Abstract
Purpose
Feeder cells from animals raise considerable concern for contamination because they are directly in contact with embryonic stem cells.
Methods
To address this issue we collected discarded foreskin tissue and prepared a fibroblast cell line. We transferred one parthenogenetic blastocyst on to these feeder cells, and later observed outgrowth. By this approach, we were able to derive a human parthenogenetic embryonic stem cell line successfully.
Results
The embryonic stem cells had normal morphology, expressed all expected cell surface markers, could differentiate to embryonic bodies upon culture in vitro, and differentiated further to derivatives of all three germ layers.
Conclusion
This study indicates that homologous human fibroblasts can be used as feeder cells to support not only the propagation, but also the derivation of ES cells, and this should facilitate studies of therapeutic cloning for research and clinical applications.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-010-9408-5) contains supplementary material, which is available to authorized users.
Keyword: Feeder cells, Human embryonic stem cells, Human foreskin fibroblast, Parthenogenetic blastocyst
Introduction
Human embryonic stem (hES) cells have tremendous potential in regenerative medicine and cell therapy because of their potential for self-renewal and multi-differentiation [1, 2]. Allogeneic ES cells have been found to elicit a vigorous immune response, hence it is important to use ES cells that are matched genetically with the recipient in clinical applications. There are two methods by which such ES cells can be derived: somatic nuclear cell transfer (SCNT) and parthogenesis. The derivation of hES cells by the SCNT method has yet to be reported, whereas the successful derivation of hES cells by parthenogenesis has been reported recently[3–6].
In general, parthenogenetic (PG) ES cells are derived from the inner cell mass (ICM) of blastocyst-stage embryos developed from metaphase II (MII) oocytes that have undergone spontaneous activation [4–6]. Lin et al. suggested an alternative strategy in which human PGES cells were obtained from haploid oocytes after routine in vitro fertilization (IVF) treatment [3]. In spite of differences in the methodology, these PGES cells display the same properties of self-renewal and multi-differentiation as normal ES cells.
Human PGES cells are still dependent on feeder cells of animal origin to maintain their undifferentiated status, e.g. mouse embryonic fibroblasts (MEFs), which are similar to human and mouse ES cells [7–9]. Feeder cells are necessary to maintain the self-renewal and growth of hES cells but the molecular factors that the feeder cells provide to the hES cells have not yet been elucidated. Martin et al. demonstrated that certain mouse glycoproteins can be identified on the surface of hES cells that are cultured on MEFs, and such glycoproteins, if maintained on the surface of the cells, could result in a severe immune reaction against any transplanted cells [10]. To resolve this issue, two strategies were suggested: a) a feeder-free culture system and b) the use of human homologous feeder cells. The normal characteristics of hES cells can be maintained using a feeder-free culture system, and successful derivation has been reported using this system [11, 12], however, this system has not yet been applied widely because of its complexity and associated difficulties. Feeder cells from human tissues, including foreskin fibroblasts [13] and adult Fallopian tube epithelial cells [14], have been used to derive and culture hES cells, and all of these types of feeder cell can support the prolonged undifferentiated growth of hES cells.
In the study described herein, we investigated whether or not human PGES cells could be derived and established using human foreskin fibroblasts as feeder cells. We describe the establishment and characteristics of human PGES cells. The results obtained should facilitate the further study and application of such cells in research and clinical settings.
Materials and methods
Informed consent
The donated oocytes were collected from the Center for Reproductive Medicine, Tianjin Central Hospital for Obstetrics and Gynecology, which is certified by the Ministry of Health of the People’s Republic of China. The egg donor was on an assisted reproductive technique (ART) cycle. No financial benefit was involved in the donation process. The egg donor was clearly informed of all the study details, including the disposition of the embryos and research destination, and they signed detailed informed consent documents voluntarily. We guaranteed that the embryos would only be used for basic scientific research and not for reproductive purposes.
Oocytes collection and embryo culture
A total of 22 oocytes were collected from the donor. However, only four oocytes were at the MII stage, the others were still at the MI stage. The matured oocytes underwent the IVF procedure, whereas the immature oocytes were cultured first in medium that contained follicle stimulating hormone (FSH) and human chorionic gonadotrophin (HCG) [15]. On the second day, 14 oocytes had matured in vitro and were fertilized by intracytoplasmic sperm injection (ICSI). The remaining four oocytes continued to be cultured in the maturation medium. After two days, one 8-cell embryo was identified among these oocytes, and was transferred into G2 medium to be cultured for a further three days. The blastocytes were observed at the expanded stage and used for the derivation of ESCs.
Preparation of human foreskin cell line
One piece of foreskin tissue was obtained from a 4-year old child after circumcision and was donated by his parents. The foreskin was washed, minced with scissors, and dissociated to single cells by trypsinization. Then the cells were grown in a culture medium that consisted of 85% Dulbecco’s modified Eagle’s medium (DMEM; high-glucose) supplemented with 15% fetal bovine serum (FBS; GIBCO), 2 mM L-glutamine, 1% MEM nonessential amino acids, and 1% penicillin-streptomycin. The human foreskin fibroblasts (hFFs) were split using 0.25% trypsin–EDTA every 4–6 days. They could be expanded for more than 40 passages using the same split ratio and rate. The foreskin fibroblasts from passages 15–25 were chosen as the feeder cells.
Isolation and culture of human parthenogenetic embryonic stem cells
The ICM of the embryo was isolated by immunosurgery as described previously [16]. After immunosurgery, the trophectoderm was removed using a pipette and the ICM was isolated. The ICM was plated on gelatin-coated tissue culture dishes on a hFF feeder layer that had been treated with 10 μg/ml mitomycin. The culture medium consisted of 80% KnockOut DMEM (GIBCO), 20% KnockOut Serum Replacement (GIBCO), 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, 1% MEM nonessential amino acids, 1% penicillin-streptomycin, and 8 ng/mL of basic fibroblast growth factor (bFGF). After 6–8 days, the ICM clump was removed mechanically with a micropipette from differentiated cell outgrowths and replated onto a fresh hFF feeder layer. The resulting colonies were further propagated mechanically in small clumps of ∼100 ES-like cells on hFF feeder layers approximately every 7 days. After the clumps had been dissociated mechanically for ten passages, they were passaged routinely using 1 mg/mL type IV collagenase every 6 days. All of the parthenogenetic stem cells were cultured at 37°C in a 5% CO2 incubator.
Embryoid body formation
To form embryoid bodies (EBs), one confluent six-well plate was used. The ES cells were passaged using 1 mg/mL of type IV collagenase, and dispersed further into small clumps using a pipette tip. The clumps were cultured under suspension culture conditions. The medium consisted of 80% KnockOut DMEM, 20% KnockOut Serum Replacement, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, 1% MEM nonessential amino acids, without bFGF.
Specific markers characterization of human parthenogenetic stem cell line
To assay alkaline phosphatase (AP) activity, hPGES cells on the hFF feeder layer were fixed with 10% formalin, and then the cells were incubated with the AP substrate BCIP/NBT (Sigma-Aldrich). The resulting precipitate was observed using an inverted microscope.
For immunofluorescence staining, the PGES cells were fixed with 4% paraformaldehyde and incubated with primary antibodies (1:100) overnight at 4°C. The primary antibodies included antibodies against stage-specific embryonic antigen (SSEA)-1, SSEA-3, and SSEA-4 (R&D), in addition to antibodies against TRA-1-60 and TRA-1-81 (BD). TX-conjugated goat anti-mouse and TRITC-conjugated goat anti-rat immunoglobulin antibodies were used as secondary antibodies (1:200).
Gene expression analysis by the reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted using Trizol Reagent (Invitrogen) from undifferentiated PGES cells that had been grown for more than 20 passages on the hFF feeder layer, according to the manufacturer’s recommended protocol. cDNA was synthesized from ∼1 mg of total RNA using SuperScript Reverse Transcriptase (Invitrogen) and was subjected to PCR amplification using primers for the OCT4, NANOG, REX1, H19, h-UBE3A, h-SNPRN, and IGF-2 coding sequences. Total RNA was extracted by the same method from day 7 EBs to analyze the expression of specific markers of the three embryonic germ layers, including AFP, HBZ, and NEUOD-1. The hES cell line H9 was used as a control. The PCR primers used are described in Supplement Table 1. The PCR products were fractionated by size on a 2% agarose gel and visualized by staining with Ethidium Bromide.
Karyotype analysis
Parthenogenetic stem cell division was blocked in mitotic metaphase by incubation with colchicine. The nuclear membrane was disrupted by hypotonic treatment with potassium chloride. Then the cells were fixed with methanol:glacial acetic acid and dropped onto glass slides. The chromosome spreads were stained with Giemsa and photographed. To visualize the chromosomes, standard G-band staining was used. At least 50 metaphase cells were examined from each sample.
Results
Derivation of a new human parthenogenetic stem cell line
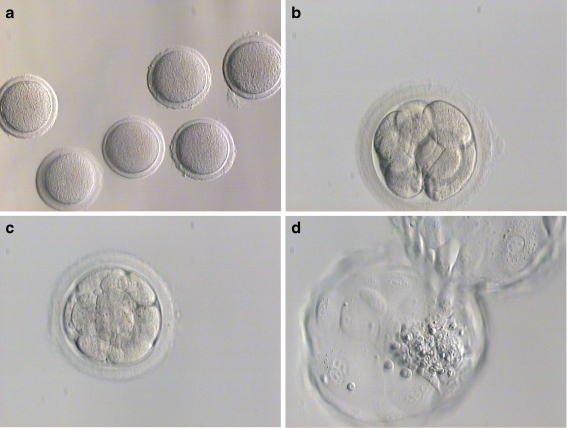
The blastocyst developed from an MII oocyte that had undergone spontaneous activation after maturation in vitro (Fig. 1). The parthenogenetic blastocyst had a visible ICM and typical fusiform trophoblast cells. The ICM was capable of forming outgrowths and was dissected mechanically after 6–8 days. The PGES cell line was propagated for more than 40 passages on a hFF feeder layer (Supplement Fig. 1).
Fig. 1.
Development of human embryos from spontaneously activated metaphase II oocytes matured in vitro
Characterization of the parthenogenetic stem cell line
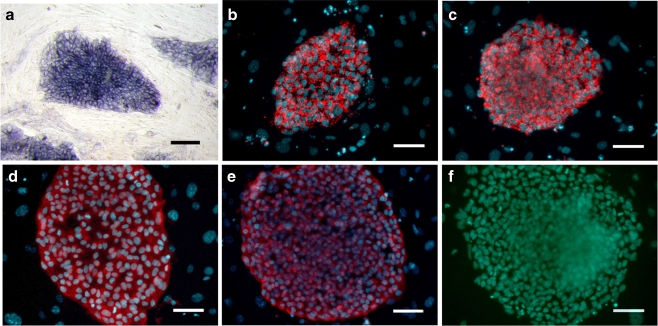
The hPGES cells had the normal morphological characteristics of hES cells, which include a high nuclear/cytoplasm ratio, formation of tightly-packed colonies, and high levels of AP activity (Fig. 2a). The hPGES colonies expressed positive cell surface markers that included SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81, but did not express the negative cell surface marker SSEA-1 (Fig. 2b, c, d, e, f), which is similar to the expression pattern of normal hES cells.
Fig. 2.
Characterization of undifferentiated human parthenogenetic embryonic stem cells (passage level 20). The line P-TJ had high alkaline phosphatase activity (a) and expressed SSEA-3 (b), SSEA-4 (c), TRA-1-60 (d), and TRA-1-81 (e) but did not express SSEA-1 (F). Bar: 100 μm
The expression of genes related to pluripotency was detected by RT-PCR, and the gene products included OCT4, NANOG, and REX1. The gene expression profile was the same as that of the control hES cell line H9 (Supplement Fig. 2).
Differentiation potential in vitro of the parthenogenetic embryonic stem cell line
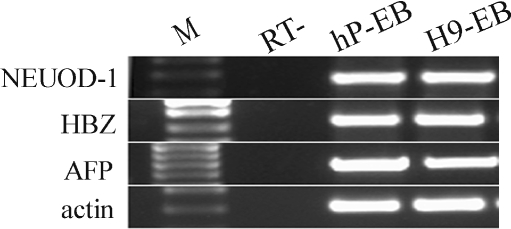
To determine whether the hPGES cells had the potential to differentiate into all three embryonic germ layers in vitro, the cells were differentiated into EBs in vitro under suspension culture conditions (Supplement Fig. 3). After spontaneous differentiation for 7 days, RNA was extracted from the EBs and RT-PCR was performed. The results showed that the expression of markers for all three embryonic germ layers could be identified successfully, including NEUOD-1 (ectoderm), HBZ (mesoderm), and AFP (endoderm) (Fig. 3).
Fig. 3.
RT-PCR analysis of the expression of specific markers of the three embryonic germ layers. The EBs derived from human parthenogenetic embryonic stem cells expressed NEUOD-1 (ectoderm), HBZ (mesoderm), and AFP (endoderm). The EBs from the control hES cell line H9 expressed all three embryonic germ layer markers
Identification of parthenogenetic origin
PGES cells only contain maternally imprinted genes and lack paternally imprinted genes, whereas both types are expressed in normal fertilized embryos. Therefore, analysis of the expression profiles of imprinted genes enables the verification of parthenogenetic embryos. In this study, the expression of imprinted genes was analyzed in both the hPGES line and the control H9 ES cell line. Transcription of the paternally expressed hSNRPN and IGF-2 genes was not detected in the hPGES cell line, whereas transcription of maternally expressed UBE3A and H19 was observed (Supplement Fig. 4). All of the above imprinted genes were expressed in the H9 control cells.
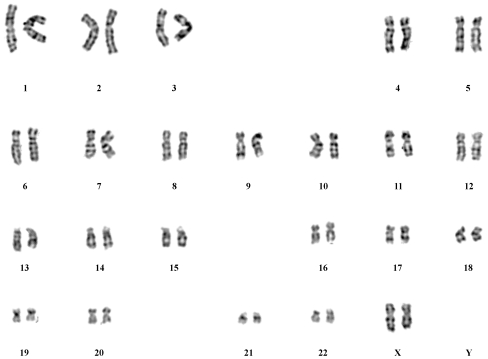
Karyotyping of the parthenogenetic embryonic stem cell line
Karyotyping was performed at passage 20. The 46, XX karyotype was consistent with the hPGES cells originating from a female donor (Fig. 4).
Fig. 4.
Karyotyping of parthenogenetic embryonic stem cells at passage level 20. The line P-TJ had a normal 46, XX karyotype
Discussion
Human ES cells have been an ongoing focus since the first successful report by Thomson et al. twelve years ago [7]. They are regarded as a promising resource for regenerative medicine and tissue engineering [17], given that they can differentiate into somatic cell lineages that include insulin-producing cells [18], neuronal cells [19], and myocytes [2]. However, the issue of rejection due to the induction of an immune response has limited the development and application of hES cells [20, 21]. Parthenogenetic ES cells are derived from the ICM of a parthenogenetic blastocyst that has developed from a MII oocyte and they only contain the maternal genome. As a result these cells have the same genetic background as, and are immune compatible with, the egg donor.
In our study, we derived and established a hPGES cell line using an embryo that had arisen due to the spontaneous activation of a metaphase II oocyte matured in vitro. Oocyte aging is a potential contribute of the spontaneous parthenogenetic activation of oocytes because the level of Maturation/M-phase promoting factor (MPF) decreases as the oocytes age. High levels of MPF induce the arrest oocytes in the metaphase stage of the cell cycle, including starfish [22], Xenopus [23], mammals [24–27], and it is inactivated in matured oocytes in response to fertilization [24, 25, 28, 29] or parthenogenetic activation [28, 30, 31]. Therefore, the gradual decrease of MPF activity is a key to resume meiosis capacity of oocytes after fertilization [32], which is also compatible with the gradual increase in the ability of aging oocytes to undergo spontaneous activation during aging.
It is well known that feeder cells could secrete factors that support the growth of ES cells. however, one of the most popular types of feeder cell, MEFs, is derived from mice, and has the potential for fertility between feeder cells and ES cells [10]. The use of homologous feeder cells is one feasible solution to this problem. Li et al. indicated that homologous somatic cells from monkey could support the growth of monkey ES cells regardless of the tissue from which the feeder cells were derived [33]. In humans, some studies also indicated that cells from the foreskin [13] or adult Fallopian tube epithelium [14] could support the propagation and self-renewal of hES cells. These cells not only support the growth of ES cells, but also enable the formation of ICM outgrowths and the derivation of ES cells with a similar efficiency to mouse feeder cells. At present, human homologous feeder cells have been used widely in the culture of hES cells, but their use has not been described for hPGES cells. In previous studies, the derivation and culture of hPGES cells both have been performed on MEF feeder cells, which limit their potential application in clinical settings [3–5]. Here we used foreskin fibroblasts as feeder cells, and derived an hPGES cell line successfully. This should facilitate the study of hPGES cells in research and clinical settings.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
PCR primers used to detect gene expression in human ES and EB cells (DOC 34 kb)
Colony of line P-TJ (passage level 15) growing on the hFF feeder layer. Bar: 100 μm (GIF 218 kb)
High resolution image file (TIFF 3614 kb)
RT-PCR analysis of the expression of pluripotent genes in line P-TJ (lanes 1–4). The hES cell line H9 was analyzed at the same time as a control (lanes 5–8). Lanes 1, 5: β-actin; lanes 2, 6: OCT4; lanes 3, 7: NANOG; lanes 4, 8: REX1 (GIF 42 kb)
Embryoid bodies at day 4 in suspension culture conditions. Bar: 100 μm (GIF 128 kb)
High resolution image file (TIFF 219 kb)
T-PCR analysis of the expression of selected imprinted genes. The human parthenogenetic embryonic stem cells expressed only maternal genes (lanes 1, 2), and not paternal genes (lanes 3, 4). The control hES cell line H9 expressed both maternal and paternal genes (lanes 5–8). Lanes 1, 5: H19; lanes 2, 6: UBE3A; lanes 3, 7: hRPNR; lanes 4, 8: IGF2 (GIF 27 kb)
High resolution image file (TIFF 195 kb)
Acknowledgements
We are grateful to Michelle Yu for the invaluable collaboration in the writing and revision of the manuscript. This work was supported by grants from the Major State Basic Research Development Program of China (973 Program) (No. 2007CB948001) and the National High Technology Research and Development Program of China (863 Program) (No. 2006AA02A101).
Footnotes
Zhenyu Lu, Wanwan Zhu and Yang Yu contributed equally to this work.
Capsule One human embryonic stem cell line from parthenogenetic embryos was derived using human foreskin feeder cells and maintained normal characteristics during long term propagation.
References
- 1.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 2.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–14. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin G, OuYang Q, Zhou X, Gu Y, Yuan D, Li W, et al. A highly homozygous and parthenogenetic human embryonic stem cell line derived from a one-pronuclear oocyte following in vitro fertilization procedure. Cell Res. 2007;17:999–1007. doi: 10.1038/cr.2007.97. [DOI] [PubMed] [Google Scholar]
- 4.Mai Q, Yu Y, Li T, Wang L, Chen MJ, Huang SZ, et al. Derivation of human embryonic stem cell lines from parthenogenetic blastocysts. Cell Res. 2007;17:1008–19. doi: 10.1038/cr.2007.102. [DOI] [PubMed] [Google Scholar]
- 5.Revazova ES, Turovets NA, Kochetkova OD, Kindarova LB, Kuzmichev LN, Janus JD, et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2007;9:432–49. doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- 6.Kim K, Ng K, Rugg-Gunn PJ, Shieh JH, Kirak O, Jaenisch R, et al. Recombination signatures distinguish embryonic stem cells derived by parthenogenesis and somatic cell nuclear transfer. Cell Stem Cell. 2007;1:346–52. doi: 10.1016/j.stem.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 8.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–6. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 9.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–8. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin MJ, Muotri A, Gage F, Varki A. Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nat Med. 2005;11:228–32. doi: 10.1038/nm1181. [DOI] [PubMed] [Google Scholar]
- 11.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–4. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 12.Klimanskaya I, Chung Y, Meisner L, Johnson J, West MD, Lanza R. Human embryonic stem cells derived without feeder cells. Lancet. 2005;365:1636–41. doi: 10.1016/S0140-6736(05)66473-2. [DOI] [PubMed] [Google Scholar]
- 13.Amit M, Margulets V, Segev H, Shariki K, Laevsky I, Coleman R, et al. Human feeder layers for human embryonic stem cells. Biol Reprod. 2003;68:2150–6. doi: 10.1095/biolreprod.102.012583. [DOI] [PubMed] [Google Scholar]
- 14.Richards M, Fong CY, Chan WK, Wong PC, Bongso A. Human feeders support prolonged undifferentiated growth of human inner cell masses and embryonic stem cells. Nat Biotechnol. 2002;20:933–6. doi: 10.1038/nbt726. [DOI] [PubMed] [Google Scholar]
- 15.Goud PT, Goud AP, Qian C, Laverge H, Elst J, Sutter P, et al. In-vitro maturation of human germinal vesicle stage oocytes: role of cumulus cells and epidermal growth factor in the culture medium. Hum Reprod. 1998;13:1638–44. doi: 10.1093/humrep/13.6.1638. [DOI] [PubMed] [Google Scholar]
- 16.Solter D, Knowles BB. Immunosurgery of mouse blastocyst. Proc Natl Acad Sci U S A. 1975;72:5099–102. doi: 10.1073/pnas.72.12.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffith LG, Naughton G. Tissue engineering—current challenges and expanding opportunities. Science. 2002;295:1009–14. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 18.Assady S, Maor G, Amit M, Itskovitz-Eldor J, Skorecki KL, Tzukerman M. Insulin production by human embryonic stem cells. Diabetes. 2001;50:1691–7. doi: 10.2337/diabetes.50.8.1691. [DOI] [PubMed] [Google Scholar]
- 19.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–33. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 20.Bradley JA, Bolton EM, Pedersen RA. Stem cell medicine encounters the immune system. Nat Rev Immunol. 2002;2:859–71. doi: 10.1038/nri934. [DOI] [PubMed] [Google Scholar]
- 21.Taylor CJ, Bolton EM, Pocock S, Sharples LD, Pedersen RA, Bradley JA. Banking on human embryonic stem cells: estimating the number of donor cell lines needed for HLA matching. Lancet. 2005;366:2019–25. doi: 10.1016/S0140-6736(05)67813-0. [DOI] [PubMed] [Google Scholar]
- 22.Labbe JC, Capony JP, Caput D, Cavadore JC, Derancourt J, Kaghad M, et al. MPF from starfish oocytes at first meiotic metaphase is a heterodimer containing one molecule of cdc2 and one molecule of cyclin B. EMBO J. 1989;8:3053–8. doi: 10.1002/j.1460-2075.1989.tb08456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drury KC, Schorderet-Slatkine S. Effects of cycloheximide on the “autocatalytic” nature of the maturation promoting factor (MPF) in oocytes of Xenopus laevis. Cell. 1975;4:269–74. doi: 10.1016/0092-8674(75)90175-0. [DOI] [PubMed] [Google Scholar]
- 24.Choi T, Aoki F, Mori M, Yamashita M, Nagahama Y, Kohmoto K. Activation of p34cdc2 protein kinase activity in meiotic and mitotic cell cycles in mouse oocytes and embryos. Development. 1991;113:789–95. doi: 10.1242/dev.113.3.789. [DOI] [PubMed] [Google Scholar]
- 25.Fulka J, Jr, Jung T, Moor RM. The fall of biological maturation promoting factor (MPF) and histone H1 kinase activity during anaphase and telophase in mouse oocytes. Mol Reprod Dev. 1992;32:378–82. doi: 10.1002/mrd.1080320411. [DOI] [PubMed] [Google Scholar]
- 26.Naito K, Daen FP, Toyoda Y. Comparison of histone H1 kinase activity during meiotic maturation between two types of porcine oocytes matured in different media in vitro. Biol Reprod. 1992;47:43–7. doi: 10.1095/biolreprod47.1.43. [DOI] [PubMed] [Google Scholar]
- 27.Naito K, Toyoda Y. Fluctuation of histone H1 kinase activity during meiotic maturation in porcine oocytes. J Reprod Fertil. 1991;93:467–73. doi: 10.1530/jrf.0.0930467. [DOI] [PubMed] [Google Scholar]
- 28.Collas P, Sullivan EJ, Barnes FL. Histone H1 kinase activity in bovine oocytes following calcium stimulation. Mol Reprod Dev. 1993;34:224–31. doi: 10.1002/mrd.1080340215. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi K, Naito K, Daen FP, Izaike Y, Toyoda Y. Histone H1 kinase activity during in vitro fertilization of pig follicular oocytes matured in vitro. Theriogenology. 1995;43:523–32. doi: 10.1016/0093-691X(94)00044-U. [DOI] [PubMed] [Google Scholar]
- 30.Kikuchi K, Izaike Y, Noguchi J, Furukawa T, Daen FP, Naito K, et al. Decrease of histone H1 kinase activity in relation to parthenogenetic activation of pig follicular oocytes matured and aged in vitro. J Reprod Fertil. 1995;105:325–30. doi: 10.1530/jrf.0.1050325. [DOI] [PubMed] [Google Scholar]
- 31.Barnes FL, Collas P, Powell R, King WA, Westhusin M, Shepherd D. Influence of recipient oocyte cell cycle stage on DNA synthesis, nuclear envelope breakdown, chromosome constitution, and development in nuclear transplant bovine embryos. Mol Reprod Dev. 1993;36:33–41. doi: 10.1002/mrd.1080360106. [DOI] [PubMed] [Google Scholar]
- 32.Carroll J. The initiation and regulation of Ca2+ signalling at fertilization in mammals. Semin Cell Dev Biol. 2001;12:37–43. doi: 10.1006/scdb.2000.0215. [DOI] [PubMed] [Google Scholar]
- 33.Li T, Wang S, Xie Y, Lu Y, Zhang X, Wang L, et al. Homologous feeder cells support undifferentiated growth and pluripotency in monkey embryonic stem cells. Stem Cells. 2005;23:1192–9. doi: 10.1634/stemcells.2004-0286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
PCR primers used to detect gene expression in human ES and EB cells (DOC 34 kb)
Colony of line P-TJ (passage level 15) growing on the hFF feeder layer. Bar: 100 μm (GIF 218 kb)
High resolution image file (TIFF 3614 kb)
RT-PCR analysis of the expression of pluripotent genes in line P-TJ (lanes 1–4). The hES cell line H9 was analyzed at the same time as a control (lanes 5–8). Lanes 1, 5: β-actin; lanes 2, 6: OCT4; lanes 3, 7: NANOG; lanes 4, 8: REX1 (GIF 42 kb)
Embryoid bodies at day 4 in suspension culture conditions. Bar: 100 μm (GIF 128 kb)
High resolution image file (TIFF 219 kb)
T-PCR analysis of the expression of selected imprinted genes. The human parthenogenetic embryonic stem cells expressed only maternal genes (lanes 1, 2), and not paternal genes (lanes 3, 4). The control hES cell line H9 expressed both maternal and paternal genes (lanes 5–8). Lanes 1, 5: H19; lanes 2, 6: UBE3A; lanes 3, 7: hRPNR; lanes 4, 8: IGF2 (GIF 27 kb)
High resolution image file (TIFF 195 kb)