and distorted leaf (vdl) (Wang et al., 2000), tomato ghost (gh) (Josse et al., 2000), as well as monocot barley albostrians (Yaronskaya et al., 2003) and maize yellow stripe 1 (ys1) (Curie et al., 2001) have demonstrated that variegated mutants are invaluable materials for studying the mechanisms of plastid and chloroplast development and maintenance.
In the variegated leaves, the white/yellow sectors are viable cells that contain undifferentiated chloroplasts (Kato et al., 2007). The defects in the white/yellow sectors do not appear to interfere with proper chloroplast development in the green sectors. When examining individual cells, only one type of plastid is observed in each cell, either normal chloroplasts or abnormal plastids. A mixture of plastid types in a single cell is rare (Wetzel et al., 1994), indicating that the cell is programmed by certain factor(s) at an early stage either to have normal chloroplasts or undeveloped plastids. Two general scenarios have been proposed to explain leaf-variegation (Wetzel et al., 1994; Yu et al., 2007). In the first case, cells in the green sectors consist of the wild-type genotype whereas cells in the white/yellow sectors consist of a mutant genotype expressing a mutant phenotype based on several possible mechanisms that include transposable element activity, RNA silencing, and mutations in the nuclear, mitochondrial, or chloroplast genome. In the second case, both green and white/yellow sectors have cells with mutant genomes, but compensation mechanisms exist that allow near normal chloroplast development in cells within the green sectors. Known examples of such compensation leading to leaf variegation are the Arabidopsis mutants var2 (Chen et al., 2000; Yu et al., 2004) and im (Wu et al., 1999) where green sectors develop in cells that accumulate threshold levels of a key component in chloroplast development. Thus, the leaf variegation phenotype may originate from different underlying mechanisms that interact with chloroplast development. Each mutant offers potentially new insights into the complex processes leading to chloroplast formation, and this argument is the basis for expanding studies of leaf variegation to include additional plant systems for comparison studies to resolve the mystery of programmed plastid types.
Variegated plants are widely present in nature, but few of them have been selected for molecular studies because in only a limited number of cases, such as Arabidopsis or other model plants, is there sufficient genetic and molecular information available. Many current studies focus on variegated Arabidopsis mutants, but are limited by the number of mutants having a variegated phenotype (Yu et al., 2007). Another challenge is the difficulty of dissecting the green and white sectors from variegated Arabidopsis leaves for protein profiling and gene expression studies because of their small leaf sizes (Kato et al., 2007). Studies of a few variegated mutants have provided novel insights into the mechanisms of chloroplast biogenesis (Yu et al., 2007), but these results are far from sufficient to understand the complex interactions of as many as 3500 chloroplast proteins encoded by both nuclear and chloroplast genes (Peltier et al., 2002). Therefore, it would be interesting to know whether previously uncharacterized variegated species can be used for the investigation of chloroplast biogenesis with current techniques.
‘Golden Pothos’ (E. aureum), a naturally variegated ornamental foliage plant (Chen et al., 2005), is native to Southeast Asia and the Solomon Islands (Huxley, 1994). It flowers rarely and there is very limited genetic information available. Green, pale yellow, and variegated lines were created by tissue culture (Hung and Xie, 2009). Regenerated plants from contrasting tissue types were subjected to suppression subtractive hybridization (SSH) (Diatchenko et al., 1996) to identify differentially expressed genes. Using cDNA SSH between regenerated pale yellow and green plants, nine down-regulated and 18 up-regulated genes in pale yellow plants were isolated. By comparing green and pale yellow sectors in naturally variegated leaves, expression of the EaZIP, a homologue gene encoding Mg-protoporphyrin IX monomethyl ester (MPE) cyclase, was identified as a factor that may contribute to the yellow sector formation.
Materials and methods
Plant material preparation
Methods for regenerating plantlets from tissue culture of E. aureum ‘Golden Pothos’ and maintaining all regenerated plants in a growth chamber or greenhouse were the same as those reported previously by Hung and Xie (2009).
Transmission electron microscopy (TEM) analysis
Leaves were cut into 1 mm3 blocks and fixed in 3% glutaraldehyde in 0.05 M KPO4 buffer, pH 5.8 at 4 °C. Samples were post-fixed in 2% OsO4 in the same buffer in the dark at 4 °C. After dehydration with a graded series of ethanol, they were infiltrated and embedded with Spurr's resin (Ladd Research Industries). Samples were sectioned by an LKB NOVA ultramicrotome (Leica Microsystems) and placed onto 200-mesh grids. The grids/block were then stained with a 4% aqueous uranyl acetate in the dark at 25 °C followed by three distilled water washes (40 °C) and 1 min in Reynold's lead citrate followed by three more distilled water washes. All sections were observed under a JEOL JEM 100S transmission electron microscope. Images were captured using Kodak 4489 EM film. After developing, the film was scanned at 1200 dpi on an Epson Perfection 4870 Photo Scanner, inverted and adjusted for printing using histogram stretch and gamma software functions.
Measurement of chlorophyll and MPE
For extracting chlorophyll, samples were resuspended in acetone as described in Hung and Xie (2009). For MPE analysis, previously published procedures (Tottey et al., 2003; Pontier et al., 2007) were adopted with some modifications. Leaves, treated with 10 mM 5-aminolevulinic acid (ALA) hydrochloride (MP Biomedicals) or control solution, were collected and ground in liquid nitrogen under dim light. Each 50 mg of powder was immediately resuspended in 375 μl of acetone:25% NH4OH (9:1 v/v) and centrifuged at 4 °C. To remove carotenoids and chlorophylls, the supernatant was diluted with 750 μl of acetone buffer and extracted with 750 μl of hexane. After centrifugation at room temperature, the lower acetone layer was collected and directly subjected to fluorescence analysis at an excitation of 420 nm and an emission peak around 595 nm was recorded under a LS-55 Fluorescence spectrometer (PerkinElmer) controlled by a software program, FL WinLab (PerkinElmer). For HPLC analysis (LC 1200 series, Agilent), the lower acetone layer was first evaporated to almost dryness under nitrogen gas, then the dry pigments were resuspended in methanol containing 2 mM KOH. Protoporphyrin IX, magnesium protoporphyrin IX, and MPE (Frontier Scientific) were used as standard pigments. Pigments were separated by chromatography on a 3.0×100 mm (3.5 μm) XTerra® MS C18 column. The mobile phase consisted of two solvents: A (0.010 M NH4HCO3 in water) and B (acetonitrile). The pigments were eluted with a linear gradient from 10% to 95% B over 10 min. Injection volume was 60 μl and flow rate was 0.500 ml min−1. The eluate was monitored with a photodiode array detector in the detection wavelength of 420 nm.
RNA isolation
RNA samples were prepared using Qiagen RNeasy® Plant Mini kit (Qiagen). All samples were treated with DNase I (Qiagen) to remove any possible DNA contamination. RNA samples were quantified by a spectrophotometer and visualized on a 1.2% agarose gel as described in Hung and Xie (2009).
Suppression subtractive hybridization (SSH)
SSH was performed using the Clontech PCR-Select™ cDNA Subtraction Kit (Clontech). All procedures were done according to the manufacturer's recommended protocol except the first strand cDNA was synthesized by SuperScript™ II Reverse Transcriptase (Invitrogen) combined with oligo(dT) primer (Clontech). For the forward subtraction, cDNAs from pale yellow tissues were the driver, and for the reverse subtraction, cDNAs from green tissues were the driver. After enrichment by the primary and secondary PCR using Advantage PCR polymerase (Clontech), the PCR products were ligated into pCR®2.1 vector of the TA cloning kit (Invitrogen) at 14 °C for 16 h. The ligation product was then transformed into Top10 competent cells (Invitrogen). All positive colonies were collected and analysed by PCR using M13 reverse (5′-CAGGAAACAGCTATGACCAT-3′) and forward (5′-GTAAAACGACGGCCAGTGA-3′) primers. The inserted sequences with size greater than 250 bp were sequenced by MWG Biotech Inc. They were used to search against all non-redundant GenBank protein databases using program BLASTX (Altschul et al., 1997) with the e-value cut off set at 1e-05. DNA sequences of all unique clones were subjected to nucleotide sequence search. Sequence alignment was done by a web-based tool, ClustalW2, from the European Molecular Biology Laboratory, European Bioinformatics Institute. The sequences reported in this paper have been deposited in GenBank with the accession nos. FJ666043 to FJ666051 for nine down-regulated genes and GO924845 to GO924862 for 18 up-regulated genes.
5′-RACE PCR
The 5′ cDNA sequence of EaZIP was obtained using SMART™ RACE cDNA Amplification kit (Clontech). The only modification was using SuperScript™ II Reverse Transcriptase (Invitrogen) to synthesize first strand cDNA. EaZIP specific primer EaZIP RACE3 (5′-GGGATAGACTTGGAATTCAGGGTTGGC-3′) was paired with the Universal Primer Mix (UPM) included in the kit to amplify the 5′ end of cDNA. PCR products were first ligated to pCR®2.1 vector and then transformed into Top10 Cells. Plasmid DNA was isolated using QIAprep® Spin Miniprep kit (Qiagen) and sequenced as described above.
qRT-PCR and RT-PCR
For qRT-PCR, total RNAs from three independent green and three independent pale yellow plants, including the same tissue line used in SSH, were prepared separately. Equal amounts of RNA were used to synthesize first-strand cDNA by MultiScribe™ Reverse Transcriptase and random primers (Applied Biosystems). The cDNAs were amplified by Power SYBR Green PCR Master mix (Applied Biosystems) and specific primers (see Supplementary Table S1 at JXB online) which were designed using Primer Express 3.0 software (Applied Biosystems). Both PCR reactions and fluorescence signal detections were performed under a platform of the 7500-Fast Real-Time PCR system (Applied Biosystems). Each sample was assayed in triplicates. Data of three independent biological samples were averaged. The calculation of Ct value was based on Pfaffl (2001). The ΔCt was a relative expression level compared to the 18S rRNA in the same sample. The fold change of transcript abundance between green and pale yellow plants was calculated by comparing their ΔCt value. Each difference of ΔCt value represents a 2-fold change.
For RT-PCR, the first strand cDNA was made by SuperScript™ II Reverse Transcriptase using random primers, and PCR reaction was carried out by Taq DNA polymerase (Sigma) and specific primers: EaZIP primers are EaZIPcDNAF 5′-ACGAAGGCTAGGCAGTACAC-3′ and EaZIPcDNAR 5′-CAGAGACTAGTGCTGCGATG-3′; EaF156 primers are EaF156cDNAF 5′-GGT-GTTGTCTCAGCTGCAGA-3′ and EaF156cDNAR 5′-ACATGGGAGGCATGCTGCTG-3′. The QuantumRNA™ 18S Internal Standard (Ambion) was used as the endogenous standard.
Protein isolation and immunoblotting analysis
Total proteins from mature leaves of regenerated ‘Golden Pothos’, tobacco, and Arabidopsis were isolated using the Plant Total Protein Extraction kit (Sigma). About 100 mg of leaf tissue was first disrupted by grinding in liquid nitrogen and followed by the methanol washes. Proteins were then precipitated in acetone at –20 °C for 5 min followed by centrifugation at 16 000 g for 5 min. To solubilize the proteins, each milligram of dried pellet was incubated with 4 μl of Reagent Type 2 Working Solution at 25 °C for 15 min. After centrifugation at 16 000 g for 30 min, the supernatant was collected and the protein concentrations were determined by the Bradford method using the Bio-Rad protein assay reagent (Bio-Rad). Protein samples were mixed with 1× sample buffer (Invitrogen) containing 50 mM dithiothreitol. After heating at 70 °C for 10 min, samples were analysed on a NuPAGE 4–12% Bis-Tris mini-gel (Invitrogen) running in MES SDS running buffer (Invitrogen) under 200 V for 35 min. For immunoblotting, proteins were transferred onto a PVDF membrane (Bio-Rad) in 1× transfer buffer (Invitrogen) under 30 V for 2 h. Membrane was then blocked with 10% dry milk in 1× 20 mM Tris-buffered saline with 1% Tween 20 for 1 h before incubating with primary antibodies: anti-MPE cyclase (CRD1) (1:3000), anti-Rubisco large subunit (RbcL) (1:50 000) and anti-Plastocyanin (PC) (1:50 000) (Agrisera). The primary antibodies were detected by secondary antibody Goat anti-Rabbit IgG horseradish peroxidase conjugate (Jackson ImmunoResearch Laboratories) and followed by incubation with SuperSignal® West Pico Chemiluminescent substrate (Thermo Scientific). The luminescent signal was captured by Kodak Biomax light film (Kodak). The blot was then stained with 0.2% (w/v) Amido Black (MP Biomedicals) in 10% (v/v) acetic acid to visualize total proteins.
Element analysis
Mature leaves that were subjected to element analysis were first dried in a 75 °C oven for 48 h and then ground into fine powder with a mortar and pestle. Total P, K, Ca, Mg, Cu, Fe, Mn, Zn, and Na concentrations were determined by inductively-coupled plasma (ICP) spectrometry conducted by the Analytical Laboratory Service at North Carolina State University and the Agronomic Division of the North Carolina Department of Agriculture and Consumer Services. All three types of regenerated plants were analysed each time with two independent biological samples. Data of three sets of experiments were subjected to statistical analysis using t test assuming equal variance.
Results
Characterization of regenerated plants
The pale yellow, green, and variegated leaves were examined previously under a light microscope and it was found that yellow leaves predominantly had colourless plastids while variegated leaves had several different types of chloroplasts (Hung and Xie, 2009). To examine these different types of chloroplasts in greater detail, TEM was used to observe their differences (Fig. 1). TEM observation showed that those chloroplasts present in leaves from green plants had typical thylakoids and grana structures (Fig. 1A). In leaves of yellow plants, cells contained plastids with undeveloped internal membrane structures (Fig. 1B) very similar to prolamellar bodies often seen in the early stage of etiolated cotyledons when exposed to light. A similar structure was observed in plastids of the white sector of var2 mutant (Sakamoto et al., 2009). In the leaves of variegated plants, a type of chloroplast was found which was light green in colour and had loose stacks of grana thylakoids (Fig. 1C), a characteristic similar to Xantha-f60 mutants deficient in magnesium chelatase (Von Wettstein et al., 1995). This type of chloroplast seems to represent most of the chloroplasts found in those variegated leaves. No obvious difference of nuclei or mitochondria was observed between cells of the green and pale yellow leaves (Fig. 1D, E).
Fig. 1.
TEMs of chloroplasts in green (A), pale yellow (B), and variegated (C) mature leaves. TEMs of nucleus and mitochondria in green (D) and pale yellow (E) leaves. Plants were grown on MS medium under a growth condition with a light intensity of 50 μmol m−2s−1 at 25 °C. Scale bar=2 μm.
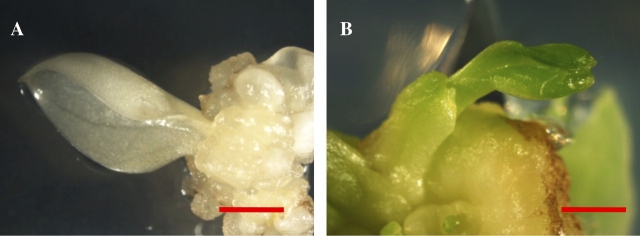
Although pale yellow plants do not have normal chloroplasts, they can be maintained and propagated on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962), and no colour change has been found after one year. Moreover, the leaf and petiole segments from pale yellow plants could be used efficiently to induce pale yellow callus and adventitious shoots (Fig. 2A) as those from green (Fig. 2B) and previously reported variegation plants (Hung and Xie, 2009). The results fully indicated that cells of pale yellow plants are viable and capable of differentiation and proliferation.
Fig. 2.
Regeneration of callus and shoots from regenerated pale yellow and green plants. A new shoot regenerated from callus derived from a pale yellow explant (A) and a new shoot regenerated from callus derived from a green explant (B). Scale bar=2 mm. (This figure is available in colour at JXB online.)
Because leaf chlorosis can be indicative of nutrient deficiency (Bloom, 1998), the contents of the nine elements found most commonly in plants were examined by ICP spectrometry in order to diagnose whether differences in nutrient availability contribute to the pale yellow colour and variegation (Fig. 3). The results showed that only Ca, Mg, and Mn were significantly lower (P <0.05) in pale yellow plants compared to green plants, whereas the other six elements had similar levels. As for variegated plants, the levels of Ca, Mg, and Mn were slightly reduced, but not significantly different from the levels found in green plants (P >0.15). Considering the fact that all regenerated plants were grown on the same MS medium and transferred to fresh medium every month, the low contents of Ca, Mg, and Mn in pale yellow plants were probably not caused by nutrient deficiencies.
Fig. 3.
Analysis of element contents in regenerated plants. An equal amount of dry leaves from green (G) (in light grey), variegated (V) (in dark grey), and pale yellow (PY) (in black) regenerated plants were analysed. Data shown here are the percentages of contents compared to that in green leaves (as 100%). Data represent an average of three independent experiments ±standard deviation. The asterisks indicate that those differences are significant (P <0.05).
SSH reveals differentially expressed genes in pale yellow plants
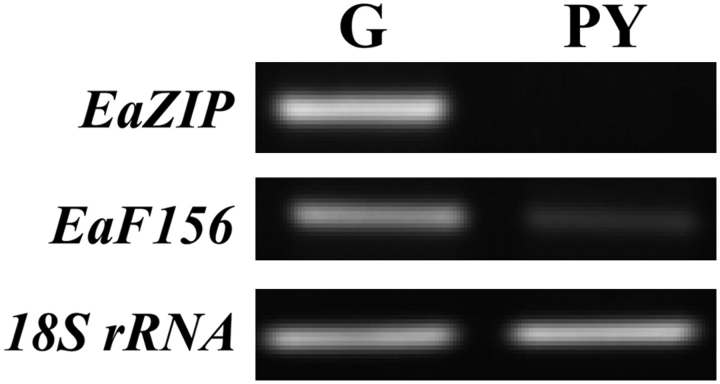
In order to discover genes that might be involved in forming variegation, SSH was used to identify differential transcript abundance between green and pale yellow plants. From the subtracted libraries, about 1000 colonies were picked and checked by PCR analysis using M13 primers. A total of 432 colonies with inserted sequences of 250 bp or greater were sequenced, and 113 unique genes were present in forward hybridization representing transcript abundance in green plants; 39 of those were present in reverse hybridization representing transcript abundance in pale yellow plants. QRT-PCR was used to confirm further whether these genes were indeed differentially expressed. This confirmation was carried out using plants generated from multiple cultures to represent better the differences between green and pale yellow plants, including the same culture used to generate subtraction libraries. The results showed that a total of nine genes were down-regulated in pale yellow plants with fold changes greater than two (Table 1). Seven of them were nuclear genes while two had no significant match from a GenBank search. Among them, the EaZIP had more than 4000-fold less transcripts in pale yellow plants than in green plants, and virtually no detectable signal was found in pale yellow plants in RT-PCR analysis (Fig. 4). The EaF156 that had a reduced transcript level of 6.2-fold in pale yellow plants was also confirmed by RT-PCR analysis (Fig. 4).
Table 1.
Down-regulated genes in pale yellow plants
| Gene | bp | Accession no. | Best match | e-value | ΔCt | Fold change | |
| G (n=3) | Y (n=3) | ||||||
| EaF7 | 561 | FJ666043 | Resistance protein Ler3 [Arabidopsis thaliana] (AAP80285) | 1e-10 | 8.4±0.6 | 10.0±0.4 | −3.0 |
| EaF82 | 544 | FJ666044 | Unknown | 6.3±1.2 | 10.0±0.6 | −13.0 | |
| EaF103 | 598 | FJ666045 | Hypothetical protein [Vitis vinifera] (CAN70903) | 1e-11 | 9.4±0.8 | 10.8±0.5 | −2.6 |
| EaZIP | 1203 | FJ666046 | ZIP [Nicotiana tabacum] (AAO89565) | 9e-47 | 6.4±0.6 | 18.5±1.5 | −4289.7 |
| EaLytB | 815 | FJ666047 | LYTB-like protein 1 [Brassica rapa subsp.] (AF398145) | 2e-99 | 6.4±0.7 | 7.8±0.5 | −2.6 |
| EaSBEIIb | 332 | FJ666048 | Starch branching enzyme IIb [Zea mays] (AAC33764) | 3e-63 | 7.5±0.8 | 9.1±0.5 | −3.1 |
| EaF156 | 793 | FJ666049 | Ferric reductase [Oryza sativa] (BAD18962) | 1e-105 | 7.1±0.2 | 9.7±0.3 | −6.2 |
| EaF207 | 532 | FJ666050 | Unnamed protein product [Vitis vinifera] (CAO67174) | 4e-8 | 13.1±0.8 | 15.4±0.5 | −4.8 |
| EaF289 | 394 | FJ666051 | Unknown | 4.8±0.2 | 6.6±0.4 | −3.5 | |
Only genes with fold changes greater than two and P <0.05 are shown. The ΔCt value is the average of three biological replicates ±standard deviation. Each replicate was assayed in triplicate. The fold change is determined as 2(ΔΔCt). The ΔΔCt value is determined by the differences in the ΔCt between pale yellow and green plants. The negative fold changes mean the pale yellow plants have reduced transcript abundance. e-Value reflects the statistically significant similarity of our novel sequences to the best matched results.
Fig. 4.
RT-PCR of selected cDNA clones identified by SSH. Equal amounts of total RNA used to make first strand cDNA were isolated from green (G) and pale yellow (PY) regenerated plants. PCR reactions were carried out using specific primers designed to target two SSH identified clones, EaZIP and EaF156. 18S rRNA was used as an internal control.
There were 18 subtracted cDNA clones that were more abundant in pale yellow plants with fold changes greater than two (see Supplementary Table S2 at JXB online). Surprisingly, all of them were to be found similar to chloroplast genes or non-coding regions in the chloroplast genome, and none of them was a nuclear gene. By nucleotide sequence comparison of 18 cDNA clones to available chloroplast genomes, the results showed that one clone (R61) contains only an intron sequence and two clones (R71 and R106) contain only an intergenic region while R18, R30, and R34 clones contain both partial exon and intron. It is not clear why these clones are abundant in pale yellow plants. Because the precursor RNAs of higher plant chloroplast genes need to undergo a series of processing events to become mature mRNA (Monde et al., 2000), these results may suggest at least some of these 18 cDNA clones were products of either an alternative splicing or an unusual transcription. Why these RNA products are abundant or sustained longer in yellow plants than in green plants will need to be investigated further.
EaZIP is a homologue of tobacco NTZIP, encoding MPE cyclase
The EaZIP transcript was uniquely undetectable in pale yellow plants. Results from sequence searches (Table 1) indicated that the EaZIP is highly homologous to the tobacco NTZIP, which encodes MPE cyclase, a key enzyme responsible for converting MPE into divinyl protochlorophyllide a in chlorophyll biosynthesis (Liu et al., 2004). To verify further whether EaZIP is a homologue of NTZIP, a full-length EaZIP gene from regenerated green plants was obtained using 5′-RACE PCR. The deduced amino acid sequence of EaZIP revealed an 86% identity to the tobacco NTZIP, 84% to the Arabidopsis CHL27 (Tottey et al., 2003) and 60% to the Chlamydomonas reinhardtii CRD1 (Moseley et al., 2000) (Fig. 5). The NTZIP, CHL27, and CRD1 have all been characterized and are known to encode MPE cyclase (Moseley et al., 2000; Tottey et al., 2003; Liu et al., 2004). The major features of the MPE cyclase, both the di-iron binding sites and leucine zipper domain, were highly conserved among EaZIP and other MPE cyclases (Fig. 5).
Fig. 5.

Alignment of deduced amino acid sequence of EaZIP. The alignment was generated by ClustalW2 (EMBL-EBI). Identical residues are marked with an asterisk (*); those which were conserved substitutions or semi-conserved substitutions are marked with a colon (:) or with a full stop (.), respectively. The carboxylate-bridged di-iron binding sites are indicated in red of regions I and II. The five amino acids representing leucine zipper domain are indicated in red of region III. The GenBank accession numbers of the sequences are: NTZIP, Nicotiana tobacum AY2211682; CHL27, Arabidopsis thaliana NM_115553; and CRD1, Chlamydomonas reinhardtii XM_001692505. (This figure is available in colour at JXB online.)
Using an antibody specific for C. reinhardtii CRD1 protein, bands around 40 kDa molecular weight were detected in total protein extracts of regenerated green ‘Golden Pothos’ plants, Arabidopsis, and tobacco, but none was detected in any pale yellow plants (Fig. 6). Conversely, two antibodies specific for chloroplast proteins, RbcL and PC, demonstrated a similar detection level between pale yellow and green plants (Fig. 6). These results indicated the presence of chloroplast proteins in the pale yellow plants. Lack of cross reaction of anti-CRD1 is due to lack of EaZIP-encoded product. This further supports that the EaZIP-encoded product is a homologue of CRD1.
Fig. 6.
Immunoblot analysis. (A) Total proteins of Arabidopsis (At), tobacco (To), and ‘Golden Pothos’ regenerated pale yellow plants (PY1 and PY2), and green plants (G1 and G2) were subjected to 4–12% SDS-PAGE, then transferred to a PVDF membrane. Primary antibodies, anti-CRD1, anti-RbcL and anti-PC, were indicated on the left. The molecular weights of each detected protein were also indicated on the right. (B) A membrane stained with a 0.2% (w/v) Amido Black solution shows protein loading. (This figure is available in colour at JXB online.)
Results from barley mutants (Rzeznicka et al., 2005) have shown that mutants Xantha-l81 and Xantha-l82 defective in MPE cyclase exhibited a yellow phenotype. The substrate of the MPE cyclase, MPE, was also accumulated when these mutants were fed with ALA, the first committed intermediate of the porphyrin synthesis pathway. To show that the pale yellow plants with reduced levels of EaZIP expression lack functional MPE cyclase, an ALA feeding experiment was carried out to detect any accumulation of MPE. Both green and pale yellow tissues were first incubated with ALA solution in the dark and then the treated leaves were subjected to acetone extraction. By adding hexane into the acetone extracts, the green chlorophylls were partitioned into the hexane layer and removed. Fluorescence analysis of the acetone extracts revealed a single emission peak at 595 nm with an excitation set at 420 nm in the ALA-treated pale yellow tissues, but not in green tissues (Fig. 7A). The HPLC separation further indicated that MPE was the main chlorophyll biosynthesis intermediate accumulated only in ALA-treated pale yellow tissues (Fig. 7B, C). This result indicated the MPE cyclase function was lost in pale yellow plants and further support that the EaZIP-encoded product is a MPE cyclase.
Fig. 7.

Accumulation of MPE in pale yellow plants. Pigments were isolated from leaf discs of green (G), or pale yellow (PY) regenerated plants treated with (+) or without (–) 10 mM ALA hydrochloride. (A) Extracted pigments were analysed by excitation at 420 nm and their fluorescence emissions were scanned from 570 to 630 nm. (B) Pigments were separated by HPLC. The eluate was monitored at 420 nm, the optimal detection for protoporphyrin IX (P), magnesium protoporphyrin IX (MP), and magnesium protoporphyrin IX monomethyl ester (MPE) (left). Spectral absorbance of the major eluate peak was measured from 200 to 600 nm (right). All peaks were compared to known standards relative to their retention times (left) and specific absorbance (right). (C) No peak was observed in extracts isolated from both without ALA-treated pale yellow and green tissues. (This figure is available in colour at JXB online.)
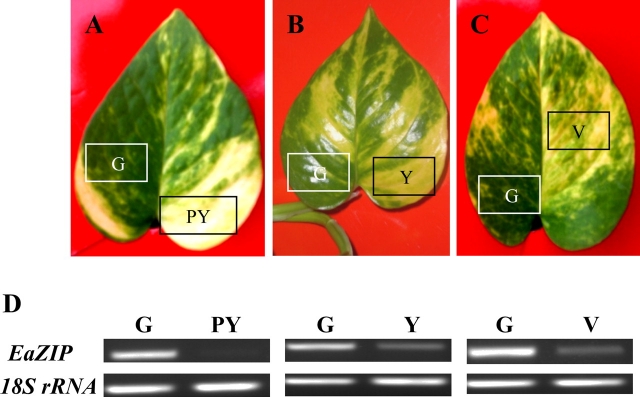
The transcript level of EaZIP was reduced in all yellow parts of naturally variegated ‘Golden Pothos’
To confirm that the low expression of EaZIP isolated by SSH is involved in the variegation of ‘Golden Pothos’, three types of variegated leaves with different degrees of yellow colours, i.e. pale yellow, yellow, and variegated (Fig. 8A–C) were selected. From each type of leaf, different colour sectors on the same leaf were separated to compare their EaZIP expression levels by RT-PCR and chlorophyll contents. Results showed that yellow sectors from all three types of plants have lower EaZIP transcript levels compared with those of their green counterparts (Fig. 8D). Also the expression level of EaZIP in pale yellow was undetectable and much lower than those of yellow and variegated sectors (Fig. 8D). To show that the EaZIP gene expression is correlated with chlorophyll contents, chlorophyll in different sectors was also examined. The total chlorophyll a and b of pale yellow, yellow, and variegated leaf sectors were 0.55, 0.62, and 1.28 mg g−1 fresh weight, which were all lower than their green counterparts of 7.76, 5.41, and 4.70 mg g−1, respectively (Table 2). Taking all the above results together, it was concluded that the EaZIP expression level of each sector correlates to its chlorophyll content, suggesting that EaZIP level may contribute to the variegation formation.
Fig. 8.
RT-PCR of EaZIP in variegated ‘Golden Pothos’ leaves. Variegated leaves (A, B, C) were harvested from plants grown in soil under the same controlled conditions. Total RNAs were isolated from green (G), pale yellow (PY), yellow (Y), and variegated (V) parts. RT-PCR was performed using specific primers targeting EaZIP and 18S rRNA (D). (This figure is available in colour at JXB online.)
Table 2.
The comparison of chlorophyll (Chl) contents in green and yellow parts separated from three different variegated ‘Golden Pothos’ leaves shown in Fig. 8
| Chl | Variegated A | Variegated B | Variegated C | |||
| (mg g−1FW) | Green (G) | Pale Yellow (PY) | Green (G) | Yellow (Y) | Green (G) | Variegated (V) |
| Chl a | 5.68±0.40 | 0.46±0.02 | 3.96±0.08 | 0.50±0.02 | 3.47±0.09 | 1.00±0.01 |
| Chl b | 2.08±0.15 | 0.09±0.02 | 1.45±0.03 | 0.12±0.02 | 1.23±0.03 | 0.28±0.01 |
| Chl a+b | 7.76 | 0.55 | 5.41 | 0.62 | 4.70 | 1.28 |
| Chl a/b ratio | 2.73 | 5.11 | 2.73 | 4.17 | 2.82 | 3.57 |
Leaves were harvested from variegated plants grown in soil under the same controlled conditions. Data represent the average of three independent assays ±standard deviation.
Discussion
Plant tissue culture provides to each cell the potential to regenerate into a new plant. Use of tissue culture techniques to separate cells from variegated leaves and further regenerate plants is a new approach for studying variegated plants that avoids the problems associated with directly dissecting different coloured sectors. Both green and pale yellow regenerated plants could be stably maintained on MS medium for over a year (Hung and Xie, 2009), indicating that the fate of chloroplast development was determined at the early stage of cell division in the original source plant as suggested previously (Sakamoto, 2003), and these regenerated green and pale yellow plants are genetically stable. With this method, the cross contamination problem can be avoided when plant materials are prepared for comparative studies. Although variegated ‘Golden Pothos’ has not been studied previously at the molecular level, SSH and sequence homology searches were successfully combined to isolate and identify a group of differentially expressed genes, including a major gene, EaZIP. Overall, the results suggest that previously uncharacterized variegated species can be utilized for the study of chloroplast biogenesis with current techniques and available nucleotide sequence information.
The cells of regenerated pale yellow plants are viable
It has been reported that cells of the white sectors in var2 variegated leaves are viable and have undifferentiated plastids (Kato et al., 2007). Using tissue culture, it was confirmed that cells of regenerated pale yellow plants from ‘Golden Pothos’ are viable. Firstly, not only can pale yellow plants be maintained on MS medium (Hung and Xie, 2009), but their tissues can be used to produce callus and regenerate pale yellow shoots (Fig. 2A). The successful tissue culture with explants derived from pale yellow plants demonstrates that pale yellow cells are capable of growth and differentiation. Secondly, although the contents of Ca, Mg, and Mn were significantly lower (P <0.05) in pale yellow plants than in green plants, there is no general reduction in micronutrient content in pale yellow plants (Fig. 3), suggesting that development of pale yellow cells was regulated to allow long-term viability. Thirdly, the TEM results also reveal a normal structure of nucleus and mitochondria (Fig. 1D, E). All these results indicate that regenerated pale yellow plants are a stable system, which can be used for molecular and other comparative studies.
The role of EaZIP in ‘Golden Pothos’
In this first study of ‘Golden Pothos’ at the molecular level, a group of differentially expressed genes was isolated using SSH to compare regenerated green and pale yellow plants. Among nine down-regulated genes (Table 1), EaZIP was the only differentially expressed gene, which had no detectable expression level in pale yellow plants. It was confirmed that the EaZIP gene encodes MPE cyclase by independent approaches including deduced amino acid sequence alignment (Fig. 5), anti-CRD1 immunoblot analysis (Fig. 6), and an ALA-feeding experiment (Fig. 7). The RT-PCR results of naturally variegated plants further confirmed the importance of EaZIP involvement in variegation formation because the EaZIP transcript not only was absent in regenerated pale yellow plants (Fig. 4A) but also was correlated with chlorophyll content (Fig. 8D; Table 2). Further research on the regulation of EaZIP expression may shed new light on the basis for variegation formation.
Whether down-regulation of the remaining eight genes is the result of downstream effects from low EaZIP expression remains unclear. A recent microarray comparison of green and white sectors in the im mutant revealed an array of genes involved in photosynthesis and carbohydrate metabolism that was repressed in white sectors (Aluru et al., 2009). Comparing the list of genes with current results, only one gene, EaSBEIIb (E. aureum starch branching enzyme IIb), was found to be homologous to SBE2.1 (AT2G36390) and shared the same reduction level of about 3-fold. A similar reduction was also found in plants subjected to photobleaching using Norflurazon treatment (Aluru et al., 2009), suggesting the reduction of EaSBEIIb expression is a consequence of lacking normal chloroplasts. From other microarray data comparison of gene expression profiles between the CHL27 knockout line (SALK_009052) and the wild type (Bang et al., 2008), EaF156, encoding a protein product similar to ferric reductase, was reported in their list of repressed genes. The EaF156 gene also had a similar 6.2-fold reduction in their study (Bang et al., 2008), suggesting that the reduction of EaF156 was probably a downstream effect of the low EaZIP. The cause of low expression levels of the remaining genes and the relationship between EaZIP and the other down-regulated genes requires further study.
Changes in EaZIP expression levels may contribute to the variegation formation
Among a selection of various variegated ‘Golden Pothos’ leaves, the EaZIP transcript levels were correlated with the chlorophyll levels in leaf sectors exhibiting a range of variegation patterns (Fig. 8; Table 2). One of the plausible explanations for the appearance of green sectors and yellow sectors in the same leaf is the differential EaZIP transcription activities between the cells of green and yellow sectors controlled by transcription factors. Two such transcription factors GOLDEN2-LIKE (GLK) (Waters et al., 2009) and NtbZIP (Yang et al., 2009) have been identified in higher plants. The former regulates a group of nuclear photosynthetic genes in Arabidopsis including CHL27 (Waters et al., 2009) while the latter specifically regulates the expression of NTZIP (Yang et al., 2009). These factors may also be present in ‘Golden Pothos’ but requires future work to confirm.
Another plausible explanation is that the EaZIP transcripts may fail to accumulate or become unstable during an early stage of plastid development, leading to a loss of the MPE cyclase and resulting in the variegated phenotype. Previous work showed that antisense tobacco lines exhibited the variegated phenotype as the result of disrupting enzymatic activities of MPE cyclase (Liu et al., 2004). In Arabidopsis, some CHL27 antisense lines also displayed a degree of variegation (Tottey et al., 2003), in contrast to a T-DNA insertion line of the CHL27 promoter region that reduced the mRNA level but showed uniformly light green colour (Bang et al., 2008). These differences between antisense (knockdown) and T-DNA insertion (knockout) lines support hypotheses that variegation is caused by factors that control the transcript abundance of genes involved in the chlorophyll biosynthesis pathway.
Support for the second proposed explanation can be found in a survey of the literature that compares phenotypic differences between antisense and T-DNA insertion lines of chlorophyll biosynthesis pathway genes. From 22 reports that include results from four species, a pattern emerged relating transformation approach with the resulting phenotype (see Supplementary Table S3 at JXB online). All nine studies of T-DNA insertion lines reported either had an albino or a uniformly pale yellow or pale green phenotype. In contrast, variegated transgenic plants were found in 12 out of 13 studies where chlorophyll genes encoding eight enzymes were interrupted by antisense technology. For the one exception, it is not clear whether the antisense transgene was inserted into a chlorophyll biosynthesis gene causing the pale yellow colour phenotype instead of leaf variegation. These findings suggest that natural leaf variegation may be the result of regulating chlorophyll gene transcript levels by post-transcriptional mechanisms that mimic antisense technology. Post-transcriptional gene silencing by small RNAs (Hamilton and Baulcombe, 1999) is one possibility. Small RNAs and microRNAs that control EaZIP transcript levels may occur naturally in ‘Golden Pothos’. To confirm the existence of such RNAs, further work will be required.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Primer sequences for qRT-PCR.
Supplementary Table S2. Clones with abundant transcripts in pale yellow plants.
Supplementary Table S3. Chlorophyll-deficient patterns of transgenic lines with antisense or T-DNA insertion in chlorophyll a and b biosynthesis pathway genes in four species.
Supplementary Material
Acknowledgments
The work was mainly carried out: North Carolina Central University and North Carolina State University. This work was supported by Award Number SC3GM088084 from the National Institute of General Medical Sciences to JX and a Startup Fund of Golden LEAF Foundation to BRITE. We thank Lisa Lentz and Dr Wayne P Robarge of the Analytical Laboratory Service at North Carolina State University, and Dr Brenda R Cleveland of the Agronomic Division Plant/Waste/Solution Laboratory at North Carolina Department of Agriculture and Consumer Services for analysing element contents. Special thanks to Valerie M Knowlton of the Center for Electron Microscopy at the North Carolina State University for help with the TEM.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru MR, Bae H, Wu D, Rodermel SR. The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiology. 2001;127:67–77. doi: 10.1104/pp.127.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru MR, Zola J, Foudree A, Rodermel SR. Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiology. 2009;150:904–923. doi: 10.1104/pp.109.135780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang WY, Jeong IS, Kim DW, et al. Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling. Plant and Cell Physiology. 2008;49:1350–1363. doi: 10.1093/pcp/pcn111. [DOI] [PubMed] [Google Scholar]
- Bloom AJ. Mineral nutrition. In: Taiz L, Zeiger E, editors. Plant physiology. Sunderland: Sinauer Associates, Inc; 1998. pp. 103–124. [Google Scholar]
- Chen J, McConnell DB, Henny RJ, Norman DJ. The foliage plant industry. Horticulture Reviews. 2005;31:47–112. [Google Scholar]
- Chen M, Choi YD, Voytas DF, Rodermel S. Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. The Plant Journal. 2000;22:303–313. doi: 10.1046/j.1365-313x.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences, USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hung CY, Xie JH. A comparison of plants regenerated from a variegated. Epipremnum aureum. Biologia Plantarum. 2009;54:610–616. [Google Scholar]
- Huxley A. The new royal horticultural society dictionary of gardening. London: Macmillan; 1994. [Google Scholar]
- Josse EM, Simkin AJ, Gaffe J, Laboure AM, Kuntz M, Carol P. A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiology. 2000;123:1427–1436. doi: 10.1104/pp.123.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Miura E, Matsushima R, Sakamoto W. White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiology. 2007;144:952–960. doi: 10.1104/pp.107.099002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Yang YT, Liu HH, Yang GD, Zhang NH, Zheng CC. NTZIP antisense plants show reduced chlorophyll levels. Plant Physiology and Biochemistry. 2004;42:321–327. doi: 10.1016/j.plaphy.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Monde RA, Schuster G, Stern DB. Processing and degradation of chloroplast mRNA. Biochimie. 2000;82:573–582. doi: 10.1016/s0300-9084(00)00606-4. [DOI] [PubMed] [Google Scholar]
- Moseley J, Quinn J, Eriksson M, Merchant S. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in. Chlamydomonas reinhardtii. The EMBO Journal. 2000;19:2139–2151. doi: 10.1093/emboj/19.10.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiology Plantarum. 1962;15:473–497. [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. The Plant Cell. 2002;14:211–236. doi: 10.1105/tpc.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Albrieux C, Joyard J, Lagrange T, Block MA. Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. Effects on chloroplast development and on chloroplast-to-nucleus signaling. Journal of Biological Chemistry. 2007;282:2297–2304. doi: 10.1074/jbc.M610286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzeznicka K, Walker CJ, Westergren T, Kannangara CG, von Wettstein D, Merchant S, Gough SP, Hansson M. Xantha-l encodes a membrane subunit of the aerobic Mg-protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Pro-ceedings of the National Academy of Sciences, USA. 2005;102:5886–5891. doi: 10.1073/pnas.0501784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W. Leaf-variegated mutations and their responsible genes in. Arabidopsis thaliana. Genes and Genetic Systems. 2003;78:1–9. doi: 10.1266/ggs.78.1. [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Tamura T, Hanba-Tomita Y, Murata M. The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes to Cells. 2002;7:769–780. doi: 10.1046/j.1365-2443.2002.00558.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Uno Y, Zhang Q, Miura E, Kato Y, Sodmergen Arrested differentiation of proplastids into chloroplasts in variegated leaves characterized by plastid ultrastructure and nucleoid morphology. Plant and Cell Physiology. 2009 doi: 10.1093/pcp/pcp127. 10.1093/pcp/pcp127. [DOI] [PubMed] [Google Scholar]
- Takechi K, Sodmergen Murata M, Motoyoshi F, Sakamoto W. The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant and Cell Physiology. 2000;41:1334–1346. doi: 10.1093/pcp/pcd067. [DOI] [PubMed] [Google Scholar]
- Tottey S, Block MA, Allen M, Westergren T, Albrieux C, Scheller HV, Merchant S, Jensen PE. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proceedings of the National Academy of Sciences, USA. 2003;100:16119–16124. doi: 10.1073/pnas.2136793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Wettstein D, Gough S, Kannangara CG. Chlorophyll biosynthesis. The Plant Cell. 1995;7:1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Duby G, Purnelle B, Boutry M. Tobacco VDL gene encodes a plastid DEAD box RNA helicase and is involved in chloroplast differentiation and plant morphogenesis. The Plant Cell. 2000;12:2129–2142. doi: 10.1105/tpc.12.11.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. GLK transcription factors co-ordinate expression of the photosynthetic apparatus in Arabidopsis. The Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel CM, Jiang CZ, Meehan LJ, Voytas DF, Rodermel SR. Nuclear-organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. The Plant Journal. 1994;6:161–175. doi: 10.1046/j.1365-313x.1994.6020161.x. [DOI] [PubMed] [Google Scholar]
- Wu DY, Wright DA, Wetzel CM, Voytas DF, Rodermel SR. The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. The Plant Cell. 1999;11:43–55. doi: 10.1105/tpc.11.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YT, Yu YL, Yang GD, Zhang JD, Zheng CC. Tissue-specific expression of the PNZIP promoter is mediated by combinatorial interaction of different cis-elements and a novel transcriptional factor. Nucleic Acids Research. 2009;37:2630–2644. doi: 10.1093/nar/gkp126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaronskaya E, Ziemann V, Walter G, Averina N, Borner T, Grimm B. Metabolic control of the tetrapyrrole biosynthetic pathway for porphyrin distribution in the barley mutant albostrians. The Plant Journal. 2003;35:512–522. doi: 10.1046/j.1365-313x.2003.01825.x. [DOI] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR. The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. The Plant Journal. 2004;37:864–876. doi: 10.1111/j.1365-313x.2003.02014.x. [DOI] [PubMed] [Google Scholar]
- Yu F, Fu AG, Aluru M, Park S, Xu Y, Liu HY, Liu XY, Foudree A, Nambogga M, Rodermel S. Variegation mutants and mechanisms of chloroplast biogenesis. Plant, Cell and Environment. 2007;30:350–365. doi: 10.1111/j.1365-3040.2006.01630.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.