Abstract
Aim
To explore cancer cell-specific phage fusion pVIII coat protein, identified using phage display, for targeted delivery of drug-loaded liposomes to MCF-7 breast cancer cells.
Material & methods
An 8-mer landscape library f8/8 and a biopanning protocol against MCF-7 cells were used to select a landscape phage protein bearing MCF-7-specific peptide. Size and morphology of doxorubicin-loaded liposomes modified with the tumor-specific phage fusion coat protein (phage–Doxil) were determined by dynamic light scattering and freeze-fraction electron microscopy. Topology of the phage protein in liposomes was examined by western blot. Association of phage–Doxil with MCF-7 cells was evaluated by fluorescence microscopy and fluorescence spectrometry. Selective targeting to MCF-7 was shown by FACS using a coculture model with target and nontarget cells. Phage–Doxil-induced tumor cell killing and apoptosis were confirmed by CellTiter-Blue® Assay and caspase-3/CPP32 fluorometric assay.
Results
A chimeric phage fusion coat protein specific towards MCF-7 cells, identified from a phage landscape library, was directly incorporated into the liposomal bilayer of doxorubicin-loaded PEGylated liposomes (Doxil®) without additional conjugation with lipophilic moieties. Western blotting confirmed the presence of both targeting peptide and pVIII coat protein in the phage–Doxil, which maintained the liposomal morphology and retained a substantial part of the incorporated drug after phage protein incorporation. The binding activity of the phage fusion pVIII coat protein was retained after incorporation into liposomes, and phage–Doxil strongly and specifically targeted MCF-7 cells, demonstrating significantly increased cytotoxicity towards target cells in vitro.
Conclusion
We present a novel and straightforward method for making tumor-targeted nanomedicines by anchoring specific phage proteins (substitute antibodies) on their surface.
Keywords: breast cancer, Doxil®, drug delivery, landscape phage, liposome, major coat protein pVIII, phage display, tumor targeting
Despite long-term efforts, conventional chemotherapeutic preparations still demonstrate dose-limiting toxicity and poor performance on reaching tumor tissues. The use of targeted drug delivery and controlled release could provide an efficient solution to overcoming these limitations. Drug carriers, such as liposomes, used for the delivery of anticancer drugs in vivo can be specifically targeted to tumors by their coupling with various tumor cell-binding ligands [1]. Monoclonal antibodies (mAbs) are frequently used [2], although their broad use is restricted by their instability and high production cost. In addition, whole mAbs show nonspecific uptake by the phagocytic cells mediated by their Fc region and are too large to efficiently penetrate solid tumor. The use of smaller antibody fragments (e.g., Fab’, scFv) lacking the Fc region improves biodistribution and tumor penetration of targeted drug carriers [3]; however, conjugation of mAbs and antibody fragments with drug-loaded nanocarriers requires special efforts. Thus, the search for alternative ligands and methods of their conjugation with nanomedicines is ongoing.
The integration of phage-display technology with a nanocarrier-based drug delivery platform is emerging as a new approach [4]. Phage technique evolved as a result of advances in combinatorial chemistry, and phage display has allowed identification of tumor-specific peptides in a high throughput fashion [5–7]. The tumor-specific phage can be affinity selected from multibillion clone libraries [8,9] by their ability to interact very specifically with cancer cell surface receptors. Using the well-established biopanning protocol, a number of phage-borne peptides specific to a broad array of tumors have been identified [10].
These peptides have already been successfully used as targeting moieties to deliver pharmaceutical nanocarriers, such as liposomes, to tumors. RGD, a tripeptide sequence selected from phage-displayed libraries against endothelial and melanoma cells, has been incorporated into liposome after being coupled with a lipid anchor [11]. Doxorubicin-loaded RGD-targeted liposomes demonstrated targeting of integrin-expressing cells in vitro and improved therapeutic efficacy in vivo in a C26 colon carcinoma mouse model. Tumor vasculature-targeting peptide PC5–52, discovered using an in vivo phage display, was modified with a hydrophobic residue and used to target doxorubicin-loaded liposomes to tumors in mice with human lung and oral cancer xenografts, thus increasing their survival rate [12]. Still, the preparation of phage peptide-targeted liposomes (or other pharmaceutical nanocarriers) requires chemical modification of targeting peptides with the hydrophobic anchor, which complicates the preparation process and can alter the properties and specificity of peptides [13].
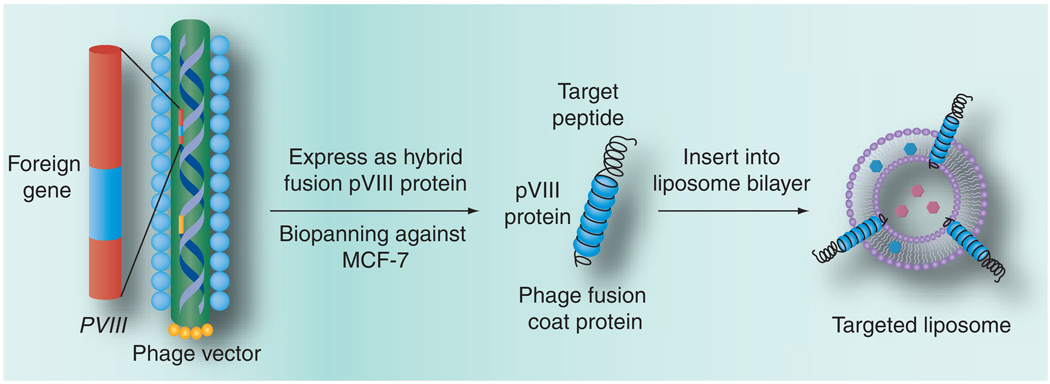
Recently, we proposed using landscape phage fusion coat proteins – easy to produce ‘substitute antibodies’ – as targeting ligands for drug-loaded pharmaceutical nanocarriers (e.g., liposomes [14]) to overcome the drawbacks associated with the chemical modification of nanocarriers with cancer-selective peptides. This approach is based on the ability of the phage major coat protein to spontaneously insert into bacterial membranes [15,16] and lipid bilayers of liposomes [17]. We assumed that the hybrid phage pVIII coat protein fused to the tumor-specific peptides would spontaneously incorporate into the liposome membrane via its C-terminal hydrophobic segment, while its water-exposed N-terminal binding segment would be exposed on the surface of drug-loaded liposomes to serve as a targeting moiety (Figure 1). Accordingly, the chemical conjugation procedure can be avoided.
Figure 1. Production of hybrid phage fusion coat protein with genetically fused target peptide and its incorporation into liposome.
In contrast to conjugation procedures used for coupling of tumor-specific peptides and mAbs, the new landscape phage-based approach relies on powerful and precise mechanisms of selection, biosynthesis and self-assembly. A culture of cells secreting filamentous phage is an efficient protein production system yielding up to 300 mg/l of pure phage, with the major coat protein constituting more than 90% of the total protein mass of the virion – a purity hardly reachable in normal synthetic and bioengineering procedures. Phage itself and its components are not toxic and have already been tested for safety in preclinical trials [18]. Furthermore, the technique of polyvalent phage display on the major coat protein pVIII [19] for construction of large (>109 clones) 8- and 9-mer landscape libraries has been developed. It relies on splicing of the corresponding degenerate coding sequence into the beginning of the pVIII coat-protein gene, replacing wild-type codons 2–4 or 2–5 [20–21]. Targeted phage probes against prostate, glial and breast tumor cells were successfully selected from these libraries using advanced biopanning protocols [8]. The new concept has been tested in a model experiment with a streptavidin-binding fusion phage protein incorporated into liposomes to impart them affinity to streptavidin-coated matrices [14].
In this article, we explore the application of the phage protein to prepare drug-loaded liposomes specifically interacting with cancer cells. Doxorubicin-loaded PEGylated liposomes (Doxil®, Ortho Biotech Products, L.P., NJ, USA) modified with phage proteins specific towards MCF-7 breast cancer cells demonstrated strong specific binding with target cells and increased cytotoxicity in vitro.
Materials & methods
Materials & reagents
Doxil was purchased from Ben Venue Laboratories Inc. (OH, USA). l-α-phosphatidylcholine (egg); 1,2-dipalmitoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (sodium salt; DPPG); 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt; DOTAP); 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)2000] (ammonium salt; PEG2000-PE); cholesterol (98%), and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt, Rh-PE) were from Avanti Polar Lipids Inc. (AL, USA). Sodium cholate, 2.5% CHAPS: 3-[(3-cholamidopropyl)dimethylammonio]-1-proanesulfonate, carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), phenylmethanesulfonyl fluoride (PMSF), proteinase K and anti-fd IgG were from Sigma (MO, USA); 16% nongradient Tris-tricine gel from Jule Inc. (CT, USA); Immobilin-P PVDF membrane from Millipore (MA, USA); NeutrAvidin™-horseradish peroxidase and BCA protein assay kits and chemiluminescent substrate solution from Pierce (IL, USA); biotinylated-SP-conjugated Affinitipure goat antirabbit IgG (1:30,000) from Jackson Immunoresearch (PA, USA); CellTiter-Blue Assay kit from Promega (WI, USA); and caspase-3/CPP32 Fluorometric Assay Kit from Bioversion (CA, USA). Fluor Mounting Medium was from Trevigen Inc. (MD, USA). MCF-7 human breast adenocarcinoma (HTB 22™) cells and control MCF-10A (CRL-10317™) cells – nontumorigenic human epithelial cells, HepG2 (HB-8065™) cells – human hepatocellular carcinoma cells and C166-GFP (CRL-2583™) – mouse yolk sac endothelial cells, NIH3T3 (CRL-1658™) – mouse fibroblasts, as well as WI-38 (CCL-75™) cells – normal human lung fibroblasts used for depletion of the phage libraries were obtained from the ATCC (VA, USA). All cells were grown as recommended by ATCC at 37°C, 5% CO2.
Phage purification & cell binding
An individual phage clone DMPGTVLP (in this study the phage is designated by the sequence of the fused foreign peptide) was propagated and purified as described [8]. Binding specificity and selectivity of the phage was determined in a phage capture assay [8] adapted for a 96-well culture plate format. Briefly, target cells (MCF-7), MCF-10A cells and HepG2 cells were cultivated in triplicate to confluence in separate wells of 96-well cell culture plates. Cell culture growth medium was incubated in separate wells in triplicates as control. The experiment commenced by aspirating cell culture growth media from wells containing the confluent cells and the control wells. Cells were washed and incubated with serum-free medium at room temperature (RT) for 1 h. Phage DMPGTVLP or control unrelated phage VPEGAFSS (streptavidin binder; 106 cfu) in 100 µl blocking buffer were added to the corresponding well and incubated for 1 h at RT or 37°C. Unbound phages were removed and cells were washed eight times with 100 µl of washing buffer for 5 min. Cells were treated with 25 µl of lysis buffer (2.5% CHAPS) for 10 min on a shaker. The concentration of the phage was measured (in triplicate) by titrating in E. coli (K91 BlueKan) host bacterial cells, and phage recovery was calculated as the ratio of input phage to output phage.
pVIII coat fusion protein
A landscape phage bearing breast cancer cell-specific peptide DMPGTVLP was selected from the 8-mer landscape library f8/8 [20,22] using biopanning against MCF-7 cells. Phage fusion 55-mer coat protein ADMPGTVLPDPAKAAFDSLQASATEYIGYAWAMVVVIVGATIGIKLFKKFTSKAS (MW 5747.72) was prepared by stripping the phage in cholate buffer [14,23]. Briefly, a mixture of 350 µl of phage in the Tris-buffered saline (~1 mg/ml) and 700 µl of 120 mM cholate in 10 mM Tris-HCl, 0.2 mM EDTA, pH 8.0 and 27 µl of chloroform, was incubated at 37°C overnight. The fusion protein was purified from the viral DNA and traces of bacterial proteins by size-exclusion chromatography on a Sepharose 6B-CL (Amersham, NJ, USA) column (1 × 45 cm) eluted with 10 mM cholate in 10 mM Tris-HCl, 0.2 mM EDTA, pH 8.0. The process was monitored by the Econo UV monitor (Bio-Rad, CA, USA); 2 ml fractions were collected and stored at 4°C. The protein was isolated as an aggregate with a molecular weight of approximately 46 kDa (8-mer) determined by chromatography on the column calibrated with standard molecular weight markers: aprotinin (6.5 kDa), cytochrome C (12.4 kDa), carbonic anhydrase (29 kDa) and BSA (66 kDa) as previously described [23]. Protein concentration was measured by a spectrophotometer, assuming that one absorbance unit AU280 corresponds to 0.7 mg protein/ml, as determined using the PROTEAN program (DNA STAR Inc., WI, USA).
Phage protein-modified Doxil (phage–Doxil)
Phage–Doxil was prepared by incubating Doxil with the cholate-stabilized phage pVIII coat fusion protein at the lipid-to-protein weight ratio of 200:1. After overnight incubation at 37°C, the crude formulation was dialyzed at 4°C overnight against the cholate-free phosphate-buffered saline (PBS) buffer to remove cholate. Size and size-distribution of the phage–Doxil were measured by dynamic light scattering with a Beckman Coulter N4 Plus Particle analyzer (Beckman Coulter, CA, USA). Entrapped doxorubicin was determined using the Labsystems Multiskan MCC/340 (Fisher, PA, USA) at 492 nm after the treatment of the sample with 1% Triton-100.
The presence of fusion pVIII coat protein in phage–Doxil was monitored by western blotting. Samples were mixed with an equal volume of Tricine sample buffer (8% sodium dodecyl sulfate, 24% glycerol, 0.1 M Tris, pH 6.8, 4% 2-mercaptoethanol, 0.01% Brilliant blue G, 2×) and heated at 95°C for 40 min. Denatured samples of 10 µl volume were loaded onto a 16% nongradient Tris-tricine gel. Electrophoresis was carried out for 90 min at 100 V. Proteins were transferred to an Immobilin-P PVDF membrane. The resulting blots were probed with anti-fd IgG (0.6 ng/ml) for the detection of the N-terminus or with affinity purified IgG against C-terminal pVIII peptide (3.6 ng/ml) for C-terminus detection (C-terminal pVIII peptide TIGIKLFKKFTSKAS was used for the generation of the affinity purified IgG). This was followed by incubation with biotinylated-SP-conjugated Affinitipure goat antirabbit IgG (1:30,000) and with NeutraAvidin-horseradish peroxidase and the blots were visualized using a chemiluminescent substrate solution.
To determine the protein topology in the lipid membrane, phage–Doxil was treated with the proteinase K at a final concentration of 50 µg/ml at RT for 1 h. The reaction was inhibited by the addition of PMSF (5 mM final). Diethyl ether (95% v/v final concentration) was added to the reaction mixture, vortexed vigorously and centrifuged for 5 min at 14,000 rpm in a microcentrifuge (Beckman Coulter). Following centrifugation, the solution separated into a lower aqueous and an upper organic phase. The sample was pipetted from the lower aqueous phase and analyzed by western blot. The proteolytic fragments were separated by SDS-PAGE and probed for the presence of the intact N-terminal and C-terminal (peptide TIGIKLFKKFTSKAS) coat sequences by binding of G-affinity purified corresponding antibodies, followed by western blotting.
Freeze-fracture electron microscopy
To investigate the structure of the Doxil liposomes after the incorporation of phage protein, the sample was quenched using the sandwich technique and liquid nitrogen-cooled propane. An estimated cooling rate of 10,000 Ks−1 was used to avoid ice crystal formation and possible artifacts of the cryofixation. The fracturing was carried out in JEOL JED-9000 freeze-etching equipment, and the exposed fracture planes were shadowed with platinum for 30 s at an angle of 25–35° and with carbon for 35 s (2 kV, 60–70 mA, 1 × 10−5 torr). The replicas were cleaned with fuming HNO3 for 24 h followed by repeated agitation with fresh chloroform/methanol (1:1 by volume) at least five times and examined with a JEOL 100 CX electron microscope.
Selective binding of phage–liposomes with targeted MCF-7 cells
MCF-7 cells were cocultured with nontarget C166 endothelial cells expressing GFP at 2:1 ratio. Nontarget NIH3T3 fibroblasts were cocultured with nontarget GFP-expressing C166 endothelial cells at 1:1 ratio. They were seeded in 25 cm2 flasks in DMEM with 10% serum. The cells were allowed to grow to 70–80% confluence before exposure to the liposome preparations. After coculture, cells were incubated with 50 µl of rhodamine-labeled phage–liposomes (EggPC:Chol:PEG-PE:Rh-PE [% molar ratio 68:30:1:1]) prepared as described previously for 1 h. Cells were detached from the flask by exposure to 2 ml 0.1% EDTA and collected by centrifugation. The cell pellets were washed three times with 1 ml of PBS and resuspended in 400 µl of PBS containing 4% paraformaldehyde. The resulting fixed cells were analyzed by flow cytometry. Four regions were created on dot plots: MCF-7 or NIH3T3 (R1), C166-GFP (R2), shifted MCF-7 or shifted NIH3T3 (R3) and shifted C166-GFP (R4) cells. Binding of the rhodamine-labeled phage–liposomes was detected as a right shift of the population on the x-axis (FL-2H, Red). The percent of the populations shifted was calculated as follows:
For MCF-7 or NIH3T3 cells: The percent of cells shifted = R3 / (R1 + R3) × 100
For C166-GFP cells: The percent of cells shifted = R4 / (R2 + R4) × 100
Cell viability
Cell viability was evaluated by the CellTiter-Blue Assay as described in the manufacturer’s manual. Target cells MCF-7 or nontarget cells NIH3T3 were seeded into a 96-well microplate at a density of 4 × 104 cells/well. After growth to 50–60% confluence, cells were treated with varying concentrations of Doxil, MCF-7-specific phage–Doxil (MCF-7-phage–Doxil), irrelevant streptavidin-targeting phage–Doxil (SA-phage–Doxil), and control drug-free liposomes modified by the MCF-7 or streptavidin phage fusion protein (drug-free MCF-7-phage–liposomes or drug-free SA-phage–liposomes) in serum-free MEM for 24 h. In addition, MCF-7 or NIH3T3 cells were exposed to Doxil, MCF-7-phage–Doxil and SA-phage–Doxil for 12 h, and further incubated for 48 h in drug-free medium before cytotoxicity determination.
Kinetics of MCF-7 cell killing was further detected at 4, 12, 24 and 48 h after treatment with 56 µM Doxil and MCF-7-phage–Doxil. Cells were then washed once with PBS, pH 7.4, and incubated with fresh complete MEM medium (100 µl/well), along with the CellTiter-Blue Assay reagent (20 µl/well), at 37°C for 1–4 h. The fluorescence intensity was measured using a multidetection microplate reader (Bio-Tek, VT, USA) with 525/590 nm excitation/emission wavelengths.
Cell viability was also assessed using the Trypan blue exclusion assay. After MCF-7 cells were grown to 70–80% confluence and treated with 8.6 µM of Doxil or MCF-7-phage–Doxil for 24 h, 100 µl of a 0.4% solution of Trypan blue in PBS were added to 100 µl of cell suspension. The aliquots were loaded onto a hemocytometer for the live- and dead-cell counts. Percent of cell survival was calculated by dividing the value obtained for the treated sample by the value obtained for the untreated sample.
Apoptosis assay
Caspase-3 activity was determined using the caspase-3/CPP32 fluorometric assay kit as described in the manufacturer’s manual. After MCF-7 cells were grown to 70–80% confluence and treated with 8.6 µM of Doxil or MCF-7-phage–Doxil for 24 h, cell lysates were prepared by resuspending 1–5 × 106 cells in 50 µl of the chilled lysis buffer and placing on ice for 10 min. The lysates were incubated with 50 µM of the synthetic substrate, DEVD-7-amino-4-trifluoromethyl coumarin (AFC) at 37°C for 1–2 h. The cleavage of the fluorogenic substrate by caspase-3 was determined in the multidetection microplate reader with 360/528 nm excitation/emission wavelengths. Caspase-3 activity of the sample was normalized for protein concentration, measured using a protein assay kit. The percent of increase-induced caspase-3 activity was calculated by dividing the value obtained for the treated cell sample by the value for the untreated cell sample.
Phage–Doxil association with MCF-7 cells
Uptake of MCF-7-phage–Doxil or Doxil by MCF-7 cells was visualized using a fluorescence microscopy. MCF-7 cells were seeded on sterile coverslips on a 6-well plate at a density of 2 × 105 cells/well, and were grown at 37°C for 24 h. Cells were incubated with 16.5 µM Doxil and MCF-7-phage–Doxil in serum-free MEM for 30 min, washed once and incubated in drug-free MEM for 2 h. Cells were then washed with PBS three times and incubated with 5 µg/ml Hoechst 33342 for 10 min. After washing with PBS, the coverslip was placed onto a glass slide over the fluorescence mounting medium. The images were acquired by a fluorescence microscope (Zeiss Co. Ltd., Germany) at ×100 magnification.
In the course of the cytotoxicity determination, cell association of Doxil or MCF-7-phage–Doxil was simultaneously monitored by a fluorescence spectrometer. After cells were treated with Doxil or MCF-7-phage–Doxil for the required time and washed with PBS, cell-associated doxorubicin was quantified using a multidetection microplate reader at 485/590 nm excitation/emission wavelengths.
Statistical analysis
The statistical significance of the results was analyzed using SPSS (version 16). Differences between experiment groups were compared using ANOVA followed by Bonferroni post hoc test. The results were considered statistically significant if the p-value was less than 0.05.
Results
Phage selectivity
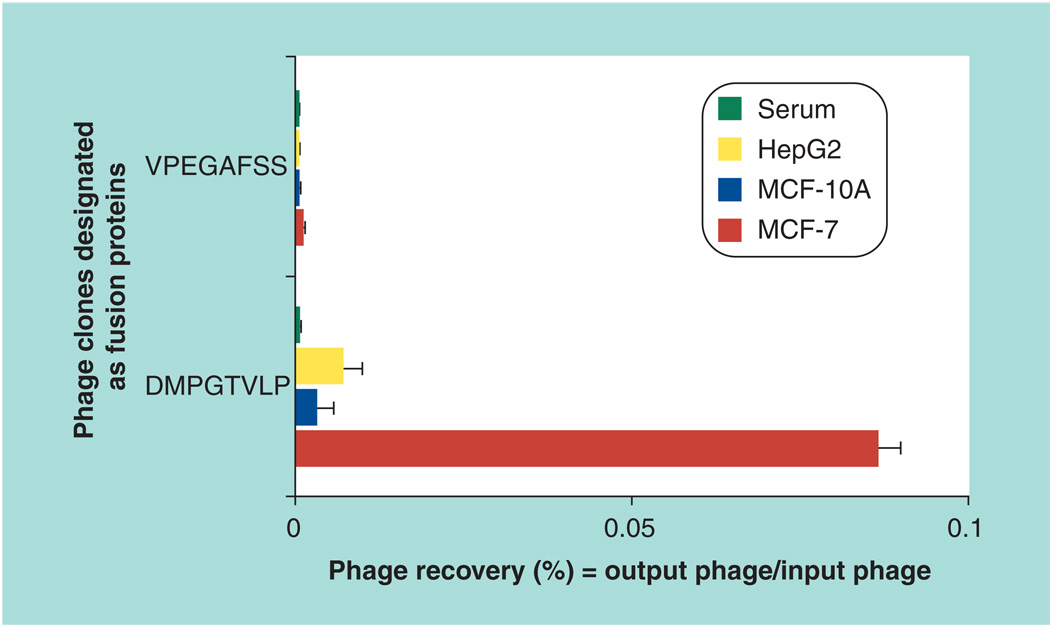
The selectivity of the phage was determined by measuring its binding to MCF-7 cells in comparison with control MCF-10A and HepG2 cells. DMPGTVLP phage demonstrated very high selectivity towards the targeted MCF-7 cells, binding them at a level 26-times higher than MCF-10A cells and 12-times higher than HepG2 cells (Figure 2). Control VPEGAFSS phage bound the same cells at a level 70-times lower than DMPGTVLP phage.
Figure 2. Phage selectivity.
Binding of the phage clone designated by the sequence of the fused foreign peptide (DMPGTVLP) with target MCF-7 cells and control MCF-10A and HepG2 cells (as percent phage recovery). The unrelated streptavidin targeting phage clone designated by the sequence of the fused foreign peptide (VPEGAFSS) was used as a control.
Phage–Doxil characterization
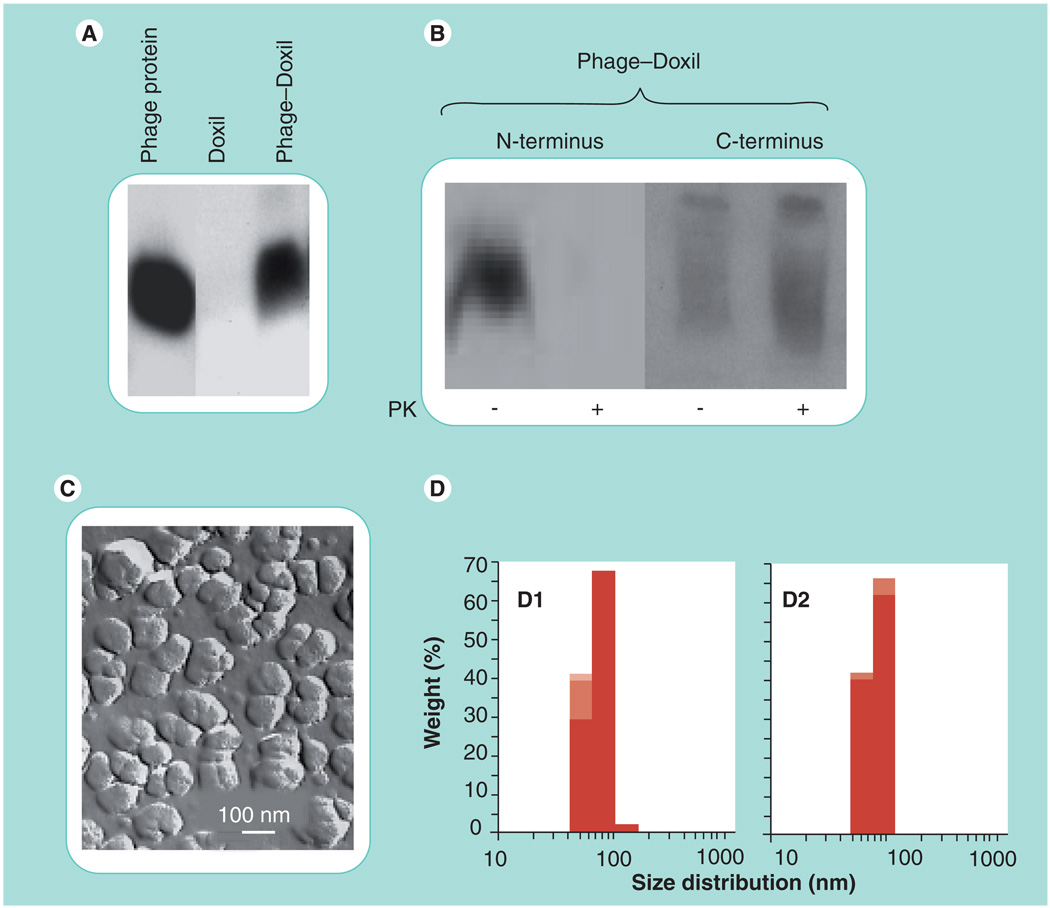
The presence of the fusion pVIII coat protein in MCF-7-phage–Doxil was confirmed by western blotting probed with anti-fd phage antibodies that specifically bind the N-terminal segment of the major coat protein (Figure 3A). To establish the orientation of the phage proteins in the liposomes, we analyzed which part of the protein was protected against digestion with proteinase K. In MCF-7-phage–Doxil, a major fraction of the N-terminal segment was lost (digested), while the C-terminal part of the protein remained intact (i.e., immersed into the membrane) as revealed with C-terminus-specific antibodies (Figure 3B). This result indicated that the intact chimeric pVIII coat fusion protein can spontaneously incorporate into the liposomal membrane via its hydrophobic part and expose the binding N-terminus on the liposome outer surface.
Figure 3. Characterization of Doxil® modified with MCF-7-specific phage protein (MCF-7-phage–Doxil).
(A) Western blotting pattern showing the presence of the phage protein in MCF-7-phage–Doxil. (B) The topology of the phage protein in the liposome membrane determined by the treatment with PK and terminus-specific antibodies: the N-terminus was significantly degraded in MCF-7-phage–Doxil, while the C-terminal part of the protein remained intact. (C) The liposomal morphology of MCF-7-phage–Doxil by freeze-fracture electron microscopy. (D) Size and size distribution of Doxil (D1) and MCF-7-phage–Doxil (D2). Samples were measured in triplicate and results are shown in stacked column charts.
PK: Proteinase K.
The incorporation of the targeting protein into Doxil liposomes did not affect their integrity and shape. The freeze fracture electron microscopy image showed that MCF-7-phage–Doxil displayed typical liposomal morphology (Figure 3C), and the average size of the MCF-7-phage–Doxil particles was within 88 ± 21 nm intervals (Figure 3D).
Doxorubicin in phage–Doxil liposomes
The results of four independent measurements showed that the final preparation of MCF-7-phage–Doxil contains 0.08 ± 0.01 mg of doxorubicin per mg of total lipid and 1.33 ± 0.13 mg of doxorubicin per ml, which corresponds to 66.2 ± 6.3% of the initial doxorubicin contents in Doxil (e.g., 0.12 mg of doxorubicin per mg of total lipid and 2 mg of doxorubicin per ml) and confirms a good drug loading in liposomes in the process of the phage–Doxil preparation.
Selectivity of phage–liposomes
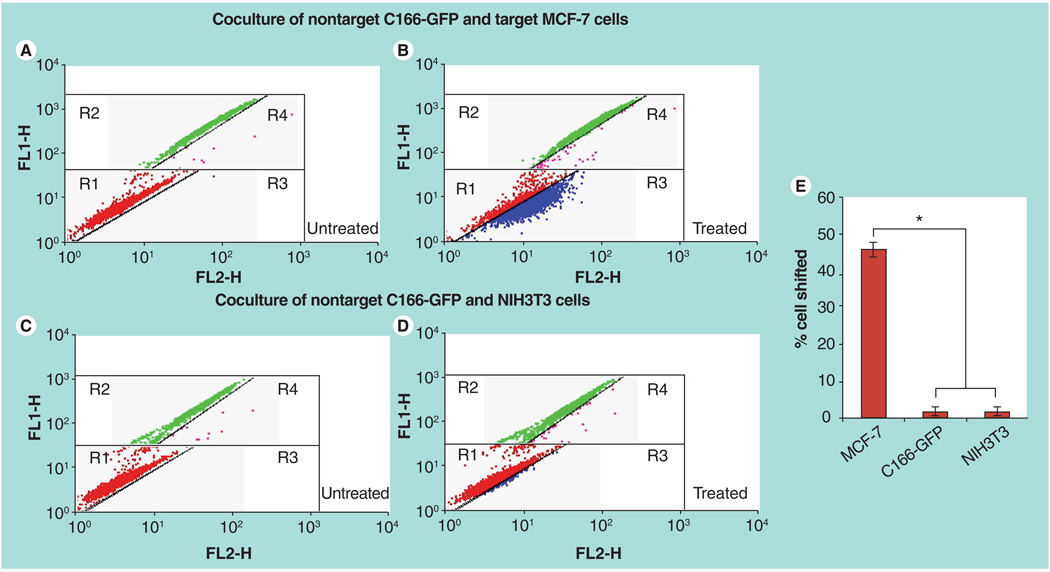
The binding activity and selectivity of the phage fusion pVIII coat protein after its incorporation into liposomes was investigated using a coculture assay followed by FACS analysis. Target MCF-7 cells were cogrown with nontarget C166-GFP cells. FACS analysis revealed two distinct coculture cell populations based on their green fluorescence intensity (FL1-H green) (Figure 4A). After treatment with rhodamine-labeled MCF-7-specific phage–liposomes in the coculture with MCF-7 and C166-GFP cells, a significantly higher percent of MCF-7 cells were shifted into region 3 (R3) compared with C166-GFP cells shifted into region 4 (R4) (Figure 4B). Statistical analysis showed that phage–liposomes bound to around 46% of MCF-7 cells and 2% of C166-GFP cells (Figure 4E), indicating significant selectivity towards the target cells. On a negative control coculture with nontarget cells NIH3T3 and C166-GFP, treatment with MCF-7-specific phage–liposomes resulted in a negligible level of shifted NIH3T3 (R3) and C166-GFP (R4) cells (Figure 4C & D), confirming the specific binding of MCF-7-phage–liposomes to target MCF-7 cells.
Figure 4. FACS analysis on binding selectivity of rhodamine-labeled MCF-7-phage–liposomes in the coculture composed of nontarget C166-GFP and target MCF-7 cells and control coculture of nontarget C166-GFP and NIH3T3 cells.
The dot plots were inserted into four regions (R1, R2, R3 and R4). FL1-H (green); FL2-H (red). (A) Untreated coculture of C166-GFP and MCF-7 cells. Red dots in R1 region showing the location of untreated MCF-7 cells and green dots in R2 region showing the location of untreated C166-GFP cells. (B) Coculture of C166-GFP and MCF-7 cells treated by rhodamine-labeled MCF-7-phage–liposomes. A number of MCF-7 cells (blue dots) were right shifted into R3 region, but a significantly less number of C166-GFP cells (pink dots) were right shifted into R4 region, indicating the preferential binding of MCF-7-phage–liposomes to target MCF-7 cells. (C) Untreated control coculture of C166-GFP and NIH3T3 cells. Red dots in R1 region showing the location of untreated NIH3T3 cells and green dots in R2 region showing the location of untreated C166-GFP cells. (D) Coculture of C166-GFP and NIH3T3 cells treated by rhodamine-labeled MCF-7-phage–liposomes showed the negligible binding of MCF-7-phage–liposomes to two nontargeted cells, indicating no selectivity between nontargeted NIH3T3 and C166-GFP cells. (E) Quantitative analysis of percent of cells shifted.
*p < 0.05, mean ± standard deviation, n = 3
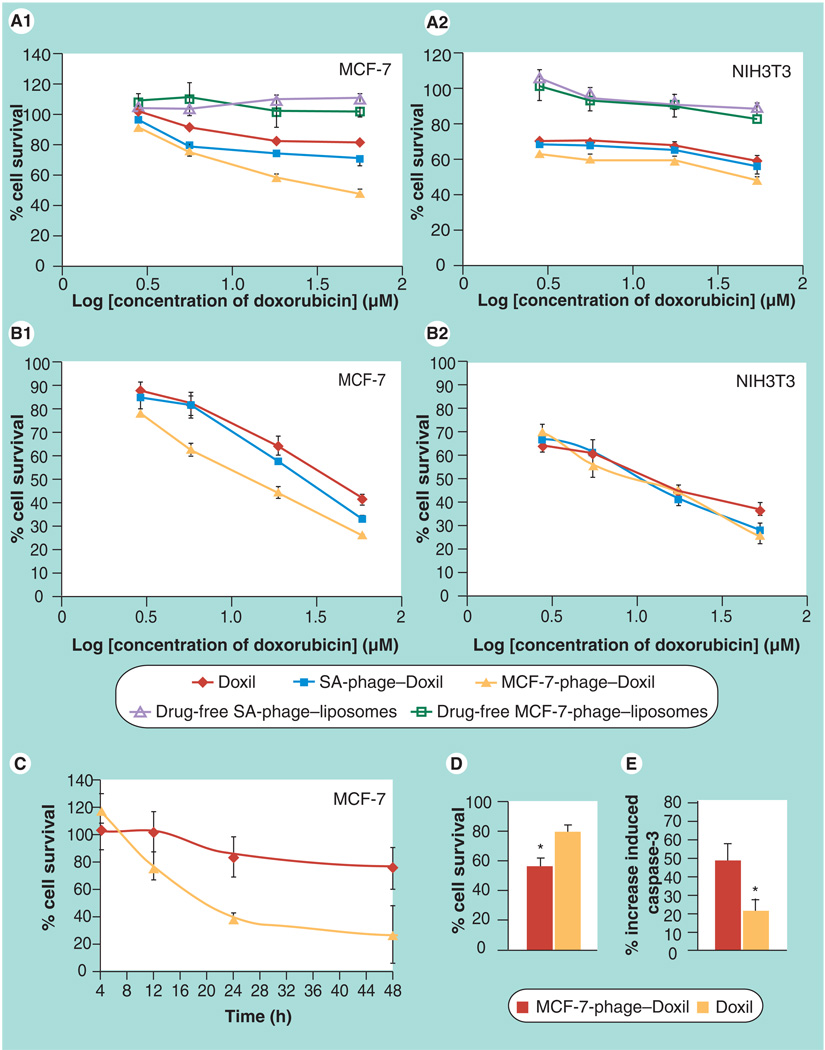
Tumor cell killing by phage–Doxil
CellTiter-Blue Assay (Figure 5A1) demonstrated a significantly higher targeted tumor cell killing by 24 h treatment of MCF-7-phage–Doxil compared with controls. While drug-free phage–liposomes did not show any noticeable cytotoxicity towards MCF-7 cells, and Doxil caused only limited tumor cell killing, MCF-7-phage–Doxil provoked a significant cell death at the same doxorubicin concentrations. Thus, MCF-7-phage–Doxil induced an approximatly 2.5-fold increase in MCF-7 cell death compared with Doxil at doxorubicin concentration of 56 µM (Figure 5A1). IC50 of MCF-7-phage–Doxil is approximately 47.8 µM at 24 h, which is much lower than the reported IC50 of Doxil (around 155 µg/ml or 267 µM) and is close to mAbs 2C5 modified Doxil (~25 µg/ml or 43 µM) [24]. The irrelevant SA-phage–Doxil triggered a slightly increased MCF-7 cell killing compared with Doxil, but still much lower than targeted MCF-7-phage–Doxil (Figure 5A1). While this result confirms the role of MCF-7 targeting phage fusion protein in MCF-7 cell death, it may imply that even nonspecific phage fusion coat proteins may bear some properties that allow them to circumvent drug-delivery barriers. Conversely, MCF-7-phage–Doxil did not show significantly higher cytotoxicity in the negative NIH3T3 (Figure 5A2) and C166 cells (data not shown) when compared with Doxil and irrelevant SA-phage–Doxil, confirming selective cytotoxicity of MCF-7-phage–Doxil only towards target cells. Similar results (Figure 5B1 & B2) were observed in a delayed viability test, in which cytotoxicity was performed after 12 h drug treatment and additional 48-h incubation in drug-free medium. Stronger time-dependent and faster MCF-7 cell killing by MCF-7-phage–Doxil was also evident (Figure 5C). These results were further confirmed by Trypan Blue exclusion assay when MCF-7 cells were treated by 8.6 µM MCF-7-phage–Doxil or Doxil for 24 h (Figure 5D).
Figure 5. Cytotoxicity of phage–Doxil.
(A) Toxicity towards MCF-7 cells (A1) and negative NIH3T3 cells (A2) after 24-h treatment with Doxil®, MCF-7-phage–Doxil, irrelevant SA-phage–Doxil, drug-free MCF-7-phage–liposomes and drug-free SA-phage–liposomes (mean ± SEM, n = 6). (B) Toxicity towards MCF-7 cells (B1) and negative NIH3T3 cells (B2) after 12-h treatment and an additional 48-h incubation in drug-free medium (mean ± SEM, n = 6). (C) Kinetics of MCF-7 cell killing in the same assay (mean ± standard deviation, n = 6). (A–C) Cytotoxicity was performed using the CellTiter-Blue® Assay. (D) Cell viability by Trypan Blue exclusion (*p < 0.05, mean ± SEM, n = 18). (E) Induction of caspase-3 in MCF-7 cells by Doxil and phage–Doxil (*p < 0.05, mean ± standard deviation, n = 3).
SA-phage: Streptavidin-targeting phage.
Triggering of apoptosis by phage–Doxil
Previous studies have shown that doxorubicin triggers the death of MCF-7 cells by inducing apoptosis. To compare the extent of apoptotic cell death in the case of MCF-7-phage–Doxil and Doxil, we tested the activity of caspase-3 (the biomarker of apoptosis) in MCF-7 cells treated by MCF-7-phage–Doxil or Doxil. Figure 5E shows that although both Doxil and MCF-7-phage–Doxil activate the caspase-3 activity, MCF-7-phage–Doxil resulted in significantly enhanced apoptosis. Control drug-free MCF-7-phage–liposomes did not show any noticeable cytotoxicity against MCF-7 cells (Figure 5A1), excluding the possibility of cell death caused by the phage coat fusion protein itself.
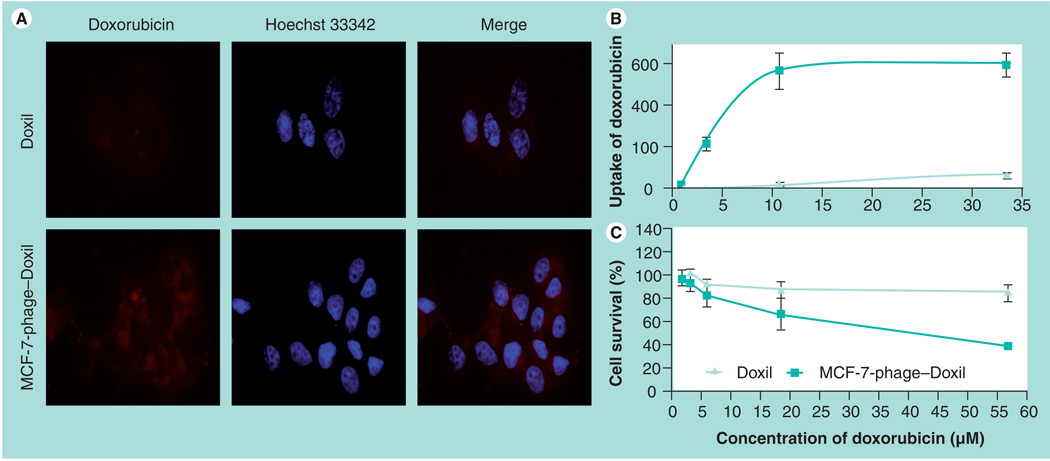
Correlation between doxorubicin uptake & cytotoxicity of Doxil & phage–Doxil
A more potent cell killing by MCF-7-phage–Doxil was the result of a more efficient delivery of doxorubicin into target cells. Cell-uptake doxorubicin was greater with MCF-7-phage–Doxil than with Doxil visualized by f luorescence microscopy (Figure 6A). A parallel quantitative analysis of the cellular uptake of doxorubicin and cytotoxicity (cell death) after 24-h treatment of MCF-7 cells with MCF-7-phage–Doxil or Doxil revealed that a significantly higher quantity of cell-associated doxorubicin in the case of MCF-7-phage–Doxil correlates well with the increased cell death (Figure 6B & C).
Figure 6. Correlation between doxorubicin uptake and cytotoxicity of Doxil® and MCF-7-phage–Doxil.
(A) Uptake of doxorubicin by MCF-7 cells was visualized by fluorescence microscopy after 30-min treatment with Doxil or MCF-7-phage–Doxil followed by 2-h incubation in drug-free medium. (B) Uptake of doxorubicin by MCF-7 cells was determined by fluorescence spectrometry after 24-h treatment with Doxil or MCF-7-phage–Doxil and was indicated as doxorubicin fluorescence intensity. (C) Cytotoxicity of Doxil and MCF-7-phage–Doxil towards MCF-7 cells after 24-h treatment (mean ± standard deviation, n = 6).
Discussion
Direct and fast screening for tumor-targeted peptides via the phage-display technology provides a promising approach to promote the targeting of various pharmaceutical agents [5]. However, the traditional phage-display approach assumes the necessity of chemical synthesis of identified tumor-selective peptides and their conjugation with pharmaceuticals or pharmaceutical nanocarriers (such as liposomes). To simplify the procedure and exclude chemical modifications, we have suggested using the intact hybrid fusion coat proteins for liposome targeting. They are easily isolated from cancer cell-specific phages selected from ‘landscape libraries’ – multibillion collections of phage with all 4000 major coat proteins pVIII fused to foreign random peptides [20,21]. Almost 90% by mass of a landscape phage is a hybrid coat protein – a chimeric polypeptide 55–56 amino acids long composed of a foreign targeting peptide segment fused to the N-terminus of the phage protein.
The ability of the major coat protein pVIII to integrate into liposomes emerges from its intrinsic function as a membrane protein. During the infection of the host Escherichia coli, the positively charged C-terminus of the protein spans the bacterial cytoplasmic membrane leaving the negatively charged N-terminus in the periplasm [25]. In infected cells, the protein is synthesized as a water-soluble cytoplasmic precursor, which contains an additional leader sequence of 23 residues at its N-terminus. When this protein is inserted into the membrane, the leader sequence is cleaved off by a leader peptidase. Later, during the phage assembly, the newly synthesized proteins are transferred from the membrane into the coat of the emerging phage. Thus, the major coat protein can change its conformation to accommodate to various distinctly different forms during phage infection and assembly. This structural flexibility of the major coat protein is determined by its unique architecture, which is studied in much detail. The structure of the major coat protein in the phage virions, micelles and bilayer membranes is well resolved [26,27].
Since the ‘fusion phage’ is constructed via in-frame insertion of the gene encoding for the targeting peptide into the gene of the phage pVIII major coat protein, the membrane nature of the pVIII major coat protein is retained even when a short foreign target peptide is fused to its N-terminus. The ‘membranophilicity’ of the fusion proteins, their ability to proceed normally during the phage infection and morphogenesis, has evolved previously during the propagation of phage libraries in the host bacteria and is ensured by the observed viability of the selected phage [21]. Thus, the pVIII major coat protein segment can serve as a membrane anchor within the liposome bilayer for the targeting peptide when exposed on/over the liposome surface.
The phage DMPGTVLP (designated by the structure of the borne foreign peptide) demonstrated a high selectivity and specificity towards the target cells versus control cells (Figure 2). Fusion protein carrying tumor cell-binding peptide DMPGTVLP inherits the major structural features of the ‘wild-type’ major coat protein pVIII. It has a positively charged C-terminus (amino acids 45–55), which can navigate the protein through the liposome membrane, a highly hydrophobic membranophilic segment (amino acids 27–40), which allows the protein to accommodate readily in the membrane and hydrophilic N-terminus (amino acids 1–26), which displays the cancer cell-binding sequence on the liposome surface.
To prepare phage fusion coat protein-targeted liposomes, we modified procedures previously developed for the insertion of membrane proteins into liposomes during their reconstitution [14,28–30], to ensure the integrity of preformed Doxil PEGylated liposomes and minimize the efflux of doxorubicin. To explore a spontaneous insertion of MCF-7-specific pVIII coat fusion protein into the preformed Doxil, we used the following procedure: solubilizing the highly hydrophobic pVIII coat fusion protein using the detergent sodium cholate at its CMC concentration, inserting the coat fusion protein into the liposome membrane by incubating mixed micelles of sodium cholate and coat fusion protein with Doxil, and finally removing sodium cholate by dialysis yielding phage–Doxil. The uniformity of liposomal nanoparticles in the protein-modified Doxil was confirmed by the freeze fracture electron microscopy and size analysis. The western blotting test also clearly demonstrated that pVIII major coat fusion protein has been associated with liposomes (Figure 3).
Since the key aspect of the technology (the topography of the associated fusion protein in the PEGylated liposomal membrane) has been previously addressed only sparingly using functional tests [14], and very scanty data existed regarding the behavior of the ‘wild-type’ major coat protein during its spontaneous insertion into non-PEGylated liposomes [17], we studied the topology of the protein in Doxil in more detail by chemical means. The topology of the membrane-bound proteins was analyzed by protease digestion experiments, in which after the binding of the coat protein to preformed vesicles, proteinase K was added into the system. Our results showing that proteinase K destroys the N-terminal segment but not the C-terminal one clearly demonstrate the preferential insertion of C-termini containing four positive lysine residues into the membrane together with a neighboring hydrophobic protein fragment. It is possible that at the initial stage, an essential portion of the fusion protein remains associated with an interfacial region of liposomes, followed by energetically unfavorable membrane translocation of the positively charged C-terminus, as was previously suggested [17].
The improved Doxil targeting with the cancer cell-specific phage protein resulting in a much better uptake of doxorubicin into target cells and more effective cell killing confirmed the preservation of specific activity by peptide fragments of the coat-fused protein associated with liposomes (Figures 4–6). Importantly, the incorporation of the phage protein into Doxil liposomes results in good retention of the initial doxorubicin by the liposomes, although further optimization of the formulation (or different incorporation procedure) is possible.
Conclusion
Our results demonstrate that the phage pVIII coat protein displaying cancer cell-targeting peptides can serve as an anchor for the integration of these peptides with drug-loaded liposomes to make them cancer cell targeted. Our findings open the way for using inexpensive and easy-to-prepare fused phage proteins as ‘substitute antibodies’ for site-specific targeting of drugs and drug-loaded pharmaceutical nanocarriers.
Future perspective
The integration of nanotechnology with combinatorial phage technique represents a paradigm shift in the development of novel, more efficient, targeted nanomedicines. The new approach, proved in this article, explores the unique propensity of phage proteins to incorporate spontaneously into lipid bilayers to form particles mimicking the structure of phage proteins in bacterial membranes. Similar to the selected parental phages, the complexes of phage proteins with nanomedicines recognize corresponding cancer cells and help to release the encapsulated drugs into the intracellular compartments. The proposed technology, based on the self-assembly of selected phage proteins and ‘stealth’ liposomes, enables development of a number of different nanomedicines targeted to a variety of cellular receptors and allows screening of their activity in a high-throughput mode. Since the landscape phage approach sets up a ‘one size fits all’ scheme, it can be readily adapted for development of combinatorial targeted nanomedicines against any cancer disease or any pathology requiring a fast turnaround time, and allows avoiding complicated chemical conjugation procedures used for preparation of peptide-targeted nanomedicines. Further use of this approach may significantly enhance the screening of specifically targeted nanomedicines, result in dramatic acceleration of the translational process and change the field of cancer research through the development of more specific, safe and efficient targeted nanotherapeutics.
Executive summary
A chimeric phage fusion coat protein specific towards MCF-7 breast cancer cells was identified from a phage landscape library.
The landscape phage fusion coat protein was directly incorporated into drug-loaded liposomes via its hydrophobic part and exposes the binding N-terminus on the liposome outer surface.
Drug-loaded liposomes modified with the tumor-specific phage fusion coat protein maintained their size and morphology.
The binding activity and selectivity of the phage pVIII fusion protein was retained after incorporation into liposomes.
Phage–Doxil strongly and specifically targeted MCF-7 cells and demonstrated significantly increased cytotoxicity towards target cells in vitro.
The phage fusion coat protein displaying cancer cell-targeting peptides can serve as an anchor for the integration of these peptides with drug-loaded liposomes to make them cancer cell targeted.
The new landscape phage-based approach overcomes the drawbacks associated with the chemical modification of nanocarriers with cancer-selective peptides.
The use of inexpensive and easy-to-prepare fused phage proteins as ‘substitute antibodies’ opens a new way for site-specific targeting of drugs and drug-loaded nanopharmaceuticals.
Acknowledgments
This work was supported by the NIH grant # 1 R01 CA125063–01 and the Animal Health and Disease Research grant 2006–9, College of Veterinary Medicine Auburn University to Valery A Petrenko.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1. Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. • Describes the potential of nanotechnology in revolutionizing cancer diagnosis and therapy.
- 2. Alexis F, Rhee JW, Richie JP, Radovic-Moreno AF, Langer R, Farokhzad OC. New frontiers in nanotechnology for cancer treatment. Urol. Oncol. 2008;26:74–85. doi: 10.1016/j.urolonc.2007.03.017. • Discusses recent advances of cancer nanotechnology with particular attention to nanoparticle systems that are in clinical practice or in various stages of development for cancer imaging and therapy.
- 3.Sofou S. Surface-active liposomes for targeted cancer therapy. Nanomedicine (Lond.) 2007;2(5):711–724. doi: 10.2217/17435889.2.5.711. [DOI] [PubMed] [Google Scholar]
- 4.Noble CO, Kirpotin DB, Hayes ME, et al. Development of ligand-targeted liposomes for cancer therapy. Expert Opin. Ther. Targets. 2004;8:335–353. doi: 10.1517/14728222.8.4.335. [DOI] [PubMed] [Google Scholar]
- 5.Sergeeva A, Kolonin MG, Molldrem JJ, Pasqualini R, Arap W. Display technologies: application for the discovery of drug and gene delivery agents. Adv. Drug Deliv. Rev. 2006;58:1622–1654. doi: 10.1016/j.addr.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol. Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 7.Mori T. Cancer-specific ligands identified from screening of peptide-display libraries. Curr. Pharm. Des. 2004;10:2335–2343. doi: 10.2174/1381612043383944. [DOI] [PubMed] [Google Scholar]
- 8.Brigati JR, Samoylova TI, Jayanna PK, Petrenko VA. Phage display for generating peptide reagents. Curr. Protoc. Protein Sci. 2008;Chapter 18(Unit 18.9) doi: 10.1002/0471140864.ps1809s51. [DOI] [PubMed] [Google Scholar]
- 9. Petrenko VA. Evolution of phage display: from bioactive peptides to bioselective nanomaterials. Expert Opin. Drug Deliv. 2008;5:825–836. doi: 10.1517/17425247.5.8.825. •• Focuses on the progress made in the development of phage-derived biorecognition nanomaterials and the use of phage as a bioselectable molecular recognition interface in medical and technical devices.
- 10. Krumpe LR, Mori T. The use of phage-displayed peptide libraries to develop tumor-targeting drugs. Int. J. Pept. Res. Ther. 2006;12:79–91. doi: 10.1007/s10989-005-9002-3. •• Good review regarding target-binding peptides isolated through the screening of phage-displayed random peptide libraries and its application in drug and nanomaterial delivery.
- 11.Hölig P, Bach M, Volkel T, et al. Novel RGD lipopeptides for the targeting of liposomes to integrin-expressing endothelial and melanoma cells. Protein Eng. Des. Sel. 2004;17:433–441. doi: 10.1093/protein/gzh055. [DOI] [PubMed] [Google Scholar]
- 12.Lee TY, Lin CT, Kuo SY, Chang DK, Wu HC. Peptide-mediated targeting to tumor blood vessels of lung cancer for drug delivery. Cancer Res. 2007;67:10958–10965. doi: 10.1158/0008-5472.CAN-07-2233. [DOI] [PubMed] [Google Scholar]
- 13.Emerich DF, Thanos CG. Multifunctional peptide-based nanosystems for improving delivery and molecular imaging. Curr. Opin. Mol. Ther. 2008;10:132–139. [PubMed] [Google Scholar]
- 14.Jayanna PK, Torchilin VP, Petrenko VA. Liposomes targeted by fusion phage proteins. Nanomedicine. 2009;5:83–89. doi: 10.1016/j.nano.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broome-Smith JK, Gnaneshan S, Hunt LA, et al. Cleavable signal peptides are rarely found in bacterial cytoplasmic membrane proteins. Mol. Membr. Biol. 1994;11:3–8. doi: 10.3109/09687689409161023. [DOI] [PubMed] [Google Scholar]
- 16. Kuhn A. Major coat proteins of bacteriophage Pf3 and M13 as model systems for sec-independent protein transport. FEMS Microbiol. Rev. 1995;17:185–190. doi: 10.1111/j.1574-6976.1995.tb00201.x. •• An important report regarding the membrane insertion mechanism of major coat proteins of bacteriophage Pf3 and M13.
- 17. Soekarjo M, Eisenhawer M, Kuhn A, Vogel H. Thermodynamics of the membrane insertion process of the M13 procoat protein, a lipid bilayer traversing protein containing a leader sequence. Biochemistry. 1996;35:1232–1241. doi: 10.1021/bi951087h. •• For the first time, a thermodynamics analysis is performed on the membrane insertion process of the M13 procoat protein.
- 18.Krag DN, Shukla GS, Shen GP, et al. Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res. 2006;66:7724–7733. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 19.Il’ichev AA, Minenkova OO, Tat’kov SI, et al. Production of a viable variant of the M13 phage with a foreign peptide inserted into the basic coat protein. Dokl. Akad. Nauk. SSSR. 1989;307:481–483. [PubMed] [Google Scholar]
- 20.Petrenko VA, Smith GP, Gong X, Quinn T. A library of organic landscapes on filamentous phage. Protein Eng. 1996;9:797–801. doi: 10.1093/protein/9.9.797. [DOI] [PubMed] [Google Scholar]
- 21.Kuzmicheva GA, Jayanna PK, Sorokulova IB, et al. Diversity and censoring of landscape phage libraries. Protein Eng. Des. Sel. 2009;22:9–18. doi: 10.1093/protein/gzn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petrenko VA, Smith GP. Phages from landscape libraries as substitute antibodies. Protein Eng. 2000;13:589–592. doi: 10.1093/protein/13.8.589. • Validates the concept of landscape phage as substituted antibodies.
- 23.Spruijt RB, Wolfs CJ, Hemminga MA. Aggregation-related conformational change of the membrane-associated coat protein of bacteriophage M13. Biochemistry. 1989;28:9158–9165. doi: 10.1021/bi00449a030. [DOI] [PubMed] [Google Scholar]
- 24.Elbayoumi TA, Torchilin VP. Enhanced cytotoxicity of monoclonal anticancer antibody 2C5-modified doxorubicin-loaded PEGylated liposomes against various tumor cell lines. Eur. J. Pharm. Sci. 2007;32(3):159–168. doi: 10.1016/j.ejps.2007.05.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webster R. Filamentous phage biology. In: Barbas CF, et al., editors. Phage Display: A Laboratory Manual. NY, USA: Cold Spring Harbor Laboratory Press; 2001. pp. 1.1–1.37. [Google Scholar]
- 26.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chemical Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stopar D, Spruijt RB, Hemminga MA. Anchoring mechanisms of membrane-associated M13 major coat protein. Chem. Phys. Lipids. 2006;141:83–93. doi: 10.1016/j.chemphyslip.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 28.Angrand M, Briolay A, Ronzon F, Roux B. Detergent-mediated reconstitution of a glycosyl-phosphatidylinositol-protein into liposomes. Eur. J. Biochem. 1997;250:168–176. doi: 10.1111/j.1432-1033.1997.t01-1-00168.x. [DOI] [PubMed] [Google Scholar]
- 29.Paternostre MT, Roux M, Rigaud JL. Mechanisms of membrane protein insertion into liposomes during reconstitution procedures involving the use of detergents. 1. Solubilization of large unilamellar liposomes (prepared by reverse-phase evaporation) by triton X-100, octyl glucoside, and sodium cholate. Biochemistry. 1988;27:2668–2677. doi: 10.1021/bi00408a006. [DOI] [PubMed] [Google Scholar]
- 30.Rigaud JL, Paternostre MT, Bluzat A. Mechanisms of membrane protein insertion into liposomes during reconstitution procedures involving the use of detergents. 2. Incorporation of the light-driven proton pump bacteriorhodopsin. Biochemistry. 1988;27:2677–2688. doi: 10.1021/bi00408a007. [DOI] [PubMed] [Google Scholar]