Abstract
Background
Diarrhea is an important cause of morbidity and mortality in all regions of the world and among all ages, yet little is known about the fraction of diarrhea episodes and deaths due to each pathogen.
Methodology/Principal Findings
We conducted a systematic literature review to identify all papers reporting the proportion of diarrhea episodes with positive laboratory tests for at least one pathogen in inpatient, outpatient and community settings that met our inclusion and exclusion criteria. We identified a total of 25,701 papers with possible etiology data and after final screening included 22 papers that met all inclusion and exclusion criteria. Enterotoxigenic Escherichia coli and V. cholerae O1/O139 were the leading causes of hospitalizations. In outpatient settings, Salmonella spp., Shigella spp., and E. histolytica were the most frequently isolated pathogens.
Conclusions/Significance
This is the first systematic review which has considered the relative importance of multiple diarrhea pathogens. The few studies identified suggest that there is a great need for additional prospective studies around the world in these age groups to better understand the burden of disease and the variation by region.
Author Summary
Diarrhea is an important cause of illness and death around the world and among people of all ages, but unfortunately we often do not know what specific bacterium or virus causes the illness. We conducted a review of the scientific literature with the goal of finding published studies that identified bacteria and viruses among patients with diarrhea in the community and in hospital settings. We initially found nearly 26,000 papers on this topic but narrowed the list to 22 studies that met all of our specific criteria for inclusion in our review. Among patients hospitalized for diarrhea, E coli and Vibrio cholerae were found in more than 49% of people living in middle income and poor countries. Among patients who sought care from their doctor on an outpatient basis, Salmonella spp., Shigella spp., and E. histolytica were most often found. In our review we focused on the differences in the distribution of pathogens between patients in inpatient vs. outpatient settings because these estimates may best approximate what we would expect to see if the distribution were applied to global estimates of diarrhea deaths vs. uncomplicated illnesses.
Introduction
Diarrhea is an important cause of morbidity and mortality in all regions of the world and among all ages [1], [2]. For children 5 years of age and older, adolescents, and adults mild to moderate diarrhea can lead to absenteeism from school or work and may require treatment by a health care provider. More severe diarrhea can lead to hospitalization; serious sequelae such as Guillain Barre' syndrome and hemolytic uremic syndrome; and in some cases death [3], [4].
Though most diarrhea episodes are self limiting and dehydration can usually be controlled with oral rehydration therapy, it would be ideal to be able to prevent diarrhea, especially the more severe episodes which have a higher likelihood of progressing to complications or death. Some prevention strategies such as improved water and sanitation and basic hygiene practices are generalizable and thus do not require knowledge of diarrhea etiology, but others such as vaccines would benefit greatly from a comprehensive understanding of the overall burden of pathogen-specific diarrheal disease.
Recent advances have led to the development of an effective rotavirus vaccine which is now recommended for young children as part of the routine immunization schedule [5]. A vaccine for cholera that could be useful in some settings in all ages has been available for several years, and is now recommended by the WHO for persons living in endemic areas [6]. The number of pathogens that are responsible for diarrheal disease goes far beyond rotavirus and Vibrio cholerae; however, the fraction of diarrhea episodes and deaths due to each pathogen is unclear, and thus uncertainty may inhibit prioritization of funding for research and disease control programs.
There have been numerous studies conducted in countries around the world to determine the presence of one or more pathogens in diarrheal stools. While isolated studies provide important pieces of information, it is difficult to draw conclusions with regard to the importance of various pathogens without looking at a complete spectrum of agents simultaneously. We conducted a systematic literature review of diarrhea etiology studies to better understand the likely distribution of pathogen-specific diarrhea episodes and deaths in older children, adolescents and adults. To our knowledge this is the first systematic review designed to compile the data from multiple pathogens which might be applied to annual incidence and mortality rates in these age groups.
Methods
We searched PubMed/Medline, CAB abstracts, System for Information on Grey Literature in Europe (SIGLE), and all World Health Organization (WHO) Regional Databases for studies published from January 1, 1980 through December 31, 2008 using all combinations of the following search and MeSH terms: “diarrhea”, “etiology”, “pathogen”, “incidence”, “mortality”, “cause of death”, and “gastroenteritis”. The objective of the search was to identify all papers reporting the proportion of diarrhea episodes with positive laboratory tests for at least one pathogen in in-patient, out-patient and community settings that met our inclusion and exclusion criteria. We included studies published in all languages and conducted in children ≥5 years, adolescents and adults with at least 12 mo of surveillance (multiples of 12 mo±1 mo for longer studies) to minimize bias due to seasonality of diarrhea pathogens. We excluded studies enrolling only patients with clinical signs of dysentery, i.e. blood in the stool, studies conducted in special populations such as travelers, patients hospitalized for other reasons, or only HIV-positive persons and all individual or outbreak case reports. Studies that did not screen for HIV status and/or did not enroll based on HIV status were included. All exclusion criteria were chosen to ensure study populations represented the general population in the study community. In addition, we assessed all laboratory methods for appropriateness and if either incorrect or inadequately described, we excluded the study.
We conducted individual searches in all databases and combined the results to eliminate duplicates using RefWorks Reference Manager [7]. We first reviewed titles for appropriateness and then all abstracts as the first steps of the screening process. For all abstracts with likely applicable data we ascertained the full text article and screened for inclusion and exclusion criteria. All papers meeting our inclusion and exclusion criteria were then abstracted by 2 trained data abstractors into a standardized database. We abstracted information with regard to study population, study setting, diarrhea definition, prevalence of each pathogen, and diarrhea definition required for inclusion in the study. After completing both abstractions we cross checked the data and rectified any differences. We initially included an extensive group of diarrhea pathogens for data abstraction (Table 1), but included only pathogens with available data in the final analyses. For inclusion in the final analyses presented here, we included only studies that adequately described where study participants were recruited from, i.e. inpatient, outpatient, or community settings.
Table 1. Pathogens included in initial abstraction.
| Enterohaemorrhagic Escherichia coli (EHEC) | Campylobacter spp. | Yersinia spp. |
| Enteroinvasive Escherichia coli (EIEC) | Aeromonas spp | Endolimax nana |
| Enterotoxigenic Escherichia coli (ETEC) | Shigella spp. | C. difficile |
| Enteropathogenic Escherichia coli (EPEC) | Salmonella spp. | Cryptosporidium spp. |
| Enteroaggregative Escherichia coli (EAEC) | Giardia spp. | E. histolytica |
| Calicivirus/Norwalk or related agents/Norovirus | C. perfringens | P. shigelloides |
Laboratory methods for each pathogen were reviewed by a laboratory expert (D. S.). Because laboratory techniques for pathogens such as diarrheagenic E. coli and E. histolytica, have changed since the mid 1990s, we included all studies with standard laboratory procedures for the time of the study [8], [9].
Analytic Methods
We first calculated the un-weighted mean, median and inter-quartile range for each pathogen for each type of patient population separately. There was only 1 community study [10] so for all analyses we combined this study with the studies conducted in outpatient settings. We then categorized inpatient and outpatient studies based on the number of pathogens reported by the authors in the methods and results sections of each published study: single pathogen studies, those reporting 2–4 pathogens, and those reporting at least 5 pathogens. For each of these categories we calculated the weighted mean for each pathogen. Among inpatient studies we removed the 1 study conducted in a high income setting to enable separate calculations for high vs. low and middle income countries separately. We stratified studies by inpatient or outpatient status and by the number of pathogens identified in each study to present the best possible summary data to approximate the most likely pathogen distributions for diarrhea mortality and all episodes, respectively.
Results
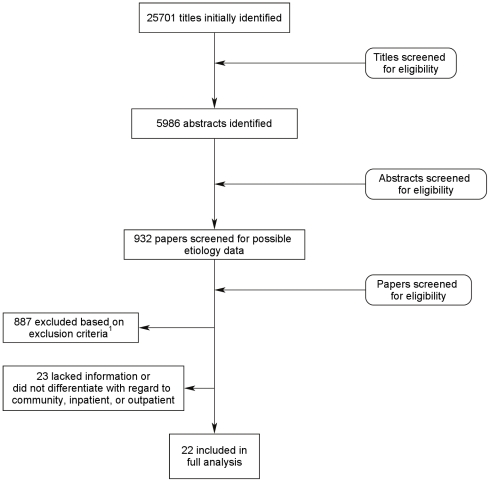
We identified 25,701 papers with possible etiology data (Figure 1). After screening 5,986 abstracts and 932 papers, we found 45 that met our inclusion and exclusion criteria. Twenty-two papers met all inclusion and exclusion criteria and described the study populations with regard to inpatient, outpatient or community study populations [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32] (Table 2). Twenty three additional papers met initial screening criteria but were subsequently excluded from the analysis presented here because they lacked information with regard to the patient population (i.e. inpatient vs. outpatient) or did not differentiate the results by population type (Table 3) [20], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53].
Figure 1. Results of systematic literature review.
Table 2. Study characteristics of community based, inpatient, and outpatient studies included in the final analyses.
| Author (ref) | Country | Date of data collection | Study duration in mos. | Age range in yrs. | Sample size | Pathogens included in study |
| Community | ||||||
| Spencer [10] | El Salvador | 1977 | 12 | ≥6 | 43 | ETEC |
| Outpatient | ||||||
| Hossain [14] | Bangladesh | 1975–1984 | 120 | ≥5 | 2488 | Shigella spp. |
| Sitbon [17] | Gabon | 1980–91 | 12 | ≥6 | 79 | Rotavirus |
| Nath [23] | India | 1994–95 | 24 | ≥5 | 916 | Cryptosporidium spp. |
| Gassama [12] | Senegal | 1997–99 | 24 | ≥18 | 121 | EPEC/EAEC, Salmonella spp., Shigella spp., Campylobacter spp., Aeromonas spp., Rotavirus, Giardia, Cryptosporidium spp., E. histolytica |
| Lau [15] | China | 2001–02 | 12 | ≥5 | 906 | Norovirus |
| MoezArdalan [21] | Iran | 2001–02 | 12 | ≥5 | 312 | Shigella spp. |
| Al-Gallas [26] | Tunisia | 2001–04 | 36 | >18 | 73 | EHEC, EIEC, ETEC, EPEC/EAEC, Salmonella spp., Shigella spp., Campylobacter spp., Aeromonas spp., Adenovirus, Rotavirus, Giardia |
| Inpatient | ||||||
| Oberle [24] | Bangladesh | 1975 | 12 | ≥5 | 1459 | Salmonella spp., Shigella spp., V. cholerae O1 |
| Zaman [32] | Bangladesh | 1978–87 | 120 | ≥5 | 17129 | Shigella spp. |
| Black [28] | Bangladesh | 1977–79 | 24 | ≥10 | 5171 | ETEC, Salmonella spp., Shigella spp., Rotavirus, Giardia, E. histolytica, V. cholerae O1, V. parahaemolytica |
| Echeverria [31] | Thailand | 1980–81 | 12 | >15 | 526 | ETEC, Salmonella spp., Shigella spp., Rotavirus, Giardia, E. histolytica, V. cholerae O1, V. parahaemolytica |
| Watson [19] | UK | 1982 | 12 | ≥12 | 515 | Salmonella spp., Shigella spp., Campylobacter spp., Aeromonas spp., P. shigelloides, C. difficile |
| Poocharoen [25] | Thailand | 1983–93 | 12 | 5–15 | 17 | Campylobacter spp. |
| Baqui [27] | Bangladesh | 1983–84 | 12 | ≥5 | 1569 | ETEC, Shigella spp., Campylobacter spp., Giardia, E. histolytica, V. cholerae O1 |
| Wasfy [18] | Egypt | 1986–93 | 96 | 5–23 | 6278 | Salmonella spp., Shigella spp., Campylobacter spp. |
| Brandonisio [29] | Italy | 1987–94 | 84 | 9–14 | 28 | Cryptosporidium spp. |
| Lim [16] | Singapore | 1989–90 | 12 | ≥6 | 5181 | Campylobacter spp. |
| Germani [13] | New Caledonia | 1990–91 | 24 | ≥15 | 420 | ETEC, EPEC/EAEC, Salmonella spp., Shigella spp., Campylobacter spp., Rotavirus, Giardia, E histolytica, C. difficile |
| Das [54] | India | 1989–91 | 24 | ≥5 | 45 | Cryptosporidium spp. |
| Vilchis-Guizar [30] | Mexico | 1995 | 12 | 39–51 | 2379 | V. cholerae O1 |
| Lau [15] | China | 2001–2002 | 12 | ≥5 | 240 | Norovirus |
| Nagamani [22] | India | 2003–06 | 48 | >5 | 906 | Cryptosporidium spp. |
Table 3. Study characteristics of studies meeting inclusion criteria but excluded from final analysis because the study population (i.e. community-based, inpatient, or outpatient) was not given or results were not stratified by group.
| Author (ref) | Country | Date of data collection | Study duration (months) | Age in yrs (mean ± range) | Sample size | Pathogens included in study |
| Echevarria [55] | Thailand | 1982–83 | 12 | ≥5 | 177 | ETEC, Shigella spp., Aeromonas spp., V. cholerae, V. parahaemolyticus |
| Chatterjee [41] | India | 1982–83 | 24 | 5–14 | 46 | EIEC, EPEC, Salmonella spp., Shigella spp., Campylobacter spp., Rotavirus, Giardia, E. histolytica, V. cholerae, V. parahaemolyticus, P. shigelloides |
| Cabrita [39] | Portugal | 1984–89 | 72 | ≥5 | 1012 | Salmonella spp., Shigella spp., Campylobacter spp. |
| Rahman [48] | Bangladesh | 1985 | 12 | ≥6 | 577 | Cryptosporidium spp. |
| Bingnan [38] | Bangladesh | 1987–89 | 24 | ≥5 | 2370 | Rotavirus |
| Zvizdic [53] | Bosnia | 1988–1991 | 48 | 5–7 | 70 | Rotavirus |
| Cassel-Beraud [40] | Madagascar | 1988–89 | 12 | 6–14 | 113 | ETEC, Salmonella spp., Shigella spp., Campylobacter spp., Rotavirus, Giardia, E. histolytica |
| Zaman [20] | Saudi Arabia | 1989–90 | 12 | ≥5 | 901 | Campylobacter spp. |
| Libanorne [47] | Italy | 1984–87 | 24 | 16–96 | 1681 | Giardia |
| Katsumata [46] | Indonesia | 1992–93 | 12 | ≥5 | 211 | Cryptosporidium spp. |
| Samonis [49] | Greece | 1992–94 | 36 | ≥15 | 1420 | EPEC, Salmonella spp., Shigella spp., Campylobacter spp., Aeromonas spp. |
| Simadibrata [50] | Indonesia | 1995–2000 | 72 | (42±14) | 207 | Shigella spp.,Giardia, E. histolytica |
| Akinyemi [34] | Nigeria | 1995–96 | 12 | ≥6 | 642 | EHEC, EIEC, ETEC, EPEC, Salmonella spp., Shigella spp., Aeromonas spp. |
| Faruque [42] | Bangladesh | 1996–2001 | 72 | ≥5 | 5779 | ETEC, EPEC, Salmonella spp., Shigella spp., Campylobacter spp., Rotavirus, E histolytica, V. cholerae |
| Bern [37] | Guatemala | 1997–98 | 12 | ≥5 | 514 | Cryptosporidium spp. |
| Gambhir [44] | India | 1997–2000 | 36 | ≥15 | 145 | Giardia, Cryptosporidium spp., E. histolytica |
| Battikhi [36] | Jordan | 1997–2001 | 48 | ≥5 | 560 | EPEC, Salmonella spp., Shigella spp., Campylobacter spp., Rotavirus |
| von Seidlein [52] | Multi-site Asia1 | 2002 | 12 | >5 | 8253 | Shigella spp. |
| Abreu-Acosta [33] | Spain | 2002–2004 | 24 | ≥13 | 17 | Cryptosporidium spp. |
| Hamedi [45] | Iran | 2003 | 12 | 5–7 | 31 | Cryptosporidium spp. |
| Uchida [51] | Nepal | 2003–04 | 12 | ≥5 | 645 | Rotavirus |
| Amarilla [35] | Paraguay | 2004–05 | 24 | 18–95 | 801 | Rotavirus |
| Feizabadi [43] | Iran | 2004–05 | 12 | 5–14 | 79 | Campylobacter spp. |
China, Thailand, Indonesia, Vietnam, Pakistan, Bangladesh.
In table 4 we present the unweighted mean and median proportion of stools which tested positive for each pathogen in both in- and out-patient settings. In this analysis V. cholerae O1/O139 and ETEC were the leading causes of hospitalization. In inpatient populations Aeromonas spp. Yersinia spp. Cryptosporidium spp., V. parahaemolyticus, P. shigelloides, and C. difficile were each found in <2% of patients. In out-patient settings, Salmonella spp., Shigella spp., and E. hystolitica were isolated the most frequently. In outpatient populations, EHEC, Campylobacter spp., Aeromonas spp. and Yersinia spp. were found in <2% of patients. Very few studies tabulated data such that the co-occurance of more than one pathogen in a diarrheal stool could be ascertained and few tested a broad enough spectrum of pathogens to be able to quantify the proportion of episodes from which no currently recognized pathogen could be identified.
Table 4. Isolation of pathogens by inpatient and outpatient/community settings.
| Inpatient (15 total studies) | Outpatient/Community (8 total studies) | |||
| Pathogen | Mean (# of studies included) | Median [IQR] | Mean (# of studies included) | Median [IQR] |
| Adenovirus | – | – | 7 (1) | 7 [NA] |
| Aeromonas spp. | 0.2 (1) | 0.2 [NA] | 0.8 (2) | 0.8 [0.4, 1.2] |
| Campylobacter spp. | 4.1 (6) | 1.4 [0.5, 5.9] | 1.5 (2) | 1.5 [1.5, 1.6] |
| C. difficile | 1.5 (2) | 1.5 [0.7, 2.2] | – | – |
| Cryptosporidia spp. | 1.9 (3) | 1.3 [0.9, 2.2] | 6.4 (2) | 6.9 [4.9, 7.0] |
| E. histolytica | 3.1 (3) | 2.3 [2.1, 3.3] | 10.7 (1) | 10.7 [NA] |
| EPEC/EAEC | 4.0 (1) | 4.0 [NA] | 4.7 (2) | 4.7 [3.7, 5.6] |
| ETEC | 14.0 (4) | 9.5 [3.4, 20.2] | 5.9 (2) | 5.7 [3.5, 8.1] |
| EHEC | – | – | 0 (1) | 0 [NA] |
| EIEC | – | – | 4 (2) | 4 [2.7, 5.3] |
| Giardia spp. | 2.5 (3) | 2.2 [2.2, 2.6] | 2.5 (2) | 2.5 [1.2, 3.7] |
| Norovirus | 10 (1) | 10 [NA] | 8.5 (1) | 8.5 [NA] |
| P. shigelloides | 0.2 (1) | 0.2 [NA] | – | – |
| Rotavirus | 3.1 (3) | 4.1 [2.0, 4.7] | 2.1 (3) | 2.3 [1.9, 2.4] |
| Salmonella spp. | 8.4 (5) | 3.3 [2.3, 11.4] | 20.4 (2) | 20.4 [13.5, 27.3] |
| Shigella spp. | 6.5 (8) | 4.3 [3.0, 10.2] | 19.6 (4) | 18.5 [10.3, 27.8] |
| V. cholerae (O1/O139) | 15.3 (5) | 14.0 [11.9, 30.2] | – | – |
| V. parahaemolyticus | 1.6 (2) | 1.6 [0.8, 2.4] | – | – |
| Yersinia spp. | 0 (1) | 0 [NA] | 0 (1) | 0 [NA] |
In Table 5 we present the analysis of inpatient studies stratified by the number of pathogens sought among those studies conducted in low and middle income countries. We separately present the results for the single analysis which included more than 4 pathogens conducted in a high income setting [19]. There were very few single pathogen studies thus it is difficult to identify a trend as one progresses from single to comprehensive studies with at least 5 pathogens. In the studies conducted in low and middle income countries which identified at least 5 pathogens, 28.1% of hospitalized patients had tested positive for ETEC and 20.7% tested positive for V. cholera O1/O139. For high income/low mortality countries, one study found that 14% of hospitalized patients tested positive for Campylobacter spp. followed by 11.5% of samples testing positive for Salmonella spp. [19].
Table 5. Isolation of single vs. multiple pathogens among inpatients.
| Weighted mean % (# of studies) for low and middle income countries | One study representing high income countries [19] | |||
| Pathogen | Single Pathogen Studies | 2–4 Pathogens Studies | >4 Pathogen Studies | Percent of patients positive for each pathogen |
| EPEC/EAEC | – | – | 4 (1) | |
| ETEC | – | – | 28.2 (4) | |
| Salmonella spp. | – | 2.7 (2) | 12.3 (2) | 11.5 |
| Shigella spp. | 0.2 (1) | 3.7 (2) | 6.7 (4) | 3.1 |
| Campylobacter spp. | 0.5 (2) | 2.3 (2) | 5.7 (2) | 14 |
| Cryptosporidia spp. | 1.3 (3) | – | – | |
| Aeromonas spp. | – | – | 0.2 | |
| Yersinia spp. | – | – | 0 (1) | |
| Giardia spp. | – | – | 2.2 (3) | |
| Rotavirus | – | – | 3.9 (3) | |
| Norovirus | 10 (1) | – | – | |
| V. cholerae O1/O139 | 11.9 (1) | 30.2 (1) | 20.7 (3) | |
| V. parahaemolyticus | – | – | 0.3 (2) | |
| E. histolytica | – | – | 3.8 (3) | |
| P. shigelloides | – | – | 0.2 | |
| C. difficile | – | – | 0.2 (1) | 2.9 |
For outpatient studies we only identified studies of single pathogens and those which looked for more than 4 pathogens (Table 6). The difference in proportion of stools testing positive for a particular pathogen is most noticeable for Shigella spp. where 34.3% of episodes were positive for Shigella in studies that sought only that pathogen, vs. only 9.4% positive among studies which looked for 5 of more pathogens.
Table 6. Isolation of single vs. multiple pathogens for outpatient studies.
| Weighted Mean (# of studies) | ||
| Pathogen | Single Pathogen | >4 Pathogens |
| EPEC/EAEC | – | 5.2 (2) |
| ETEC | 4.7 (1) | 4.6 (1) |
| EHEC | – | 0 (1) |
| EIEC | – | 2.6 (2) |
| Salmonella spp. | – | 17 (2) |
| Shigella spp. | 35.4 (2) | 9.3 (2) |
| Campylobacter spp. | – | 1.5 (2) |
| Cryptosporidia spp. | 6.9 (1) | 2 (1) |
| Aeromonas spp. | – | 1 (2) |
| Yersinia spp. | – | 0(1) |
| Giardia spp. | – | 3.1 (2) |
| E. histolytica | – | 10.7 (1) |
| Rotavirus | 2.3 (1) | 2.1 (2) |
| Norovirus | 8.5 (1) | – |
| Adenovirus | – | 2.6 (1) |
Discussion
This is the first systematic review which has considered the relative importance of multiple diarrhea pathogens for all regions of the world among children 5 years and older, adolescents, and adults using studies published in the peer reviewed literature. We stratified our results by inpatient vs. outpatient settings because it is likely that the distribution of pathogens differs by diarrhea severity. We found ETEC and V. cholerae O1/O139 to be the most frequently isolated pathogens among patients hospitalized for diarrhea; together they were observed in more than 49% of samples from patients in low and middle income countries. Because these studies were conducted in cholera endemic areas this is not surprising; the importance of cholera will depend on whether the study was done in an endemic or epidemic area thus these results are not possible to generalize to all countries. Rotavirus, which is known to be a leading cause of death among young children, was not found to be as important among older persons providing additional evidence suggesting immunity with increasing age.
In outpatient settings, Salmonella spp., Shigella spp., and E. histolytica were the most frequently isolated pathogens. Because little is known about the care-seeking behavior for community-acquired diarrhea among children 5 years of age and older and adults, additional data are needed in this age group to determine the distribution of pathogens in the community. Because blood in the stool is common for illnesses due to Shigella spp., Campylobacter spp., and E. histolytica and may occur with Salmonella spp. it is possible that the isolation of these pathogens would be higher than in a true community-based setting due to an increase in care-seeking behavior for illnesses with the presence of blood in the stool. We only identified one community-based study; thus, separate estimates for outpatient and community studies were not possible.
The overall scarcity of the data used to produce these estimates is a major limitation. This is particularly concerning when generalizing across regions and when making assumptions about variations which are likely among low, middle, and high income countries based on variation in geography and risk factors. Given the few studies meeting our criteria for inclusion in the review, it is not possible to account for the additional differences in study populations by region or over time which might have also influenced the spectrum of pathogens due to changes in pathogens chosen for isolation, pathogens circulating in a community, and baseline characteristics of the study population.
An additional limitation of this review is the time span of the included studies and thus heterogeneity of laboratory methods for some key pathogens. In the last 30 years, diagnostic methods have evolved for many pathogens, such as diarrheagenic E. coli and E. histolytica. New laboratory methods, including PCR, and antigen detection assays have increased sensitivity and decreased risk of misclassification substantially. Because some reports included in this review used older laboratory methods there is a risk that data from these may under- or over-represent the prevalence of selected pathogens. However, because of the overall paucity of data we chose to include these studies however caution should be taken when interpreting the results for these selected pathogens for which laboratory methods have improved dramatically over the past 30 years.
In this review we stratified studies by those that sought a single pathogen and those that considered multiple pathogens. Because single pathogen studies often pick study sites based on a known prevalence of a particular pathogen it can be expected that the observed rates would be higher than in studies where multiple pathogens are being isolated. This was especially true for Shigella spp. where we found the weighted mean dropped from 35.3% in the single pathogen studies to 9.4% in the multiple pathogen studies. In addition, outpatient studies did not look specifically for some pathogens such as V. cholerae O1/O139 thus limiting the inference about non severe episodes.
We recognize that we did not capture the true burden of every possible pathogen that might cause diarrhea because many pathogens occur in outbreaks and these may not have been included in these ongoing disease surveillance studies. For example, we only found one study that included detection of norovirus [15] meeting our study inclusion criteria of at least 12 mo of surveillance. Norovirus is known to be seasonal and a frequent cause of epidemics, so may be underestimated in our review. Similarly because we did not include outbreak data, pathogens that are more typically observed in outbreaks may have been missed if they were not known to be endemic in the study area.
Because we identified very few studies that tested for 5 pathogens or more and most were from South Asia, we were not able to assess regional differences in pathogen importance. For pathogens that are not known to be prevalent globally such as V. cholerae O1/O139 this is especially problematic. Ideally unique pathogen distributions would be developed for each region and for large countries, such as Brazil, China, and India. National level community-based surveillance and inpatient reports would enable countries to better understand the local burden of disease by pathogen and better design prevention programs.
In this analysis we have treated all data as isolated proportions yet we recognize that this is not the case for many diarrhea episodes. Many patients have multiple pathogens and likewise for some patients, no pathogen is found. Because we had very few studies seeking multiple pathogens and even fewer reporting mixed infections, we were not able to conduct a more complex analysis to control for the role of multiple infections. We also recognize that the identification of a pathogen in the stool does not necessary mean that it is the cause of the illness. Many patients are asymptomatic carriers and thus the prevalence of some pathogens might be found at the similar proportions in healthy individuals. These pathogens have a lower pathogenicity than those that are never or rarely identified in the stools of asymptomatic individuals. Only one study in our final data set provided data for asymptomatic controls, thus a full analysis to control for asymptomatic carriage was not possible.
This study is the first to systematically review the literature on the etiology of diarrhea in children ≥5 years of age, adolescents and adults and provides an important overview of the distribution of pathogens responsible for both infection and possible death. The few studies identified suggest that great caution must be taken when interpreting these limited data. Many limitations have been identified suggesting the need for additional prospective studies around the world in these age groups. Understanding the burden of pathogen specific diarrheal disease and the variation by region is important for planning effective control programs for the overall reduction of diarrhea disease among persons of all ages.
Supporting Information
PRISMA checklist.
(0.07 MB DOC)
Acknowledgments
This work was undertaken as a part of the Global Burden of Diseases, Injuries, and Risk Factors Study. The authors thank the members of the WHO Foodborne Disease Burden Epidemiology Reference Group (FERG) for their invaluable feedback and assistance identifying unpublished data sources. We also thank the Melinda Munos, Hilda Ndirangu, and Ramya Amyakutty for their help with the initial literature review and data abstraction. The results in this paper are prepared independently of the final estimates of the Global Burden of Diseases, Injuries, and Risk Factors Study.
Footnotes
The authors have declared that no competing interests exist.
This work was undertaken as a part of the Global Burden of Diseases, Injuries, and Risk Factors Study. A grant from the Bill and Melinda Gates Foundation supported the Study's core activities and partially supported the epidemiologic reviews in this paper. This work was jointly funded by the World Health Organization - Foodborne Disease Burden Epidemiology Reference Group. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Boschi-Pinto C, Velebit L, Shibuya K. Estimating child mortality due to diarrhoea in developing countries. Bull World Health Organ. 2008;86:710–717. doi: 10.2471/BLT.07.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. The global burden of disease: 2004 update. Geneva: World Health Organization; 2008. [Google Scholar]
- 3.Amirlak I, Amirlak B. Haemolytic uraemic syndrome: an overview. Nephrology (Carlton) 2006;11:213–218. doi: 10.1111/j.1440-1797.2006.00556.x. [DOI] [PubMed] [Google Scholar]
- 4.Allos BM. Campylobacter jejuni Infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 5.Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, et al. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med. 2006;354:23–33. doi: 10.1056/NEJMoa052664. [DOI] [PubMed] [Google Scholar]
- 6.Cholera vaccines: WHO position paper. Weekly Epidemiological Reference. 2010;85:117–128. [PubMed] [Google Scholar]
- 7.RefWorks. RefWorks: Scopus Edition. 2nd edition ed. North America: RefWorks; 2009. [Google Scholar]
- 8.Murray PR, Baron E, Jorgensen JH, Landry M, Pfaller MA, editors. Washington, D.C.: ASM Press; 2007. Manual of clinical microbiology; [Google Scholar]
- 9.WHO. Manual for Laboratory Investigations of Acute Enteric Infections. Geneva: World Health Organization; 1987. [Google Scholar]
- 10.Spencer HC, Wells JG, Gary GW, Sondy J, Puhr ND, et al. Diarrhea in a non-hospitalized rural Salvadoran population: the role of enterotoxigenic Escherichia coli and rotavirus. Am J Trop Med Hyg. 1980;29:246–253. doi: 10.4269/ajtmh.1980.29.246. [DOI] [PubMed] [Google Scholar]
- 11.Cause-specific adult mortality: evidence from community-based surveillance–selected sites, Tanzania, 1992–1998. MMWR Morb Mortal Wkly Rep. 2000;49:416–419. [PubMed] [Google Scholar]
- 12.Gassama A, Sow PS, Fall F, Camara P, Gueye-N'diaye A, et al. Ordinary and opportunistic enteropathogens associated with diarrhea in Senegalese adults in relation to human immunodeficiency virus serostatus. Int J Infect Dis. 2001;5:192–198. doi: 10.1016/S1201-9712(01)90069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Germani Y, Morillon M, Begaud E, Dubourdieu H, Costa R, et al. Two-year study of endemic enteric pathogens associated with acute diarrhea in New Caledonia. J Clin Microbiol. 1994;32:1532–1536. doi: 10.1128/jcm.32.6.1532-1536.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hossain MA, Albert MJ, Hasan KZ. Epidemiology of shigellosis in Teknaf, a coastal area of Bangladesh: a 10-year survey. Epidemiol Infect. 1990;105:41–49. doi: 10.1017/s0950268800047622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau CS, Wong DA, Tong LK, Lo JY, Ma AM, et al. High rate and changing molecular epidemiology pattern of norovirus infections in sporadic cases and outbreaks of gastroenteritis in Hong Kong. J Med Virol. 2004;73:113–117. doi: 10.1002/jmv.20066. [DOI] [PubMed] [Google Scholar]
- 16.Lim YS, Tay L. A one-year study of enteric Campylobacter infections in Singapore. J Trop Med Hyg. 1992;95:119–123. [PubMed] [Google Scholar]
- 17.Sitbon M, Lecerf A, Garin Y, Ivanoff B. Rotavirus prevalence and relationships with climatological factors in Gabon, Africa. J Med Virol. 1985;16:177–182. doi: 10.1002/jmv.1890160210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasfy MO, Oyofo BA, David JC, Ismail TF, el-Gendy AM, et al. Isolation and antibiotic susceptibility of Salmonella, Shigella, and Campylobacter from acute enteric infections in Egypt. J Health Popul Nutr. 2000;18:33–38. [PubMed] [Google Scholar]
- 19.Watson B, Ellis M, Mandal B, Dunbar E, Whale K, et al. A comparison of the clinico-pathological features with stool pathogens in patients hospitalised with the symptom of diarrhoea. Scand J Infect Dis. 1986;18:553–559. doi: 10.3109/00365548609021662. [DOI] [PubMed] [Google Scholar]
- 20.Zaman R. Campylobacter enteritis in Saudi Arabia. Epidemiol Infect. 1992;108:51–58. doi: 10.1017/s0950268800049499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MoezArdalan K, Zali MR, Dallal MM, Hemami MR, Salmanzadeh-Ahrabi S. Prevalence and pattern of antimicrobial resistance of Shigella species among patients with acute diarrhoea in Karaj, Tehran, Iran. J Health Popul Nutr. 2003;21:96–102. [PubMed] [Google Scholar]
- 22.Nagamani K, Pavuluri PR, Gyaneshwari M, Prasanthi K, Rao MI, et al. Molecular characterisation of Cryptosporidium: an emerging parasite. Indian J Med Microbiol. 2007;25:133–136. doi: 10.4103/0255-0857.32719. [DOI] [PubMed] [Google Scholar]
- 23.Nath G, Choudhury A, Shukla BN, Singh TB, Reddy DC. Significance of Cryptosporidium in acute diarrhoea in North-Eastern India. J Med Microbiol. 1999;48:523–526. doi: 10.1099/00222615-48-6-523. [DOI] [PubMed] [Google Scholar]
- 24.Oberle MW, Merson MH, Islam MS, Rahman AS, Huber DH, et al. Diarrhoeal disease in Bangladesh: epidemiology, mortality averted and costs at a rural treatment centre. Int J Epidemiol. 1980;9:341–348. doi: 10.1093/ije/9.4.341. [DOI] [PubMed] [Google Scholar]
- 25.Poocharoen L, Bruin CW. Campylobacter jejuni in hospitalized children with diarrhoea in Chiang Mai, Thailand. Southeast Asian J Trop Med Public Health. 1986;17:53–58. [PubMed] [Google Scholar]
- 26.Al-Gallas N, Bahri O, Bouratbeen A, Ben Haasen A, Ben Aissa R. Etiology of acute diarrhea in children and adults in Tunis, Tunisia, with emphasis on diarrheagenic Escherichia coli: prevalence, phenotyping, and molecular epidemiology. Am J Trop Med Hyg. 2007;77:571–582. [PubMed] [Google Scholar]
- 27.Baqui AH, Yunus MD, Zaman K, Mitra AK, Hossain KM. Surveillance of patients attending a rural diarrhoea treatment centre in Bangladesh. Trop Geogr Med. 1991;43:17–22. [PubMed] [Google Scholar]
- 28.Black RE, Merson MH, Rahman AS, Yunus M, Alim AR, et al. A two-year study of bacterial, viral, and parasitic agents associated with diarrhea in rural Bangladesh. J Infect Dis. 1980;142:660–664. doi: 10.1093/infdis/142.5.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandonisio O, Marangi A, Panaro MA, Marzio R, Natalicchio MI, et al. Prevalence of Cryptosporidium in children with enteritis in southern Italy. Eur J Epidemiol. 1996;12:187–190. doi: 10.1007/BF00145505. [DOI] [PubMed] [Google Scholar]
- 30.Vilchis-Guizar AE, Uribe-Marquez S, Perez-Sanchez PL. [The clinico-epidemiological characteristics of cholera patients in Mexico City]. Salud Publica Mex. 1999;41:487–491. [PubMed] [Google Scholar]
- 31.Echeverria P, Blacklow NR, Cukor GG, Vibulbandhitkit S, Changchawalit S, et al. Rotavirus as a cause of severe gastroenteritis in adults. J Clin Microbiol. 1983;18:663–667. doi: 10.1128/jcm.18.3.663-667.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaman K, Yunus M, Baqui AH, Hossain KM. Surveillance of shigellosis in rural Bangladesh: a 10 years review. J Pak Med Assoc. 1991;41:75–78. [PubMed] [Google Scholar]
- 33.Abreu-Acosta N, Quispe MA, Foronda-Rodriguez P, Alcoba-Florez J, Lorenzo-Morales J, et al. Cryptosporidium in patients with diarrhoea, on Tenerife, Canary Islands, Spain. Ann Trop Med Parasitol. 2007;101:539–545. doi: 10.1179/136485907X193798. [DOI] [PubMed] [Google Scholar]
- 34.Akinyemi KO, Oyefolu AO, Opere B, Otunba-Payne VA, Oworu AO. Escherichia coli in patients with acute gastroenteritis in Lagos, Nigeria. East Afr Med J. 1998;75:512–515. [PubMed] [Google Scholar]
- 35.Amarilla A, Espinola EE, Galeano ME, Farina N, Russomando G, et al. Rotavirus infection in the Paraguayan population from 2004 to 2005: high incidence of rotavirus strains with short electropherotype in children and adults. Med Sci Monit. 2007;13:CR333–337. [PubMed] [Google Scholar]
- 36.Battikhi MN. Epidemiological study on Jordanian patients suffering from diarrhoea. New Microbiol. 2002;25:405–412. [PubMed] [Google Scholar]
- 37.Bern C, Hernandez B, Lopez MB, Arrowood MJ, De Merida AM, et al. The contrasting epidemiology of Cyclospora and Cryptosporidium among outpatients in Guatemala. Am J Trop Med Hyg. 2000;63:231–235. [PubMed] [Google Scholar]
- 38.Bingnan F, Unicomb LE, Tu GL, Ali A, Malek A, et al. Cultivation and characterization of novel human group A rotaviruses with long RNA electropherotypes, subgroup II specificities, and serotype 2 VP7 genes. J Clin Microbiol. 1991;29:2224–2227. doi: 10.1128/jcm.29.10.2224-2227.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabrita J, Pires I, Vlaes L, Coignau H, Levy J, et al. Campylobacter enteritis in Portugal: epidemiological features and biological markers. Eur J Epidemiol. 1992;8:22–26. doi: 10.1007/BF02427387. [DOI] [PubMed] [Google Scholar]
- 40.Cassel-Beraud AM, Morvan J, Rakotoarimanana DR, Razanamparany M, Candito D, et al. [Infantile diarrheal diseases in Madagascar: bacterial, parasitologic and viral study]. Arch Inst Pasteur Madagascar. 1990;57:223–254. [PubMed] [Google Scholar]
- 41.Chatterjee BD, Thawani G, Sanyal SN. Etiology of acute childhood diarrhoea in Calcutta. Trop Gastroenterol. 1989;10:158–166. [PubMed] [Google Scholar]
- 42.Faruque AS, Malek MA, Khan AI, Huq S, Salam MA, et al. Diarrhoea in elderly people: aetiology, and clinical characteristics. Scand J Infect Dis. 2004;36:204–208. doi: 10.1080/00365540410019219. [DOI] [PubMed] [Google Scholar]
- 43.Feizabadi MM, Dolatabadi S, Zali MR. Isolation and drug-resistant patterns of Campylobacter strains cultured from diarrheic children in Tehran. Jpn J Infect Dis. 2007;60:217–219. [PubMed] [Google Scholar]
- 44.Gambhir IS, Jaiswal JP, Nath G. Significance of Cryptosporidium as an aetiology of acute infectious diarrhoea in elderly Indians. Trop Med Int Health. 2003;8:415–419. doi: 10.1046/j.1365-3156.2003.01031.x. [DOI] [PubMed] [Google Scholar]
- 45.Hamedi Y, Safa O, Haidari M. Cryptosporidium infection in diarrheic children in southeastern Iran. Pediatr Infect Dis J. 2005;24:86–88. doi: 10.1097/01.inf.0000148932.68982.ec. [DOI] [PubMed] [Google Scholar]
- 46.Katsumata T, Hosea D, Wasito EB, Kohno S, Hara K, et al. Cryptosporidiosis in Indonesia: a hospital-based study and a community-based survey. Am J Trop Med Hyg. 1998;59:628–632. doi: 10.4269/ajtmh.1998.59.628. [DOI] [PubMed] [Google Scholar]
- 47.Libanore M, Bicocchi R, Rossi MR, Montanari P, Sighinolfi L, et al. [Incidence of giardiasis in adults patients with acute enteritis]. Minerva Med. 1991;82:375–380. [PubMed] [Google Scholar]
- 48.Rahman M, Shahid NS, Rahman H, Sack DA, Rahman N, et al. Cryptosporidiosis: a cause of diarrhea in Bangladesh. Am J Trop Med Hyg. 1990;42:127–130. doi: 10.4269/ajtmh.1990.42.127. [DOI] [PubMed] [Google Scholar]
- 49.Samonis G, Maraki S, Christidou A, Georgiladakis A, Tselentis Y. Bacterial pathogens associated with diarrhoea on the island of Crete. Eur J Epidemiol. 1997;13:831–836. doi: 10.1023/a:1007318003083. [DOI] [PubMed] [Google Scholar]
- 50.Simadibrata M, Tytgat GN, Yuwono V, Daldiyono, Lesmana LA, et al. Microorganisms and parasites in chronic infective diarrhea. Acta Med Indones. 2004;36:211–214. [PubMed] [Google Scholar]
- 51.Uchida R, Pandey BD, Sherchand JB, Ahmed K, Yokoo M, et al. Molecular epidemiology of rotavirus diarrhea among children and adults in Nepal: detection of G12 strains with P[6] or P[8] and a G11P[25] strain. J Clin Microbiol. 2006;44:3499–3505. doi: 10.1128/JCM.01089-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Seidlein L, Kim DR, Ali M, Lee H, Wang X, et al. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 2006;3:e353. doi: 10.1371/journal.pmed.0030353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zvizdic S, Telalbasic S, Beslagic E, Cavaljuga S, Maglajlic J, et al. Clinical characteristics of rotaviruses disease. Bosn J Basic Med Sci. 2004;4:22–24. doi: 10.17305/bjbms.2004.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das P, Sengupta K, Dutta P, Bhattacharya MK, Pal SC, et al. Significance of Cryptosporidium as an aetiologic agent of acute diarrhoea in Calcutta: a hospital based study. J Trop Med Hyg. 1993;96:124–127. [PubMed] [Google Scholar]
- 55.Echeverria P, Seriwatana J, Taylor DN, Yanggratoke S, Tirapat C. A comparative study of enterotoxigenic Escherichia coli, Shigella, Aeromonas, and Vibrio as etiologies of diarrhea in northeastern Thailand. Am J Trop Med Hyg. 1985;34:547–554. doi: 10.4269/ajtmh.1985.34.547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA checklist.
(0.07 MB DOC)