Summary
Bacterial flagella play an essential role in the pathogenesis of numerous enteric pathogens. The flagellum is required for motility, colonization, and in some instances, for the secretion of effector proteins. In contrast to the intensively studied flagella of Escherichia coli and Salmonella typhimurium, the flagella of Campylobacter jejuni, Helicobacter pylori, and Vibrio cholerae are less well characterized and composed of multiple flagellin subunits. This study was performed to gain a better understanding of flagellin export from the flagellar type III secretion apparatus of C. jejuni. The flagellar filament of C. jejuni is comprised of two flagellins termed FlaA and FlaB. We demonstrate that the amino-termini of FlaA and FlaB determine the length of the flagellum and motility of C. jejuni. We also demonstrate that protein-specific residues in the amino-terminus of FlaA and FlaB dictate export efficiency from the flagellar T3SS of Yersinia enterocolitica. These findings demonstrate that key residues within the amino-termini of two nearly identical proteins influence protein export efficiency, and that the mechanism governing the efficiency of protein export is conserved amongst two pathogens belonging to distinct bacterial classes. These findings are of additional interest because C. jejuni utilizes the flagellum to export virulence proteins.
Keywords: flagellum, motility, protein secretion, virulence determinants, T3SS
Introduction
C. jejuni is a leading food-borne cause of bacterial gastroenteritis. An estimated 400-500 million cases of campylobacteriosis occur each year worldwide (Ruiz-Palacios, 2007). Infection with C. jejuni usually occurs from the mishandling and consumption of undercooked poultry products (Konkel et al., 2001). C. jejuni infections are usually self-limiting, with symptoms including abdominal cramps, fever, and diarrhea with blood or leukocytes. Certain serotypes of C. jejuni are linked to the development of Guillain-Barré syndrome, an acute demyelinating neuropathy characterized by flaccid paralysis (Konkel et al., 2001).
C. jejuni possesses a flagellum that functions in both motility and protein secretion (Wassenaar et al., 1991, Grant et al., 1993, Konkel et al., 2004, Guerry, 2007). The flagellum consists of a basal body, hook, and filament (Konkel et al., 2004). The basal body consists of a conduit spanning the inner and outer membranes of the cell, as well as the motor that drives flagellar rotation. The hook section of the flagellum is composed primarily of the protein FlgE, and connects the flagellar filament to the basal body. The flagellar filament is composed of two proteins, FlaA and FlaB, which are exported through the flagellum and polymerize to form a helical conduit 1-3 μm in length (Guerry et al., 1991). Expression of the flagellar apparatus components are regulated by three sigma factors: σ70 (RpoD), σ54 (RpoN), and σ28 (FliA) (Jagannathan et al., 2001). σ70 controls expression of some basal body components, while σ54 regulates the other components needed for a functional flagellar secretory apparatus, including the filament protein FlaB. σ28 regulates expression of the FlaA filament protein and other flagellar-related secreted proteins (Jagannathan et al., 2001, Carrillo et al., 2004).
Structural analysis of the Salmonella typhimurium filament protein FliC by electron microscopy and X-ray crystallography enabled the 495 residue protein to be divided into four linearly connected domains designated D0 - D3 (Samatey et al., 2001, Yonekura et al., 2003). The D0 domain consists of the amino- (residues 1-45) and carboxy-terminal (residues 456-495) ends that fold into a coiled-coil tertiary structure. The amino-terminal portion of the D0 domain facilitates protein secretion, whereas the entire domain is important for filament polymerization (Gugolya et al., 2003, Vegh et al., 2006). FliS, the chaperone for FliC, binds to 40 amino acids in the carboxy-terminal D0 domain (Ozin et al., 2003, Muskotal et al., 2006). Domain D1 consists of residues 46-180 and 408-455, and together with the D0 domain, forms the inner core of the flagellar conduit (Namba et al., 1989, Yonekura et al., 2003). Domains D2 (residues 181-190 and 285-407) and D3 (residues 191-284) form the surface-exposed outer regions of the filament (Namba et al., 1989, Yonekura et al., 2003). Deletions within the D2 and D3 region of the S. typhimurium FliC and E. coli FliC filament proteins have revealed that it is unnecessary for the formation of full-length flagella and bacterial motility (Kuwajima, 1988, Yoshioka et al., 1995). Because the structure of C. jejuni flagellin has not yet been determined, comparisons of the primary sequence of S. typhimurium FliC with those of C. jejuni FlaA and FlaB were used in this study to predict the location and function of domains in C. jejuni flagellin.
Type III secretion systems (T3SS) allow bacteria to secrete proteins from the cytosol to the extracellular environment, through a hollow conduit that spans both the inner and outer membranes (Cornelis, 2006). The bacterial flagellum is a T3SS, and is the only T3SS present in C. jejuni (Konkel et al., 2004, Minamino & Namba, 2004, Desvaux et al., 2006). C. jejuni uses its flagellar T3SS to secrete virulence proteins, termed Campylobacter invasion antigens (Cia), that are required for invasion of epithelial cells (Konkel et al., 1999, Konkel et al., 2004, Larson et al., 2008). Previous research has revealed that both flagellar and classical T3SS recognize secreted proteins through a common mechanism involving the amino-terminal amino acid sequence (Sory et al., 1995, Cornelis, 2006, Vegh et al., 2006, Badea et al., 2009). Information about the secretion of flagellin through the flagellar T3SS of C. jejuni can thus be used to further our understanding of virulence protein secretion through other T3SS.
Previous work has revealed that FlaA is the major constituent of the flagellar filament (Guerry et al., 1991, Wassenaar et al., 1991). A C. jejuni flaB mutant (flaA+B−) produces a filament that is equivalent in length to a wild-type isolate, and these bacteria are motile. In contrast, a C. jejuni flaA mutant (flaA−B+) produces a severely truncated filament, and these bacteria are non-motile (Guerry et al., 1991, Wassenaar et al., 1991, Konkel et al., 2004). The goal of this study was to determine why C. jejuni flaB mutants produce full-length flagellar filaments while C. jejuni flaA mutants produce truncated filaments. Based on the fact that expression of flaA is regulated by a σ28 promoter and flaB is regulated by a σ54 promoter (Jagannathan et al., 2001, Carrillo et al., 2004), we hypothesized that the contrasting phenotypes of C. jejuni flaA or flaB mutants were due to differences in gene expression levels. However, we found that filament length and motility were only partially dependent on flagellin gene expression levels. Amino acid sequence analysis of C. jejuni flagellar filament proteins revealed that the D0 domain of the FlaA and FlaB proteins contain highly-conserved residues. Additional experiments were then performed to determine whether these unique residues could affect the secretion efficiency of flagellin monomers. Our results revealed that flagellar filament length is determined by flagellin gene expression levels and the amino-terminal sequences of FlaA and FlaB. The latter finding was supported using an alternative approach whereby we examined FlaA and FlaB protein export from Yersinia enterocolitica. Thus, the amino-terminal sequence of proteins dictates the efficiency of export through the flagellar T3SS.
Results
C. jejuni FlaA and FlaB share homology with S. typhimurium FliC in the amino- and carboxy-termini
Sequence analysis of the filament proteins from 8 different C. jejuni strains revealed that each strain possesses two filament proteins, ranging in size from 1716 nucleotides (572 amino acids) to 1740 nucleotides (580 amino acids). At the amino acid level, the two filament proteins share greater than 90% identity within a strain (Supplemental Table 1). Alignments of the C. jejuni NCTC 11168 FlaA and FlaB amino acid sequences with S. typhimurium LT2 FliC were conducted to estimate the spatial organization of domains in FlaA and FlaB (Supplemental Figure 1). Putative domain boundaries were assigned solely to approximate the size of the amino- and carboxy-terminal domains, which are more homologous than the central domains. The amino acid identity was greatest in the D0 domain (> 40% identity), followed by the D1 domain (> 20% identity), with the D2 and D3 domains being highly variable (< 20% identity) (Table 1). Using the domain boundaries determined in FliC by X-ray crystallography, putative domain boundaries were assigned to FlaA and FlaB based upon the amino acid alignment. Comparisons of C. jejuni FlaA (Table 2A) and FlaB (Table 2B) amino acid sequences revealed the greatest sequence conservation is within the D0 and D1 domains (> 90% identical). The D3 domain is less conserved (70% to 100% identical) than the D0 and D1 domains, and the D2 domain is the most variable (50% to 100% identical). These results indicate that the D0 and D1 terminal domains, which function in secretion and subunit polymerization, are more conserved than the surface-exposed central domains, which are targets of adaptive immune defenses.
Table 1.
Alignment of S. typhimurium FliC with C. jejuni NCTC 11168 FlaA and FlaB
| Domain | Residues within domaina | % Identity with FliCb | ||
|---|---|---|---|---|
| FliC | FlaA/FlaB | FlaA | FlaB | |
| D0 (N) | 1-45 | 1-45 | 48.9 | 48.9 |
| D1 (N) | 46-180 | 46-181 | 27.2 | 28.9 |
| D2 (N) | 181-190 | 182-192 | 18.2 | 30.0 |
| D3 | 191-284 | 193-335 | 21.5c | 24.2c |
| D2 (C) | 285-407 | 336-473 | 19.5c | 18.7c |
| D1 (C) | 408-455 | 474-532 | 31.2c | 29.2c |
| D0 (C) | 456-495 | 533-572 | 55.0 | 50.0 |
The domain boundaries within the C. jejuni FlaA and FlaB proteins were assigned based on alignment with S. typhimurium FliC.
Percent identity was determined using Megalign version 7.1.0, Clustal W algorithm with a gap penalty of 10.0 and a gap length penalty of 0.2.
The values shown were based on a sub-alignment of a specific domain, and not on the alignment of the entire protein.
Table 2.
In silico analysis of FlaA and FlaB amongst C. jejuni strains
|
A. FlaA amino acid % identity compared with C. jejuni NCTC 11168 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | D0 (N) | D1 (N) | D2 (N) | D3 | D2 (C) | D1 (C) | D0 (C) | Whole |
| NCTC 11168 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 81116 | 95.6 | 94.9 | 63.6 | 71.3 | 62.8 | 98.3 | 95.0 | 81.1 |
| 81-176 | 95.6 | 94.9 | 63.6 | 71.3 | 62.8 | 98.3 | 95.0 | 81.1 |
| Rm1221 | 100 | 99.3 | 81.8 | 93 | 77.7 | 100 | 100 | 92.1 |
| F38011 | 91.1 | 93.4 | 63.6 | 76.2 | 58.1 | 93.2 | 95.0 | 81.2 |
| CF93-6 | 100 | 100 | 100 | 100 | 99.3 | 100 | 100 | 99.8 |
| 260.94 | 91.1 | 93.4 | 63.6 | 73.4 | 56.1 | 96.6 | 95.0 | 79.2 |
| 84-25 | 100 | 100 | 100 | 100 | 99.3 | 100 | 100 | 99.8 |
| B. FlaB amino acid % identity compared with C. jejuni NCTC 11168 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | D0 (N) | D1 (N) | D2 (N) | D3 | D2 (C) | D1 (C) | D0 (C) | Whole |
| NCTC 11168 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 81116 | 95.6 | 95.6 | 54.5 | 73.4 | 60.7 | 98.4 | 97.5 | 81.5 |
| 81-176 | 95.6 | 94.1 | 54.5 | 73.4 | 60.7 | 98.4 | 97.5 | 81.1 |
| Rm1221 | 100 | 98.5 | 90.9 | 97.2 | 77.9 | 100 | 100 | 93.4 |
| F38011 | 97.8 | 91.9 | 45.5 | 73.4 | 56.6 | 91.9 | 97.5 | 80.2 |
| CF93-6 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 260.94 | 97.8 | 92.6 | 54.5 | 74.1 | 53.8 | 98.4 | 97.5 | 79.5 |
| 84-25 | 100 | 100 | 90.9 | 100 | 100 | 100 | 100 | 99.8 |
Considerations of plasmid copy number and flagellar gene phase variation
A C. jejuni F38011 flaAB mutant was generated via homologous recombination to replace the flaA and flaB genes with a tetracycline resistance marker. To study the individual roles of the flagellin genes and promoters, the flaAB mutant was transformed with the E. coli-C. jejuni shuttle vector pRY111 harboring flaA or flaB expressed from the flaA or flaB promoters. Because pRY111 is a multi-copy plasmid of unknown copy number, we compared the flagellin levels in the C. jejuni F38011 wild-type strain, F38011 flaAB mutant, and F38011 flaAB mutant harboring pRY111 containing flaA under control of the flaA promoter (Supplemental Figure 2). Similar levels of flagellin were detected in the whole cell lysates of the C. jejuni F38011 wild-type strain and flaAB mutant harboring flaA, whereas flagellin was not detected in the whole cell lysate of the flaAB mutant (Supplemental Figure 2A). Examination of the supernatants from both the C. jejuni F38011 wild-type strain and flaAB mutant harboring flaA revealed that most of the exported FlaA and FlaB proteins were incorporated into the filament. While neither the FlaA and FlaB proteins were detected in the supernatant fraction of the C. jejuni F38011 wild-type strain and flaAB mutant, a faint band was observed in the lane containing the flaAB mutant harboring flaA (Supplemental Figure 2B). These results demonstrate that similar amounts of flagellin are produced whether the flagellin genes are expressed from a single copy on the chromosome or the multi-copy plasmid pRY111. Collectively, these findings suggested that expression of the flagellin genes from the multi-copy plasmid did not significantly alter the amount of flagellin produced, and did not alter the composition or function of the flagellar filament.
Another factor that could potentially influence the amount of flagellin produced and the composition of the flagellar filament is phase variation of flagellar regulatory and structural genes. Phase variation can occur via strand slippage, recombination, or gene inversion, resulting in activation/inactivation of genes that would affect the regulation of flagellar assembly or motility. To determine the frequency at which wild-type and the flaAB mutant harboring pRY111 spontaneously undergo phase variation to become non-motile, cultures of C. jejuni were diluted and plated in MH soft agar (0.4% agar), such that each plate contained ~50 CFUs (Supplemental Figure 3). After 48 h, 500 individual colonies were examined for motility. The percentage of non-motile colonies observed for F38011 wild-type and the flaAB mutant harboring pRY111 containing flaA expressed from the flaA promoter were 2% and 1.4%, respectively. These results demonstrated that the rate of phase variation in wild-type and the flaAB mutant harboring pRY111 was low enough to allow us to accurately examine motility, filament length, and flagellin synthesis.
Expression of flaA or flaB from the flaA promoter results in increased motility and filament length compared to the flaB promoter
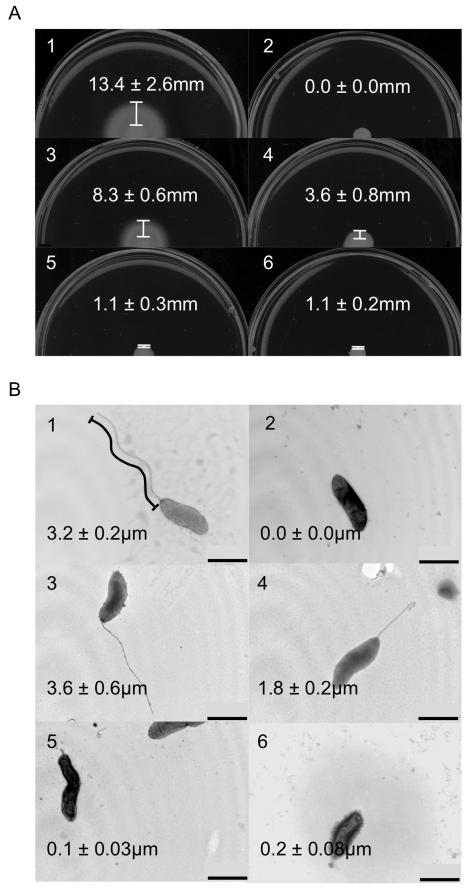
After confirming that expression of flaA in trans from pRY111 in the flaAB mutant did not overtly affect flagellin synthesis or phase variation compared to F38011 wild-type, we sought to determine why full-length flagella are produced by C. jejuni strains that possess a flaA gene, while C. jejuni strains that encode only flaB produce severely truncated filaments and are non-motile (Guerry et al., 1991, Wassenaar et al., 1991, Konkel et al., 2004). We hypothesized that gene expression, dictated by the promoter, was responsible for the phenotypic differences between flaA and flaB mutants. To determine the effects of the promoter on flagellar filament length, a C. jejuni F38011 flaAB mutant was transformed with shuttle vectors harboring flaA and flaB expressed from the flaA and/or flaB promoter. Motility assays were performed using the C. jejuni wild-type strain, flaAB mutant, and flaAB mutant transformed with vectors containing the flaA or flaB genes (Figure 1A). The wild-type strain displayed the greatest zone of motility, measuring 13.4 ± 2.6 mm, while the C. jejuni flaAB mutant was non-motile. When the C. jejuni flaAB mutant was transformed with the shuttle vector harboring flaA or flaB expressed from the flaA promoter, the width of the motility zones were 8.3 ± 0.6 mm and 3.6 ± 0.8 mm, respectively. When the C. jejuni flaAB mutant was transformed with the shuttle vector harboring flaA or flaB expressed from the flaB promoter, the width of the motility zones were reduced to 1.1 ± 0.3 mm and 1.1 ± 0.2 mm, respectively. TEM was also performed to determine the length of the flagellar filament in each of the different strains (Figure 1B). The number of flagellated bacteria observed varied depending on the sample preparation, and not the genotype being examined. Results were recorded only from sample preparations in which the majority of bacteria were flagellated. The length of the flagellum was very similar among flagellated bacteria, while a small population of flagellated bacteria displayed shortened flagella, presumably due to partial shearing of the flagellum during sample preparation (many intact flagella were observed detached from cells). Hundreds of individual bacteria from multiple sample preparations were observed, and the trends described were highly consistent. The length of intact flagella from ten individual bacteria were measured from each sample. The C. jejuni wild-type strain had a filament length of 3.2 ± 0.2 μm, while the C. jejuni flaAB mutant did not have an observable flagellum. Transformation of the C. jejuni flaAB mutant with flaA or flaB driven by the flaA promoter resulted in longer flagellar filaments (3.6 ± 0.6 and 1.8 ± 0.2 μm, respectively) versus when flaA or flaB were driven by the flaB promoter (0.1 ± 0.03 and 0.2 ± 0.08 μm, respectively). The motility and filament length phenotypes of the flaAB mutant containing pRY111 expressing flaA or flaB from their native promoters are in agreement with the results obtained by others for flaB and flaA mutants, respectively (Guerry et al., 1991, Wassenaar et al., 1991). Taken together, the motility assays and TEM data indicate that expression of the filament protein genes from the flaA promoter is required for maximal motility and filament length. However, it was clear that the promoter was not the only factor determining filament length and motility, as flaB expressed from the flaA promoter did not result in a full-length flagellar filament.
Figure 1. Motility assays and TEM of promoter experiments.
Motility assays and TEM of C. jejuni F38011 wild-type and a flagellar flaAB mutant transformed with a vector harboring either the flaA and flaB genes. In this experiment, the C. jejuni NCTC 11168 flaA and flaB genes were expressed from both the flaA and flaB promoters (PflaA or PflaB). C. jejuni strains were cultured on MH 0.4% agar plates, and the distance from the edge of the culture spot to the haze of motility was determined (Panel A). Bacteria were stained with 1% phosphotungstic acid, TEM performed, and the length of the flagellum was measured (Panel B). The images are of the: 1) C. jejuni F38011 wild-type strain; 2) C. jejuni F38011 flaAB mutant; 3) C. jejuni F38011 flaAB mutant harboring PflaA-flaA; 4) C. jejuni F38011 flaAB mutant harboring PflaA-flaB; 5) C. jejuni F38011 flaAB mutant harboring PflaB-flaA; and 6) C. jejuni F38011 flaAB mutant harboring PflaB-flaB. The values shown represent the mean ± standard deviation of 6 motility assays and 10 TEM images. One representative image is shown for each strain. Bar = 1 μm.
The amino-terminal D0 domain of FlaA results in increased motility and filament length compared to the D0 domain of FlaB
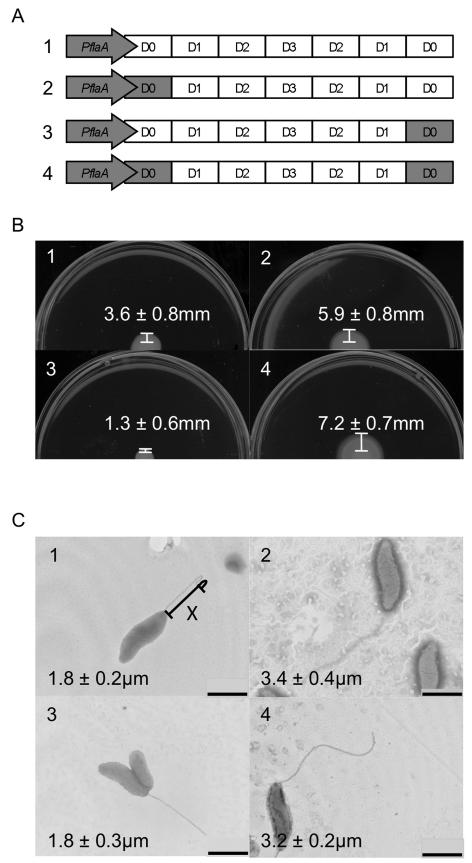
Previous studies have revealed that the amino-terminus of a protein exported via a T3SS harbors the export sequence (Sory et al., 1995, Ramamurthi & Schneewind, 2002, Vegh et al., 2006). To determine whether the D0 domains of FlaA and FlaB affect bacterial motility and filament length, three domain swap constructs were generated in which the amino- and carboxy-termini of FlaB were substituted with those of FlaA (Figure 2A). The amino-termini and carboxy-termini were swapped both individually and together to generate three modified versions of flaB. The flaB domain-swap genes were expressed from the flaA promoter, to allow for maximum filament length. The domain swap constructs were transformed into a C. jejuni flaAB mutant and assayed for motility and filament length.
Figure 2. Motility assays and TEM of domain swaps.
Panel A shows a linear schematic of the domain organization of the C. jejuni flaB gene and three constructs containing the domain swaps (not drawn to scale). Regions in gray correspond to C. jejuni flaA gene, while regions in white correspond to the C. jejuni flaB gene. Motility assays and TEM of flagellar mutants transformed with a vector harboring flaB, which had been modified to include the D0 domain of FlaA. C. jejuni strains were cultured on MH 0.4% agar plates, and the distance from the edge of the culture spot to the haze of motility was determined (Panel B). Bacteria were stained with 1% phosphotungstic acid, TEM performed, and the length of the flagellum was measured (Panel C). The images are of the: 1) C. jejuni F38011 flaAB mutant harboring PflaA-flaB; 2) C. jejuni F38011 flaAB mutant harboring PflaA-flaAD0BD123BD0; 3) C. jejuni F38011 flaAB mutant harboring PflaA-flaBD0BD123AD0; and 4) C. jejuni F38011 flaAB mutant harboring PflaA-flaAD0BD123AD0. The values shown represent the mean ± standard deviation of 6 motility assays or 10 TEM images. One representative image is shown for each strain. X = Example of line drawn to measure flagellar length. Bar = 1 μm.
When the C. jejuni flaAB mutant was transformed with a shuttle vector containing either the amino-terminal D0 domain swap construct (PflaA-flaAD0BD123BD0) or the amino and carboxy-terminal D0 domain swap construct (PflaA-flaAD0BD123AD0), the width of the motility zones were 5.9 ± 0.8 mm and 7.2 ± 0.7 mm, respectively (Figure 2B). Replacement of the carboxy-terminal D0 domain with that of flaA (PflaA-flaBD0BD123AD0) resulted in a zone of motility 1.3 ± 0.6 mm in width, while the C. jejuni flaAB mutant transformed with PflaA-flaB had a motility zone width of 3.6 ± 0.8 mm. The filament length of the C. jejuni flaAB mutant was also increased when the isolate was transformed with a vector containing either the PflaA-flaAD0BD123BD0 (3.4 ± 0.4 μm) or PflaA-flaAD0BD123AD0 (3.2 ± 0.2 μm) constructs (Figure 2C). Both the PflaA-flaB and PflaA-flaBD0BD123AD0 constructs resulted in intermediate length filaments of 1.8 ± 0.2 μm and 1.8 ± 0.3 μm, respectively. In summary, replacement of the amino-terminal D0 domain of flaB with that of flaA results in a more motile bacterium with a longer filament versus a C. jejuni flaAB mutant expressing a wild-type copy of the flaB gene.
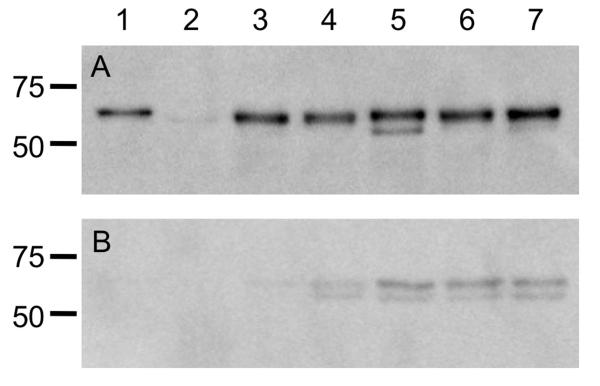
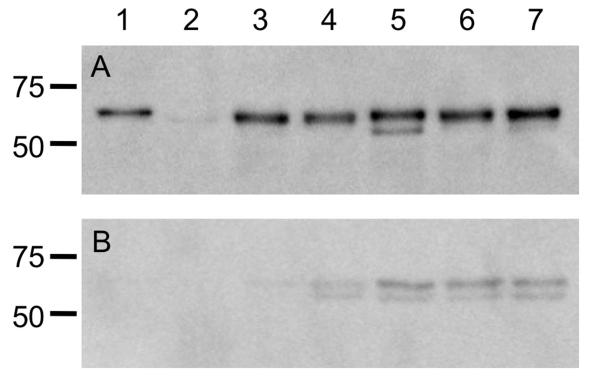
To ensure that the level of protein synthesis was dependent only on the promoter and was not affected by the sequence of flaA or flaB, immunoblot analysis was performed using a polyclonal antibody that recognizes both FlaA and FlaB. Whole cell lysates of each C. jejuni flaAB mutant transformed with a vector harboring flaA, flaB, or one of the three domain-swap constructs, which were all expressed from the flaA promoter, contained similar amounts of flagellin protein (Figure 3A). Immunoblots performed using serial two-fold dilutions of C. jejuni whole-cell lysates confirmed that the difference in protein concentrations between the samples was less than two-fold (not shown). A reactive band was sometimes observed below the flagellin band; we believe this is a flagellin degradation product, as it was observed in multiple lanes in different immunoblots. The immunoblot analysis data also confirmed that replacement of the flaB amino- and carboxy-terminal D0 domains did not significantly affect protein synthesis. Culture supernatants were also probed for the presence of flagellin (Figure 3B). The flaAB mutant strains harboring the three domain swap constructs had more flagellin in the supernatant fraction than the flaAB mutant strains harboring flaA or flaB. However, the quantity of flagellin in the supernatants was equal among the three domain swap construct strains, despite the reduced filament length and motility phenotype seen in the strain harboring the PflaA-flaBD0BD123AD0 construct. Collectively, these results demonstrate that flagellin synthesis is similar in each of the strains examined, and that the majority of flagellin produced is found in the whole-cell lysate and not the supernatant fractions.
Figure 3. Immunoblots of mutants and transformed strains.
Analysis of FlaA and FlaB protein synthesis in the C. jejuni F38011 wild-type strain, F38011 flaAB mutant, and transformed mutants. Whole cell lysates were prepared from broth cultures of C. jejuni in mid-exponential growth phase, and separated in 12% SDS-polyacrylamide gels. Panels: A) Whole cell lysates; and B) Supernatants were electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane and the immunoblots were probed with a polyclonal serum that recognizes both FlaA and FlaB. The samples in each lane are the: 1) C. jejuni F38011 wild-type strain; 2) C. jejuni F38011 flaAB mutant; 3) C. jejuni F38011 flaAB mutant harboring PflaA-flaA; 4) C. jejuni F38011 flaAB mutant harboring PflaA-flaB; 5) C. jejuni F38011 flaAB mutant harboring PflaA-flaAD0BD123BD0; 6) C. jejuni F38011 flaAB mutant harboring harboring PflaA-flaBD0BD123AD0; and 7) C. jejuni F38011 flaAB mutant harboring PflaA-flaAD0BD123AD0. The proteins loaded in the immunoblots (Panels A and B) are normalized according to culture density, so the levels of flagellin in the whole cell lysates and supernatants can be directly compared.
The D0 domain of FlaA and FlaB contain protein-specific conserved residues
Based on the results obtained from the promoter and domain swapping experiments, we hypothesized that differences in the amino acid sequence between the two proteins resulted in different filament lengths. To investigate this possibility, alignments were performed using the FlaA and FlaB sequences from the eight C. jejuni strains. While the central domains of the filament proteins are variable, the terminal D0 and D1 domains are more conserved (Table 2A and 2B). Despite the high degree of conservation in the D0 domain, certain residues within this domain were unique to FlaA or FlaB, but they were identical among the eight strains (Figure 4). The seven amino-terminal protein specific residues include 9, 10, 15, 19, 20, 24, and 27, which are V, A, K, D, L, S, and A in FlaA, and I, G, H, V, V, E, and K in FlaB, respectively. We also noted that the five carboxy-terminal protein specific residues include 543, 546, 558, 562, and 569, which are Y, A, A, S, and R in FlaA, and F, Y, S, A, and K in FlaB, respectively. These twelve residues within the amino- and carboxy-terminal D0 domain are specific to FlaA or FlaB. Regardless, the domain swap experiments indicated that the difference in length of the flagellum is due to specific residues in the amino-terminus of the FlaA protein.
Figure 4. Alignment of FlaA and FlaB D0 domains.
Amino acid sequence alignment of the D0 domain of FlaA and FlaB from 8 different strains of C. jejuni performed using MegAlign and the ClustalW algorithm. The amino- (Panel A) (residues 1-45) and carboxy-terminal (Panel B) (residues 533-572) D0 domain sequences of FlaA (above black line) and FlaB (below black line) are shown. Residues shaded in gray/black are conserved, while residues in white are divergent.
The amino-terminal D0 domain of FlaA facilitates enhanced secretion of a YplA fusion protein from Yersinia enterocolitica compared to the D0 domain of FlaB
We chose to utilize a Yersinia enterocolitica phospholipase (YplA) reporter secretion assay to directly examine the efficiency of T3 secretion conferred by the amino-termini of FlaA and FlaB, as this system was not affected by flagellin polymerization, flagellar regulation/assembly, or the sequence upstream of the flagellin gene (i.e., promoter and 5′ untranslated region). This assay is based on the principle that the amino-terminal sequence of a protein is required for its recognition and export by the flagellar apparatus (Christensen et al., 2009). Y. enterocolitica also recognizes and exports C. jejuni proteins that contain T3S amino-terminal sequences (Christensen et al., 2009). Secretion of the phospholipase YplA into TYE media containing Tween 80 results in a zone of fatty-acid precipitation surrounding the bacterial colony that can be easily quantified.
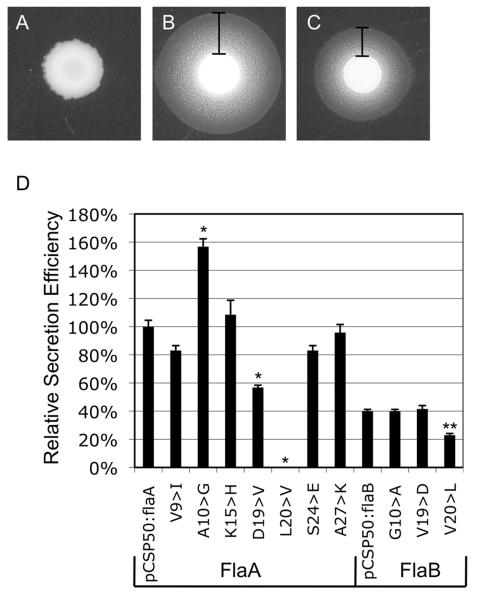
A Y. enterocolitica yplA mutant transformed with pCSP50 harboring a 5′ truncated yplA gene did not secrete YplA, as the T3S amino-terminal sequence was removed (Figure 5A). When the end of the flaA or flaB genes (i.e., which encode the first 36 amino acids) were fused to the 5′ truncated yplA gene, secretion zone widths of 2.6 ± 0.12 mm and 1.7 ± 0.06 mm were observed end of cysM, which encodes a cytoplasmic at 24 h, respectively (Figure 5B and C). The 5′ protein from C. jejuni, served as a negative control and did not result in secretion of YplA (not shown). Immunoblot analysis of the Y. enterocolitica strains with an α-YplA antibody confirmed that the amino-terminal sequences did not affect YplA synthesis (not shown). Different mixtures (ratios) of the Y. enterocolitica YplAB mutant transformed with the flaA-yplA fusion gene and the Y. enterocolitica YplAB mutant transformed with empty pCSP50 vector were assayed for YplA secretion to generate a standard curve of relative percent secretion efficiency (i.e., 100% secretion efficiency was defined by a culture containing only the Y. enterocolitica YplAB mutant transformed with FlaA-YplA fusion gene vector). Based on the standard curve, the FlaB-YplA fusion protein was secreted only 40% as efficiently as the FlaA-YplA protein. The increased zone of precipitation resulting from the FlaA-YplA fusion relative to the FlaB-YplA fusion demonstrated that the amino-terminus of FlaA is secreted more efficiently. These results are consistent with the observed increase in filament length of the C. jejuni flaAB mutant transformed with the PflaA-flaAD0BD123BD0 or PflaA-flaAD0BD123AD0 vectors relative to C. jejuni flaAB mutant harboring the PflaA-flaB vector.
Figure 5. YplA secretion assays.
Secretion of flagellin-YplA fusion proteins from Y. enterocolitica. The 5′ end of the flaA or flaB genes, gene. Panels: A) which encode the first 36 residues, were fused to the truncated yplA Secretion assay performed using Y. enterocolitica yplA mutant harboring pCSP50 empty vector; B) Secretion assay performed using Y. enterocolitica yplA mutant harboring flaA-yplA; C) Secretion assay performed using Y. enterocolitica yplA mutant harboring flaB-yplA; and D) Relative secretion efficiency of Y. enterocolitica yplA mutant harboring modified flaA-yplA and flaB-yplA fusion gene constructs. The values shown represent the mean zone of secretion from six individual assays, and the error bars represent ± one standard deviation. The width of the zone of secretion was determined by measuring from the edge of colony growth to the outer edge of precipitation. A standard curve was created by assaying varying combinations of pCSP50:flaA and pCSP50 containing Y. enterocolitica.
Mutation of specific residues in the amino-terminus of FlaA and FlaB alters secretion efficiency
To determine the effect of individual amino-terminal protein-specific conserved residues on secretion efficiency, seven of the residues within the FlaA amino-terminal sequence were mutated to the respective residues found within FlaB (Figure 5D). The mutated residues were V9I, A10G, K15H, D19V, L20V, S24E, and A27K. While the mutations V9I, K15H, S24E, and A27K did not significantly affect secretion (P-value > 0.01), mutation D19V reduced secretion by 43%. Surprisingly, the mutation of L20V completely abolished secretion of the fusion protein, and the A10G mutation increased secretion by 57%. To investigate the role of the three residues at positions 10, 19, and 20 in determining secretion efficiency of FlaB, these residues were mutated in the FlaB-YplA fusion protein to the corresponding amino acids in FlaA. Interestingly, the G10A and V19D mutations did not significantly affect the secretion efficiency relative to the FlaB-YplA fusion protein, while the V20L mutation reduced secretion efficiency nearly 50%. To determine whether the alteration of codon 20 disrupts secretion by altering the RNA sequence, FlaA-YplA fusion constructs were created using the 6 different codons for leucine. While the alternative codons alter the predicted RNA structure (RNAfold, http://rna.tbi.univie.ac.at/), secretion of the FlaA-YplA fusion proteins was not abolished, and all were secreted at levels greater than 80% of the native flaA sequence (not shown). Collectively, these data indicate that the T3S amino-terminal sequences of FlaA and FlaB contain protein-specific residues that alter the efficiency of protein secretion.
Discussion
It has been previously reported that mutations in flaA or fliA (σ28) result in severely truncated flagellar filaments, indicating that flaA is the predominant constituent of the flagellar filament (Guerry et al., 1991, Wassenaar et al., 1991, Jagannathan et al., 2001). In this study, we sought to determine whether the phenotypic differences between the C. jejuni flaA and flaB mutants were due to the regulation of flaA and flaB (i.e., the filament of a C. jejuni flaA mutant is truncated due to low level flaB expression from the σ54 promoter). Due to the fact that FlaA and FlaB are greater than 90% identical within a strain, it appeared unlikely that they differed other than in regulation of expression. Our initial experiments revealed that expression of flaB from the flaA promoter did not form a full-length filament. The fact that FlaA and FlaB are nearly identical, yet appeared to be secreted at different levels, allowed us the unique opportunity to compare the proteins and investigate their differences in secretion. Additionally, the flagellum is the only T3SS in C. jejuni, and to date there have been no other studies comparing two T3S proteins that are so similar in sequence. Therefore, we performed domain swap experiments to locate the region of the filament proteins that alter protein secretion. Replacement of the amino-terminus of FlaB with that of FlaA resulted in increased filament length and motility, while replacement of the carboxy-terminus did not appear to increase filament length or motility. Secretion assays performed with FlaA-YplA and FlaB-YplA fusion proteins in Y. enterocolitica confirmed the results seen in C. jejuni, and also allowed us to identify residues that affect secretion efficiency. In summary, these data indicate that the protein-specific residues within the amino-termini of FlaA and FlaB affect the level of protein secretion through the flagellum. As previously mentioned, the flagellum is a T3SS used to secrete flagellar structural components as well as effector proteins (Konkel et al., 1999, Konkel et al., 2004, Minamino & Namba, 2004, Desvaux et al., 2006). The mechanisms of T3S protein recognition and export are conserved between classical “injectisome” T3SS and flagellar T3SS, as it has been demonstrated that several proteins can be secreted through both systems. Salmonella typhimurium uses the Salmonella Pathogenicity Island 1 (SPI-1) T3SS to secrete flagellin into the host cell cytosol (Sun et al., 2007), and enteropathogenic Escherichia coli utilizes the locus of enterocyte effacement (LEE) encoded T3SS to secrete flagellin in the absence of a functional flagellar T3SS (Badea et al., 2009). S. typhimurium normally secretes SptP and SopE from the SPI-1 T3SS, however, when the chaperone binding domains of these proteins are deleted, they are instead secreted through the flagellar T3SS (Lee & Galan, 2004). Thus, it is expected that the mechanism by which the amino-terminus of FlaA and FlaB modulate secretion efficiency through the flagellar T3SS may apply to non-flagellar T3SS.
C. jejuni motility is dependent on the length of the flagellar filament, and motility assays can be used to indirectly observe changes in the filament length of a large population of bacteria. Wild-type C. jejuni F38011 are highly motile and possess full-length flagellar filaments. C. jejuni F38011 flaA mutants have extremely truncated flagella, and are only weakly motile (not shown). When the C. jejuni F38011 flaA mutant is complemented with flaA under control of the native promoter, filament length and motility are restored to that of wild-type (not shown). The C. jejuni flaAB mutant used in this study did not produce a flagellar filament, and was non-motile. Transformation of a C. jejuni flaAB mutant with a vector harboring flaB under control of the native promoter resulted in a phenotype equivalent to the C. jejuni flaA mutant. When the C. jejuni flaAB mutant was transformed with a vector containing the flaA gene under control of the native promoter, a filament was produced that was similar in length to wild-type isolate, but the motility was still reduced compared to wild-type. These contrasting results imply that both the FlaA and FlaB filament proteins are necessary for full motility. One possible reason for this observation is that the heterogeneity of flagellin monomers results in a flagellar structure that generates more thrust.
To examine the effects of flagellin gene expression on the function and length of the flagellar filament, TEM and motility assays were performed with promoter swap constructs. Expression of flaB from the flaA promoter resulted in an intermediate length flagellar filament, which was approximately half the length of a filament comprised solely of FlaA. Based on this finding, it was evident that regulation of gene expression was not the only factor affecting the length and motility of flagellar filaments. Other factors that we explored to explain the differences in flagellar filament length and motility included levels of protein synthesis and protein export efficiency.
In silico analyses were performed to determine whether FlaA and FlaB contain sequence variations in specific functional/structural domains. The domains of the C. jejuni FlaA and FlaB proteins were assigned by alignment with the S. typhimurium FliC protein. The purpose of determining the location of these domains in FlaA and FlaB was to be able to compare conservation or divergence within a domain to the function of that domain. The amino- and carboxy-terminal D0 and D1 domains of FlaA and FlaB were found to be highly conserved among C. jejuni strains, and shared the greatest similarity/identity with the S. typhimurium FliC protein. Despite the similarity of the three flagellin proteins in the D0-D1 region, one noteworthy difference is that S. typhimurium FliC is a potent activator of Toll-like receptor 5 (TLR5), while C. jejuni FlaA and FlaB have evolved such that they do not activate TLR5 signaling (Andersen-Nissen et al., 2005, Galkin et al., 2008).
We hypothesized that the D0 domain played a role in determining secretion efficiency because it is known to affect flagellin secretion in S. typhimurium (Vegh et al., 2006). Moreover, the amino-terminus of a protein secreted from a T3SS is known to harbor a sequence for export (Sory et al., 1995, Lloyd et al., 2001, Ramamurthi & Schneewind, 2002). We conducted the domain swap experiments with both the amino- and carboxy-termini of the filament proteins because they combine to form a coiled-coil structure (Yonekura et al., 2003). We chose to modify the flaB gene, when expressed from the flaA promoter, because the phenotype produced by this strain allowed us to observe an increase or decrease in filament length and motility versus the C. jejuni wild-type strain and flaAB mutant. The amino- and carboxy-terminal portions of the D0 domain of FlaB were replaced individually and in combination with the termini of FlaA to generate three new flagellin constructs: PflaA-flaAD0BD123BD0, PflaA-flaBD0BD123AD0, or PflaA-flaAD0BD123AD0. Motility assays and TEM of the C. jejuni flaAB mutant transformed with a vector containing the PflaA-flaAD0BD123BD0, PflaA-flaBD0BD123AD0, or PflaA-flaAD0BD123AD0 constructs revealed that only the amino-terminus is required for increased motility and filament length relative to PflaA-flaB. This is consistent with previous research, which indicates that the sequence required for flagellar secretion is located in the amino-terminus (Sory et al., 1995, Lloyd et al., 2001, Ramamurthi & Schneewind, 2002, Vegh et al., 2006). Immunoblot analysis confirmed that the protein levels among the strains were similar, and that the sequence of the flagellin gene does not affect protein synthesis. Interestingly, there was more flagellin detected in the supernatant fractions of the flaAB mutant strains containing the domain swap constructs, possibly due to issues with polymerization or flagellum stability in the recombinant proteins. However, similar levels of flagellin were seen in the supernatant fractions for strains expressing the three domain swap-constructs; thus flagellar breakage or deficient polymerization of flagellin monomers cannot account for the shortened flagella produced by strains expressing the PflaA-flaBD0BD123AD0 construct. The slight increase in motility observed for the PflaA-flaAD0BD123AD0 construct relative to the PflaA-flaAD0BD123BD0 construct could be due to the mismatching of the FlaA amino-terminus with the FlaB carboxy-terminus in the PflaA-flaAD0BD123BD0 construct.
Knowing that the amino-terminus of the filament protein dictates protein secretion, we sought to more closely examine this region of the FlaA and FlaB proteins. While others have used a flgK mutant to examine secretion of the FliC flagellin from Salmonella typhimurium, we chose not to use this approach, as regulation of flagellar gene expression (including fliC) is altered in a flgK mutant (Komoriya et al., 1999, Brown et al., 2008). Thus, the secretion efficiency of the amino-terminal sequences of FlaA and FlaB were determined using the Y. enterocolitica YplA secretion assay. The advantages of using the YplA secretion assay are: 1) All flagellin genes are expressed from the cat promoter for equivalent expression and protein synthesis; 2) The 5′ untranslated regions of flaA and flaB have been removed, so they cannot confound the results; 3) The FlaA-YplA and FlaB-YplA fusion proteins cannot polymerize into the flagellum; and 4) The secretion of FlaA-YplA and FlaB-YplA are not affected by regulation of flagellar assembly. Equivalent levels of protein synthesis amongst each of the strains was confirmed by immunoblot analysis performed using an anti-YplA antibody (not shown).
The amino-termini of both FlaA and FlaB facilitated export of the YplA-fusion protein through the flagellum of Y. enterocolitica, although the secretion efficiency of the FlaB-YplA fusion was reduced by 60% compared to FlaA-YplA. Interestingly, previous work revealed that the FlaA-YplA fusion protein is secreted at levels greater than wild-type YplA (Christensen et al., 2009). Collectively, these findings support the hypothesis that the amino-termini of FlaA and FlaB affect secretion efficiency and not gene expression/protein synthesis or filament polymerization. Additionally, the mechanism governing flagellar secretion efficiency is conserved between C. jejuni and Y. enterocolitica, as the amino-terminus of FlaA results in enhanced secretion relative to FlaB in both species.
We noted that the amino-termini of FlaA and FlaB contained residue differences specific to FlaA or FlaB. Moreover, certain residues, such as V9 in FlaA and I9 in FlaB have similar biochemical properties, while other residues, such as 19D in FlaA and 19V in FlaB have different biochemical properties. To determine if these residues affected secretion efficiency, seven protein-specific conserved residues located within the first 36 residues of the FlaA-YplA fusion protein were mutated individually to the corresponding amino acid in FlaB. The terminal domains of flagellin are known to be disordered in solution, and form coiled-coil tertiary structures composed of alpha helices when polymerized into the filament (Vonderviszt et al., 1989, Yonekura et al., 2003). Amino-terminal coiled-coil structures are common among T3S proteins, and have been suggested to play a role in secretion (Gazi et al., 2009). Interestingly, mutations in the amino-terminus of FlaA that incorporate residues favoring beta sheets (D19V, L20V) reduce secretion efficiency. The V19D mutation in the amino-terminus of FlaB had no effect on secretion, while V20L reduced secretion efficiency. Substitution of a helix-favoring alanine with glycine, which disrupts secondary structure, greatly enhanced secretion efficiency of the FlaA-YplA fusion protein. However, in the amino-terminus of FlaB, the reciprocal mutation did not affect secretion. Collectively, these results suggest that residues favoring alpha helix formation do not enhance secretion. Silent mutations in codon 20 of the FlaA-YplA fusion did not significantly reduce secretion, despite altering the RNA sequence and the predicted structure. These results support the hypothesis that secretion of FlaA from the flagellar T3SS is dependent on the peptide sequence and not the RNA sequence, but do not rule out the possibility that RNA structure may influence secretion. While additional studies are required to determine how sequence parameters affect secretion efficiency, the secretion data suggest that multiple residues work in concert to facilitate protein secretion.
The novelty of this study lies in the fact that the secretion efficiency of T3S proteins is modulated by the amino-terminal sequence, demonstrating an additional level of regulation of protein secretion. This finding was demonstrated both in C. jejuni as well as Y. enterocolitica. Thus, the difference in the amino-termini of secreted proteins provides one possible explanation for how a pathogen exports various toxins and other proteins at different levels, even when the proteins are synthesized at similar levels. Further studies will need to be performed to determine how the efficiency of protein export can influence the assembly of the T3SS export apparatus or alter the virulence of a pathogen. In the case of C. jejuni, the differences in secretion of the two filament proteins likely influence the composition of the filament. Additional work is in progress, using C. jejuni as a model, to dissect the amino-terminal secretion signal and the mechanism of T3S protein recognition to gain a better understanding of protein export, the T3SS, and bacterial pathogenesis.
Experimental procedures
In silico analyses
All genomic sequences, except for C. jejuni F38011, were obtained from microbesonline.org, compiled by the Virtual Institute for Microbial Stress and Survival. The sequences for C. jejuni F38011 FlaA and FlaB have been deposited in GenBank, accession numbers (pending) and (pending), respectively. All sequence analyses were performed using the DNASTAR Lasergene software suite, version 7.1.0. Amino acid alignments were performed with the Megalign program, using the CLUSTAL W algorithm. The gap penalty was set to 10.0, the gap length penalty was 0.2, and a Gonnet series was used for the protein weight matrix.
Bacterial strains and culture methods
C. jejuni strains F38011 (Raphael et al., 2005) and NCTC 11168 (ATCC type isolate) were used in this study. Bacteria were cultured either on Mueller-Hinton agar plates supplemented with 5% citrated bovine blood (MHB), or in Mueller-Hinton (MH) broth with constant shaking. Cultures were incubated in a microaerobic environment (85% N2, 10% CO2, 5% O2), at 37°C. The C. jejuni cultures were passaged onto fresh media every 48 h. The E. coli Inv-α (Invitrogen) and S17-1 λ-pir (Tascon et al., 1993) strains were used in this study. Both strains were cultured on Luria-Bertani agar plates (LB) or in LB broth with constant shaking. The cultures were incubated at 37°C in an aerobic environment. All cloning steps were performed in E. coli Inv- α, and the complementation vectors were transformed into E. coli S-17 for conjugation into C. jejuni.
Generation of C. jejuni flagellar mutant
The experiments described were all performed in C. jejuni F38011, a human clinical isolate. The FlaA and FlaB genes of C. jejuni F38011 were PCR amplified using HiFi Taq DNA polymerase (Invitrogen) with primers derived from the genome sequence of C. jejuni NCTC 11168, and sequenced using dideoxynucleotide sequencing. The final sequences were assembled using the Seqman program from the Lasergene software suite. A flaAB mutant of C. jejuni F38011 was generated via double crossover recombination and subsequent insertion of a tetracycline resistance cassette. Approximately 1.5 kilobases of the upstream and downstream regions flanking flaA and flaB were PCR amplified using the primer pairs MEK 1011 / MEK 1001 and MEK 1851 / MEK 1844, respectively (Supplemental Table 2). The two fragments were cloned sequentially into pBSK-Kan2, using the sites SstI, SstII, BamHI and XhoI. The tetO tetracycline resistance gene was inserted between the two homologous flanking regions using the SstII and BamHI sites. pBSK-Kan2 is identical to the pBluescript vector (Stratagene), except that the original kanamycin resistance cassette was replaced with the apha-3 kanamycin resistance cassette, which functions in both E. coli and C. jejuni. The plasmid was introduced into the C. jejuni F38011 strain via electroporation. Transformants were selected for on MHB agar plates supplemented with 2 μg/ml tetracycline or 50 μg/ml kanamycin (MHBTet2 or MHBKan50). Tetracycline resistant colonies were streaked onto MHBTet2 and MHBKan50 plates to confirm that the transformants were kanamycin sensitive. Deletion of flaA and flaB was confirmed by PCR (not shown) and immunoblot analyses.
Generation of C. jejuni flagellar strains
The flaA and flaB genes of C. jejuni NCTC 11168 were used to transform the C. jejuni F38011 flaAB mutant. Both flaA and flaB were PCR amplified using HiFi Taq DNA polymerase (Invitrogen) from NCTC 11168 chromosomal DNA using the primer pairs MEK 1332 / MEK 2015 and MEK 1334 / MEK 2025, respectively. Each gene was cloned into the E. coli-C. jejuni shuttle vector pRY111 (Yao et al., 1993), using the SstI and EcoRI sites present in the MCS. The promoter regions for flaA and flaB were PCR-amplified using the primer pairs MEK 1611 / MEK 1612 and MEK 1613 / MEK 1614, respectively. Both promoters were inserted upstream of each filament protein gene to produce four different flagellin complementation constructs.
Domain swap constructs were generated by removing the amino- and carboxy-terminal portions of flaB and replacing them with the complementary portions of flaA. The amino-termini, spanning residues 1-114, and the carboxy-termini, spanning residues 495-572, were swapped both individually and together to generate three modified versions of flaB. Cloning was performed using a native EcoRV site at +342 basepairs (residue 114), and a recombinant XbaI site at +1,484 basepairs (residue 495). The XbaI site was generated by inverse PCR of the PflaA-flaB pRY111 construct using the primer pairs MEK 1858 / MEK 1859 and MEK 1860 / MEK 1861, followed by blunt-ended ligation of the PCR product. Although the portions of the gene that were exchanged extended beyond the D0 domain, the amino acid sequences external to the D0 domain are identical in FlaA and FlaB. The flagellin gene constructs were transformed into E. coli S-17 via electroporation. The inserts contained within each vector were sequenced using the dideoxynucleotide method for confirmation. The vectors were then introduced into the C. jejuni F38011 flaAB mutant by conjugation.
Phase variation motility assays
C. jejuni F38011 wild-type and the flaAB mutant harboring flaA expressed from the flaA promoter were grown for 48 h on MHB agar plates and resuspended in MH broth. The OD540 was determined, and the sample was diluted in MH soft agar (0.4% agar) to a density that would yield ~30 colony-forming units (CFU) when plated. After 48 h incubation, the motility of 500 individual CFU was observed for each strain.
Motility assays
Cultures of C. jejuni were grown to mid-log phase overnight in MH broth, supplemented with 8 μg/ml of chloramphenicol for strains containing pRY111 derivatives. The cultures were normalized to an OD540 = 1.0 by centrifugation and resuspension in MH broth, and 5 μl of each culture was applied to the center of a MH plate containing 0.4% agar, and incubated for 48 h. The zone of motility was determined by measuring from the edge of the inoculation spot to the end of the visible migration. The image analysis program ImageJ (www.nih.gov) was used to measure scanned images of each motility assay. Six replicates of each assay were performed.
Transmission electron microscopy
C. jejuni were harvested from 48 h MHB plates supplemented with the appropriate antibiotics and resuspended in distilled water to an OD540 = 0.1. Four μl of each suspension was applied to a formvar-coated nickel grid, and combined with 4 μl of 1% phosphotungstic acid. After 60 seconds, the liquid was removed from each grid via capillary action using Whatman filter paper. Transmission electron microscopy was performed using a Jeol 1200EX electron microscope, at a magnification of 25,000x. Images were taken of bacteria possessing full-length flagella, as determined by observing the flagellated population to approximate maximal flagellar length. Bacteria lacking visible flagella or with severely truncated flagella were not included in the measurements, as it was obvious that sample preparation results in flagellar shearing. We know flagellar shearing is occurring due to the fact that many intact flagella are observed detached from cells, and the phase variation motility assay demonstrates that 98% of the bacteria are motile. Only sample preparations in which the majority of bacteria were flagellated were used for measurements. The lengths of the flagella were determined using the segmented line tool of ImageJ (Abramoff, 2004).
Yersinia enterocolitica YplA fusion protein secretion assays
To determine whether FlaA and FlaB were exported at different efficiencies in a heterologous system, we utilized the YplA fusion protein secretion assay (Christensen et al., 2009). In this assay, the first 108 bp of the amino terminal regions of both flaA and flaB genes were PCR amplified from C. jejuni NCTC 11168 chromosomal DNA with primers containing 5′ NdeI and 3′ BglII restrictions sites for directional cloning into the E. coli - Y. enterocolitica shuttle vector pCSP50. The pCSP50 vector includes a tetracycline resistance gene (tetO), a constitutive promoter (cat), a 5′ truncated phospholipase gene (yplA) lacking 150 nucleotides encoding the native T3S signal, and yplB, which is located immediately downstream of yplA (Schmiel et al., 1998). The NdeI and BglII sites facilitate directional cloning of C. jejuni T3S amino-terminal sequences as fusions with the truncated yplA. The pCSP50 derived vectors were transformed into E. coli S17-1 λ-pir, and confirmed by PCR fragment size and sequence analysis. The vectors were conjugated into a Y. enterocolitica yplAB mutant, and confirmed by agarose gel electrophoresis of restriction digested plasmid preparations.
Secretion of the YplA fusion proteins was assayed by inoculation onto phospholipase assay (PLA) media. Briefly, Y. enterocolitica strains were grown overnight in LB broth with shaking at 26°C. Secretion via the flagellar T3SS was induced by spotting 1.5 μl of culture on TYE PLA medium (1% tryptone, 0.5% yeast extract, 1.5% agar, 1% Tween 80, and 1 mM CaCl2), and incubation at 26°C. Each isolate was tested for secretion a minimum of three times from two independent PLA plate assays to ensure reproducible phenotypes. Sixteen conjugates were inoculated on each PLA plate, in addition to the Y. enterocolitica yplAB mutant expressing wild-type YplA as a positive control. All plates were scanned at 300 dpi resolution (12, 24, and 48 h), and secretion zone widths were measured manually from digital images using select tools in Adobe Photoshop CS2 version 9.0.2 (Adobe Systems Incorporated, USA).
Immunoblots
Whole cell lysates of C. jejuni were probed for FlaA and FlaB using a polyclonal antibody that recognizes both of the filament proteins. Broth cultures of C. jejuni were grown to mid-exponential growth phase, and an equivalent amount of cells (OD540 = 2) were harvested by centrifugation. The cell pellet was resuspended in 200 μl 1X sample buffer and boiled for 5 min to prepare whole-cell lysates (WCL). Culture supernatants were collected and filtered through a 0.22μm filter to remove any cells. 10 μl of each WCL was separated by SDS-PAGE (12% polyacrylamide) at 100 volts for 1 h. The gel was stained in Coomassie Blue dye for 20 min, and de-stained overnight. A second 12% SDS-polyacrylamide gel was loaded with 10 μl of a 1:10 dilution of each WCL and electrophoresed at 100 V for 1 h. A third 12% SDS-polyacrylamide gel was loaded with volumes of the supernatant fractions normalized to the culture density, so they could be directly compared to the whole-cell lysate fractions. After electrophoresis, the proteins were transferred to a polyvinylidene fluoride PVDF membrane (Immobilon P; Millipore Corp., Bedford, MA) by electrophoresis. FlaA and FlaB were detected with a 1:1000 dilution of a rabbit α-FlaA/FlaB antibody K6932, and bound primary antibodies were detected with 1:1000 α-rabbit IgG conjugated to horseradish peroxidase. The immunoblots were developed by chemiluminescence (Western Lightning, PerkinElmer Life Sciences, Inc.) and images taken using a FujiFilm LAS-2000 multi-imager.
Mutation of residues in the FlaA-YplA and FlaB-YplA fusion proteins
Seven residues in the amino-terminus of the FlaA-YplA (9, 10, 15, 19, 20, 24 and 27) and three residues in the FlaB-YplA fusion proteins (10, 19, and 20) were mutated to the corresponding amino acid in the other filament protein. The mutations were introduced into the pCSP50 based FlaA-YplA and FlaB-YplA vectors by inverse PCR using 5′ phosphorylated primers to facilitate blunt-ended ligation of the inverse PCR product. The mutations in the FlaA-YplA (9, 10, 15, 19, 20, 24 and 27) and FlaB-YplA (10, 19, and 20) vectors were performed using the primer pairs MEK 1655/1656, 1657/1658, 1659/1660, 1661/1662, 1663/1664, 1665/1666, 1667/1668, 2255/2256, 2257/2258, and 2259/2260, respectively (Table 4). The nucleotide sequence of each codon was mutated to match the corresponding codon in the other filament protein gene. The constructs were sequence confirmed, transformed into E. coli S-17, and conjugated into the Y. enterocolitica yplAB mutant.
Silent mutation of leucine 20 in the FlaA-YplA fusion construct
Codon 20 of the FlaA-YplA fusion construct, which encodes leucine (TTA), was mutated to the five alternative codons (CTT, TTG, CTA, CTC, and CTG) using the inverse PCR technique described above. The primer pairs used for inverse PCR were MEK 1664/2391 (CTT), 1664/2392 (TTG), 1664/2393 (CTA), 1664/2394 (CTC), and 1664/2395 (CTG). The constructs were sequence confirmed, transformed into E. coli S-17, and conjugated into the Y. enterocolitica yplAB mutant. Secretion efficiency was determined as describe above.
Supplementary Material
Acknowledgements
We thank the staff of the Washington State University Franceschi Microscopy and Imaging Center, particularly Dr. Michael Knoblauch, Dr. Christine Davitt, and Valerie Lynch-Holm for assistance with transmission electron microscopy sample preparation and imaging, and Derek Pouchnik for assistance with real-time PCR and DNA sequencing. We also thank Dr. Daelynn R. Buelow, Dr. Tri Duong, and Charles L. Larson for critical review of the manuscript. This work was supported from funds awarded to MEK from the National Institute of Health, Department of Health and Human Services under contract number NO1-AI-30055.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with Image J. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- Badea L, Beatson SA, Kaparakis M, Ferrero RL, Hartland EL. Secretion of flagellin by the LEE-encoded type III secretion system of enteropathogenic Escherichia coli. BMC Microbiol. 2009;9:30. doi: 10.1186/1471-2180-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Saini S, Aldridge C, Herbert J, Rao CV, Aldridge PD. The rate of protein secretion dictates the temporal dynamics of flagellar gene expression. Mol Microbiol. 2008;70:924–937. doi: 10.1111/j.1365-2958.2008.06455.x. [DOI] [PubMed] [Google Scholar]
- Carrillo CD, Taboada E, Nash JH, Lanthier P, Kelly J, Lau, et al. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J Biol Chem. 2004;279:20327–20338. doi: 10.1074/jbc.M401134200. [DOI] [PubMed] [Google Scholar]
- Christensen JE, Pacheco SA, Konkel ME. Identification of a Campylobacter jejuni secreted protein required for maximal invasion of host cells. Mol Microbiol. 2009;73:650–662. doi: 10.1111/j.1365-2958.2009.06797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- Desvaux M, Hebraud M, Henderson IR, Pallen MJ. Type III secretion: what’s in a name? Trends Microbiol. 2006;14:157–160. doi: 10.1016/j.tim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Gazi AD, Charova SN, Panopoulos NJ, Kokkinidis M. Coiled-coils in type III secretion systems: structural flexibility, disorder and biological implications. Cell Microbiol. 2009 doi: 10.1111/j.1462-5822.2009.01297.x. [DOI] [PubMed] [Google Scholar]
- Grant CC, Konkel ME, Cieplak W, Jr., Tompkins LS. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun. 1993;61:1764–1771. doi: 10.1128/iai.61.5.1764-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P. Campylobacter flagella: not just for motility. Trends Microbiol. 2007;15:456–461. doi: 10.1016/j.tim.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Guerry P, Alm RA, Power ME, Logan SM, Trust TJ. Role of two flagellin genes in Campylobacter motility. J Bacteriol. 1991;173:4757–4764. doi: 10.1128/jb.173.15.4757-4764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugolya Z, Muskotal A, Sebestyen A, Dioszeghy Z, Vonderviszt F. Interaction of the disordered terminal regions of flagellin upon flagellar filament formation. FEBS Lett. 2003;535:66–70. doi: 10.1016/s0014-5793(02)03859-0. [DOI] [PubMed] [Google Scholar]
- Jagannathan A, Constantinidou C, Penn CW. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J Bacteriol. 2001;183:2937–2942. doi: 10.1128/JB.183.9.2937-2942.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoriya K, Shibano N, Higano T, Azuma N, Yamaguchi S, Aizawa SI. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol Microbiol. 1999;34:767–779. doi: 10.1046/j.1365-2958.1999.01639.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol Microbiol. 1999;32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, Mickelson J. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol. 2004;186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Monteville MR, Rivera-Amill V, Joens LA. The pathogenesis of Campylobacter jejuni-mediated enteritis. Curr Issues Intest Microbiol. 2001;2:55–71. [PubMed] [Google Scholar]
- Kuwajima G. Construction of a minimum-size functional flagellin of Escherichia coli. J Bacteriol. 1988;170:3305–3309. doi: 10.1128/jb.170.7.3305-3309.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CL, Christensen JE, Pacheco SA, Minnich SA, Konkel M. Campylobacter jejuni secretes proteins via the flagellar type III secretion system that contribute to host cell invasion and gastroenteritis. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. ASM Press; Washington, DC: 2008. pp. 315–332. [Google Scholar]
- Lee SH, Galan JE. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol Microbiol. 2004;51:483–495. doi: 10.1046/j.1365-2958.2003.03840.x. [DOI] [PubMed] [Google Scholar]
- Lloyd SA, Norman M, Rosqvist R, Wolf-Watz H. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol Microbiol. 2001;39:520–531. doi: 10.1046/j.1365-2958.2001.02271.x. [DOI] [PubMed] [Google Scholar]
- Minamino T, Namba K. Self-assembly and type III protein export of the bacterial flagellum. J Mol Microbiol Biotechnol. 2004;7:5–17. doi: 10.1159/000077865. [DOI] [PubMed] [Google Scholar]
- Muskotal A, Kiraly R, Sebestyen A, Gugolya Z, Vegh BM, Vonderviszt F. Interaction of FliS flagellar chaperone with flagellin. FEBS Lett. 2006;580:3916–3920. doi: 10.1016/j.febslet.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Namba K, Yamashita I, Vonderviszt F. Structure of the core and central channel of bacterial flagella. Nature. 1989;342:648–654. doi: 10.1038/342648a0. [DOI] [PubMed] [Google Scholar]
- Ozin AJ, Claret L, Auvray F, Hughes C. The FliS chaperone selectively binds the disordered flagellin C-terminal D0 domain central to polymerisation. FEMS Microbiol Lett. 2003;219:219–224. doi: 10.1016/S0378-1097(02)01208-9. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Schneewind O. Yersinia enterocolitica type III secretion: mutational analysis of the yopQ secretion signal. J Bacteriol. 2002;184:3321–3328. doi: 10.1128/JB.184.12.3321-3328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raphael BH, Pereira S, Flom GA, Zhang Q, Ketley JM, Konkel ME. The Campylobacter jejuni response regulator, CbrR, modulates sodium deoxycholate resistance and chicken colonization. J Bacteriol. 2005;187:3662–3670. doi: 10.1128/JB.187.11.3662-3670.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Palacios GM. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin Infect Dis. 2007;44:701–703. doi: 10.1086/509936. [DOI] [PubMed] [Google Scholar]
- Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature. 2001;410:331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- Schmiel DH, Wagar E, Karamanou L, Weeks D, Miller VL. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect Immun. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sory MP, Boland A, Lambermont I, Cornelis GR. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci U S A. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YH, Rolan HG, Tsolis RM. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem. 2007;282:33897–33901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- Tascon RI, Rodriguez-Ferri EF, Gutierrez-Martin CB, Rodriguez-Barbosa I, Berche P, Vazquez-Boland JA. Transposon mutagenesis in Actinobacillus pleuropneumoniae with a Tn10 derivative. J Bacteriol. 1993;175:5717–5722. doi: 10.1128/jb.175.17.5717-5722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegh BM, Gal P, Dobo J, Zavodszky P, Vonderviszt F. Localization of the flagellum-specific secretion signal in Salmonella flagellin. Biochem Biophys Res Commun. 2006;345:93–98. doi: 10.1016/j.bbrc.2006.04.055. [DOI] [PubMed] [Google Scholar]
- Vonderviszt F, Kanto S, Aizawa S, Namba K. Terminal regions of flagellin are disordered in solution. J Mol Biol. 1989;209:127–133. doi: 10.1016/0022-2836(89)90176-9. [DOI] [PubMed] [Google Scholar]
- Wassenaar TM, Bleumink-Pluym NM, van der Zeijst BA. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 1991;10:2055–2061. doi: 10.1002/j.1460-2075.1991.tb07736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Alm RA, Trust TJ, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Aizawa S, Yamaguchi S. Flagellar filament structure and cell motility of Salmonella typhimurium mutants lacking part of the outer domain of flagellin. J Bacteriol. 1995;177:1090–1093. doi: 10.1128/jb.177.4.1090-1093.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.