Abstract
p53 is arguably the most intensively studied protein to date, yet there is much that we ignore about its function as a transcription factor. The p53-dependent transcriptional program is remarkably flexible, as it varies with the nature of p53-activating stimuli, the cell type and the duration of the activation signal. This flexibility may allow cells to mount alternative responses to p53 activation, such as cell cycle arrest or apoptosis. Here, I organize the available data into two alternative models to explain how this regulatory diversity is achieved.
Keywords: p21, ASPP, HZF, hCAS, serine 46, lysine 120
p53, a jack-of-all-trades
p53 is a multifaceted actor capable of playing distinct roles in different scenarios. Cells may die, arrest or senesce in response to increased p53 activity. The same cell that undergoes p53-dependent apoptosis upon stimulus X triggers p53-dependent cell cycle arrest in response to stimulus Y. In turn, not all cell types respond equally to stimulus X, as some arrest whereas others die. Examples of this diversity abound in the literature. Stimulus-specific effects were demonstrated early on by the Vogelstein lab when showing that, whereas p53 protects colon carcinoma cells from the apoptotic effects of the drug doxorubicin by enforcing a cell cycle arrest response, it mediates the apoptotic effects of the drug 5-fluorouracil (5-FU) in the same cell types (Bunz et al., 1999). Cell type-specific responses are clearly illustrated by recent work from the Vassilev group, which treated many cell lines with Nutlin-3, a small molecule inhibitor of the p53 repressor MDM2. Whereas some carcinoma cell lines can undergo reversible cell cycle arrest without signs of cell death for several days, others undergo apoptosis (Tovar et al., 2006). Furthermore, mouse cells of fibroblast origin undergo senescence after only 24 h of Nutlin-3 treatment (Efeyan et al., 2007). How can these stimulus- and cell type-specific p53-dependent responses be explained? p53 is a sophisticated transcription factor with complex functional domains (reviewed by Laptenko and Prives (2006)). A plausible explanation is that p53 regulates different subsets of downstream target genes in different scenarios, and that said subsets mediate different biological responses.
The p53 transcriptional program is astonishingly flexible. In early microarray experiments, the Levine group demonstrated that p53 provokes distinct gene expression signatures depending on the activating stimuli utilized (that is, ionizing radiation versus non-ionizing radiation versus mere overexpression) (Zhao et al., 2000). In fact, the subset of p53 target genes activated in all experimental conditions tested was very small. Tissue-specific induction of p53 target genes is also evident in vivo (Fei et al., 2002). These observations raise the question: how are specific subsets of p53 target genes selected in different scenarios? A significant amount of data has been generated toward answering this question. The aim of this review is to organize the available evidence into two alternative models. In one model, referred hereto after as the ‘The Selective Binding Model’, target gene selectivity is achieved at the level of p53 binding to DNA. In the alternative model, termed ‘The Selective Context Model’, p53 binds to all accessible genomic-binding sites, and specificity is achieved at subsequent regulatory steps. I apologize in advance for the many good reports that I will not cite in this review. Instead of covering numerous reports lightly, I have decided to explore a few of them in greater detail, which forces me to leave aside many important works.
The Selective Binding Model: to bind or not to bind?
Several reports support the idea that the ability of p53 to recognize its genomic-binding sites can be modulated in a gene-specific manner. Since DNA binding is a prerequisite for transcription factor action, it is easy to envision how selectivity could be achieved at this early biochemical step. Specifically, certain p53 cofactors and p53 post-translational modifications have been demonstrated to enable p53 to discriminate among its target genes. Before I discuss these reports in detail, I need to introduce the key player of this model: the p53 response element (p53RE).
The p53RE: it is all in the DNA?
An underlying assumption of The Selective Binding Model is that not all p53REs are equal. In order for p53 to discriminate among its target genes at the level of DNA binding, the corresponding p53REs must be somehow different.
Keyword: sequence
The p53RE DNA consensus comprises two palindromic 10 bp repeats or ‘half-sites’ (RRRCWWGYYY) (R is purine, Y is pyrimidine, W is A or T), which are infrequently separated by a spacer of a few nucleotides (el-Deiry et al., 1992; Hoh et al., 2002; Wei et al., 2006). To the transcription factor aficionado, two features of the p53RE catch the eye, that is, length and degeneracy. At 20 bp, the p53RE is relatively long. In contrast, MYC binds to 6 bp E boxes, E2Fs bind to 8 bp motifs and nuclear hormone receptors recognize 12 bp sequences. It could be argued that the only reason for the long length of the p53RE is that p53 binds DNA as a tetramer, thus the need for the four 5-mer repeats. But then, why does p53 bind DNA as a tetramer? Quadrupling the size of a monomer-bound ‘primordial’ p53RE would have exponentially increased the number of possible distinct p53REs, perhaps to create regulatory diversity. Without considering the spacer, a simple mathematical calculation says that there are > 65 000 different sequences that fit the consensus. The fact that many validated p53REs deviate from the consensus at one or more positions indicates that the aforementioned number is an underestimation of the possible (Gohler et al., 2002; Horvath et al., 2007). Is there a reason for this diversity? Many lines of evidence suggest so.
Keyword: affinity
It is well demonstrated that p53 has different affinities for distinct p53REs, which would allow the p53 transcriptional program to be modulated by the nuclear concentration of p53 (Resnick-Silverman et al., 1998; Szak et al., 2001). At low concentrations of p53, only target genes carrying high affinity p53REs would be regulated. At high concentrations, the repertoire of target genes will increase. In turn, differential affinities should translate into distinct kinetics of activation. Using microarray analysis and an inducible p53 system, Zhao et al. (2000) clearly established that low levels of p53 modulate only a subset of those target genes regulated by high levels of p53. Furthermore, they identified five distinct groups of p53 target genes based on their different kinetics of induction. It remains to be determined if differential kinetics correlate with the affinity of the corresponding p53REs.
Keyword: topology
As mentioned above, functional genomic p53REs rarely fit perfectly to the consensus, with one or more noncompliant bases being the norm (Gohler et al., 2002). One interpretation of this puzzle is that there are other determinants for p53 binding beyond the linear DNA sequence, such as DNA topology. Work by the Deppert group clearly demonstrates that architecture is an important determinant of sequence-specific DNA binding by p53 (Gohler et al., 2002). They notice that many p53REs deviating from the consensus display an internal twofold symmetry that allows for intra-strand pairing and formation of stem-loop structures. When presented in linear conformation, these sites are poorly recognized by p53. When presented in stem-loop conformation, these sites are bound with high affinity. Significantly, the effects of topology vary among p53REs of different sequence. An extreme illustration on the role of topology is provided by work from the Jovin lab showing that p53 can specifically bind to cruciform-forming sequences having no resemblance to the p53RE consensus whatsoever (Jett et al., 2000). Furthermore, p53-dependent activation of the Pig3 gene is driven by a (TGYCC)n microsatellite repeat with little resemblance to the consensus p53RE (Contente et al., 2002).
Variations in sequence, affinity and topology persuade one to think that the p53RE is a huge repository of regulatory diversity within the p53 network, but before we fully embrace this idea, we should test some of the predictions it generates. First, one would expect that meaningful differences among p53REs would be evolutionarily conserved. Second, one would expect that the p53REs within genes of different functions (for example, cell cycle arrest versus apoptosis) could be discriminated from each other on the basis of sequence, affinity or topology. Extensive bioinformatics analysis of validated p53REs carried out by Horvath et al. (2007) indicates that these expectations are not met (Horvath et al., 2007). First of all, in contrast to the REs for the transcription NFκB and NRF2, the p53REs are notorious for their lack of conservation among mammals, which somewhat discourages the idea that the differences observed have functional relevance. Second, their careful analysis fails to distinguish a defining sequence feature between cell cycle arrest and apoptotic p53REs. Instead, they found that, as a group, cell cycle p53REs are more conserved than apoptotic p53REs, suggesting that the cell cycle arrest program may have a more ancient root within the network and that the pro-apoptotic module may be too young for us to detect identifying features. Despite these caveats, and if one does accept that diversity in the cellular pool of p53REs is relevant, how is then this diversity decoded? How do differences among p53REs lead to selective regulation? It is possible that these differences impose a gene-specific requirement for p53-binding partners and/or post-translational modifications. Accordingly, several reports describe such phenomena.
Apoptosis-stimulating proteins of p53
The apoptosis-stimulating proteins of p53 (ASPP) family comprises ASPP1, ASPP2 and inhibitory ASPP (iASPP) (Slee and Lu, 2003). These proteins share sequence similarity in their C-terminal domains, which contain four ankyrin repeats and a SH3 domain, two motifs involved in protein–protein interactions. ASPP1 and ASPP2 bind to p53 through their C termini and stimulate the p53 apoptotic but not cell cycle arrest activity, whereas iASPP is an inhibitor of p53-dependent apoptosis conserved from worms to humans (Samuels-Lev et al., 2001; Slee and Lu, 2003; Bergamaschi et al., 2004, 2006). How do ASPPs achieve their selective effects? Chromatin immunoprecipitation (ChIP) analysis indicates that ASPP1 and ASPP2 can selectively stimulate the binding of p53 to the p53REs in Bax and Pig3, but not to those in p21waf1 or Mdm2, thus preferentially promoting apoptosis (Samuels-Lev et al., 2001). The mechanism by which ASPPs stimulate DNA binding selectively is unknown. One could envision that ASPP binding would induce a conformational change in p53 to affect its DNA-binding properties in a gene-specific fashion. The co-crystal structure of the C terminus of ASPP2 bound to the p53 DNA-binding domain (DBD) obtained by Gorina and Pavlevitch reveals that the SH3 domain of ASPP2 binds to the L3 loop of the DBD, whereas one of the ankyrin repeats of ASPP2 binds to the L2 loop (Gorina and Pavletich, 1996). Importantly, the contact residues in p53 are highly conserved and commonly mutated in cancer, suggesting that disruption of the p53–ASPP interaction may be an oncogenic event. Interestingly, mutation of the contact residue R181 in p53’s L2 loop blocks p53-dependent apoptosis but not cell cycle arrest, agreeing with the notion that ASPPs play gene-specific effects (Ludwig et al., 1996). However, the crystal structure reveals that the site of ASPP2 binding overlaps the site of DNA binding, and analysis of tumor-derived p53 mutations falling in this region of p53 indicates that they disrupt both ASPP2 binding and DNA binding. This led Gorina and Pavlevitch to conclude that disruption of ASPP2 binding by tumorigenic mutations may be secondary to disruption of p53 DNA binding activity, and that loss of ASPP2 binding may have little or no functional consequence for tumorigenesis. Nevertheless, they also note that mutations in residues involved in ASPP2 binding, but not in DNA binding, are also found in cancers, albeit at lower rates. Intriguingly, the structure of the p53 DBD is identical in the complex with ASPP2 as compared to the free or DNA-bound DBD, thus discouraging the idea of allosteric regulation by ASPP2. Furthermore, the fact that DNA and ASSPs use many common contact surfaces within the p53 DBD argues that, if anything, ASPPs should block p53 binding to DNA. Clearly, additional mechanistic studies are necessary to fully understand how ASPPs modulate p53 DNA-binding activity. It would be useful to determine to what extent the DNA-binding properties of p53 are affected by ASPPs. What other p53REs are positively affected by ASPPs beyond Bax and Pig3? Are there p53REs whose binding is actually inhibited by ASPPs function? Can the effects of ASPP2 on p53 DNA binding be recapitulated in a defined system with recombinant proteins and pure DNA or do these effects require additional cellular factors and/or a chromatin environment? Can the effects of ASPPs be modulated by post-translational modifications on p53 or ASPPs? Expression analysis of human tumor samples and genetic studies in mice sustain the notion that ASPP2 is a tumor suppressor that collaborates with p53 (Vives et al., 2006a, b). Bringing answers to the questions formulated above will help us understand the exact mechanism driving these tumor protective effects.
The p53 family members
In 2002, a report by Flores et al. (2002) concluded that ‘a combined loss of p63 and p73 results in the failure of cells containing functional p53 to undergo apoptosis in response to DNA damage’. Using knockouts, Flores et al. (2002) showed that E1A-transformed p63−/− p73−/− murine embryonic fibroblasts (MEFs) were as resistant to apoptosis induced by doxorubicin, cisplatin or ionizing radiation as p53−/− MEFs. Furthermore, in the absence of p63 and p73, p53 failed to activate expression of the apoptotic genes Perp and Bax, yet it efficiently activated expression of p21waf1 and Mdm2. Based on results from ChIP assays, Flores et al. concluded that p63 and/or p73 protein products are required for p53 binding to the p53REs in Perp, Bax and Noxa, but not to those in p21waf1 and Mdm2. Furthermore, they reported that p63 bound exclusively to the apoptotic p53REs. Therefore, the authors concluded that p63 and p73 are required for the recruitment of p53 to apoptotic promoters. How could p63/p73 mediate recruitment of p53 to some but not all p53REs? As mentioned above, this type of phenomena can only be understood if subsets of p53REs have some, yet unidentified, fundamental difference. At first, it seems paradoxical that proteins with similar DBDs would promote binding of each other to a common RE, rather than competing each other out. Nonetheless, plausible scenarios could be imagined. For example, p63/p73 may bind to REs not efficiently bound by p53 and recruit the latter via protein–protein interactions. However, oligomerization studies discourage this possibility, as p63 and p73 seem capable of heterotypic interactions with each other but not with p53 (Davison et al., 1999). Alternatively, a ‘priming’ model could be envisioned, in which p63/p73 could bind to a given chromatin-embedded RE not accessible to p53, and then modify said RE in such a way that it would now become ‘visible’ to p53. For example, this priming effect could be due to recruitment of nucleosome remodeling activities. Additional complexity is revealed by the fact that the phenomena reported by Flores et al. have not been observed in a different experimental paradigm. Using genetically defined, untransformed murine T lymphocytes, Senoo et al. (2004) demonstrated that p63 and p73 are fully dispensable for p53-dependent apoptosis in response to ionizing radiation. These opposing observations could be explained in different ways. First, it is possible that E1A transformation alters MEF behavior in such a way that the p53-dependent apoptotic program becomes abnormally reliant on p63 and p73. Additionally, tissue-specific effects may be involved. In either case, and assuming that the mechanism proposed by Flores et al. (2002) is correct, how could p63/p73 be required for the recruitment of p53 to apoptotic promoters in some cell types but not others, or in the same cell type under different signaling scenarios? Once again, exhaustive mechanistic studies will be required to resolve this conundrum. Finally, a more straightforward model could be envisioned, in which ΔN p63/p73 isoforms lacking the canonical transactivation domain behave as gene-specific repressors of the p53 transcriptional program by mere competition with p53 for common REs. For example, ΔNp63α, the p63 isoform predominantly expressed in epidermis, could bind with high affinity to some, but not all, p53REs, and thus exert dominant-negative effects in a gene-specific manner. In fact, it is well established that ΔNp63α is required for proliferation and survival of keratinocytes, and these effects are due at least in part to attenuation of p53 activity (Westfall et al., 2003; Truong et al., 2006; Perez et al., 2007). Similarly, ΔNp73 may repress p53 function in sympathetic neurons, as deduced from the loss of these cells in p73−/− null mice (Yang et al., 2000; Moll and Slade, 2004).
The hematopoietic zinc-finger cofactor
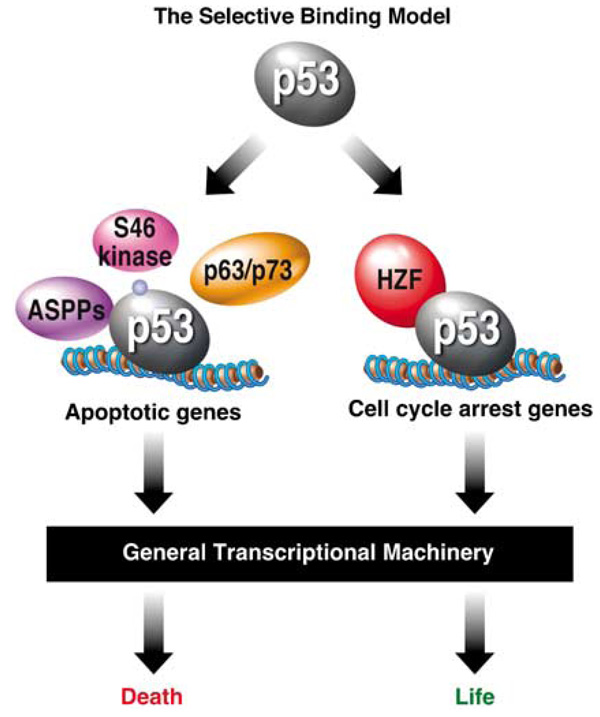
In a recent report, a novel p53 cofactor, named hematopoietic zinc-finger (HZF), has been characterized as a key determinant of gene selectivity within the p53 network (Das et al., 2007). The Hzf gene itself is transcriptionally activated by p53 and the protein product directly associates with the p53 DBD. Das et al. show that HZF facilitates p53 binding to the p53REs in p21waf1 and 14-3-3σ, inhibits p53 binding to those in Bax, Perp and Noxa, while having no effect on p53 binding to the Mdm2 and Hzf promoters. These conclusions arise from a combination of both ChIP and electrophoretic mobility shift assays (EMSA). As a consequence of such profound alteration of p53 DNA-binding properties, HZF strongly favors p53-dependent cell cycle arrest over apoptosis, as evidenced by the fact that Hzf−/− MEFs are impaired in their ability to undergo cell cycle arrest in response to p53 overexpression or DNA damage by etoposide, and displays enhanced apoptosis upon such stimuli. Finally, the authors report that sustained p53 activation promotes ubiquitination and degradation of HZF, likely via activation of an unknown ubiquitin ligase, which leads to loss of p53 binding to p21waf1, increased p53 binding to Bax and subsequent cell death. These results portray an elegant circuitry that ensures that cell cycle arrest precedes apoptosis, and that cell fate choice is determined by the duration of the p53-activating stimuli, all via mere modulation of p53 DNA-binding activity. By acting in an antagonistic fashion to ASPPs, HZF completes the Selective Binding Model, which is shown in Figure 1. However, the model for HZF action conflicts with what we currently know about p53 DNA-binding properties. First, a key feature of the model is that p53 binding to the RE in the p21waf1 promoter requires HZF. Using protein–DNA binding assays other than EMSA, independent reports amply demonstrate that pure recombinant p53 is capable of specific, high-affinity binding to the p53REs in the p21waf1 promoter in the absence of any auxiliary factors (Cain et al., 2000; Espinosa and Emerson, 2001). Second, a key prediction of the model is that, under conditions of cell cycle arrest (HZF present), levels of p53 binding should be higher at cell cycle arrest genes than apoptotic genes, whereas under conditions of apoptosis (HZF absent), the opposite behavior should be observed. Such oscillations in p53 occupancy have not been observed in independent reports. Using ChIP assays, Kaeser and Iggo tested this possibility by analysing p53 occupancy to the REs in p21waf1, Mdm2, Puma, Bax, Pig3 and p53aip1 (Kaeser and Iggo, 2002). Their results demonstrate that high affinity sites (p21waf1, Mdm2, Puma) are efficiently bound by p53 in conditions leading to either cell cycle arrest or apoptosis, whereas low affinity sites (Bax, Pig3, p53aip1) are poorly bound regardless of the ultimate cellular outcome adopted. Based on this, Kaeser and Iggo concluded that the level of chromatin-bound p53 at any given site is a mere reflection of the total levels of p53 in the cell, and that ‘the distinction between cell cycle arrest and apoptosis induction is not taken at the level of p53 binding’. Once again, additional mechanistic studies seem to be required. In particular, detailed kinetic studies of in vivo p53 binding to various p53REs should shed light onto these issues. Additionally, it will be important to determine if the effects of HZF on p53 DNA-binding activity can be reproduced in defined assays other than EMSA.
Figure 1.
The Selective Binding Model. In this model, the ability of p53 to bind to the response elements present in different target loci is modulated by cofactors and/or post-translational modifications. Cofactors such as apoptosis-stimulating proteins of p53 (ASPPs) and p63/p73 would selectively promote binding to pro-apoptotic genes, whereas cofactors such as hematopoietic zinc-finger (HZF) would promote binding to cell cycle arrest genes. The selective action of these cofactors is supported by the great diversity in sequence, affinity and topology observed among p53 response elements and by the flexible nature of the p53 DNA-binding domain (DBD). Additionally, specific p53 post-translational modifications, such as Ser46 phosphorylation, would promote binding to apoptotic genes, thus providing Ser46 kinases with key functions in cell fate choice. Once p53 binding to chromatin has been altered by cofactors or post-translational modifications, the general transcription machinery reacts to different levels of p53 at different loci to generate a transcriptional program that elicits p53-dependent cell death or cell cycle arrest. Based on this model, the cell fate choice adopted in response to p53 activation could be predicted from p53 chromatin-binding profiles.
The Serine 46 connection
I refer to it as ‘The Barcode Hypothesis’. This hypothesis postulates that there is not one p53 isoform inside cells, but many. By isoform, I am not referring here to the different protein products of the p53 gene arising by alternative promoter usage and alternative splicing (Bourdon et al., 2005). Instead, I am referring to multiple isoforms produced by post-translational modifications. p53 is phosphorylated, acetylated, methylated and ubiquitylated at multiple residues (Wahl and Carr, 2001). If all combinations of p53 post-translational modifications are indeed possible, they could serve as barcode, a source of immense regulatory diversity. Deciphering the role of p53 post-translational modifications has become a huge challenge for the field as well as an area of controversy. This area has been recently reviewed by others (Wahl and Carr, 2001; Toledo and Wahl, 2006). Do p53 post-translational modifications play a role in gene-selective regulation within the network? The Barcode Hypothesis would support this notion, yet this hypothesis does not necessarily favor one or the other of the two models formulated here, as post-translation modifications may contribute to gene selectivity at both pre- and post-DNA-binding steps. A report by the Taya group indicates that phosphorylation of Serine 46 (Ser46) favors the apoptotic activity of p53 by promoting binding to and activation of the p53aip1 gene (Oda et al., 2000). Ser46 phosphorylation and concomitant p53aip1 induction were shown to occur later than p53 stabilization and phosphorylation of other residues (for example, Ser15, Ser20), and to require higher doses of DNA-damaging agents. Furthermore, alanine replacement of Ser46 selectively impaired p53 transactivation of p53aip1, but not of p21waf1, Mdm2, Pig3, p53R2 and Noxa. How does Ser46 phosphorylation drive these gene-selective effects? Using EMSA assays with nuclear extracts from transfected cells expressing wild type or S46A p53, Oda et al. (2000) observed that the mutation inhibited p53 binding to the p53aip1 RE, but not to the p21waf1 RE. They conclude that ‘Ser 46 on p53 regulates promoter selectivity through altered sequence-specific DNA binding’. Intriguingly, Ser46 is located away from the p53 DBD, just downstream of the canonical N-terminal transactivation domain. How could phosphorylation of p53 N terminus affect p53 DNA binding? One could envision that phospho-Ser46 may regulate the DBD in an allosteric fashion within the tetramer or promote the interaction with a p53 cofactor selectively required for binding to the p53aip1 RE. It would be useful to test the effects of Ser46 phosphorylation in defined biochemical assays other than EMSA. Interestingly, antibodies directed against p53 N terminus are known to allosterically modulate p53 DNA-binding activity in DNase I footprinting assays (Cain et al., 2000), opening the possibility that phosphorylation of this domain may have similar effects.
Despite its strong appeal, the notion of a ‘p53 code’ generated by post-translational modifications has been recently challenged by studies using Nutlin-3. As described before, Nutlin-3 is a non-genotoxic activator of p53 that bypasses the need for stress-induced signaling by directly blocking MDM2 binding to p53 (Vassilev et al., 2004). Importantly, Nutlin-3 triggers p53-dependent apoptosis in some cell types without inducing p53 phosphorylation (Thompson et al., 2004; Vassilev et al., 2004; Kojima et al., 2005; Tovar et al., 2006). In their comparison of cells treated with DNA damaging agents versus Nutlin-3, Thompson et al. (2004) demonstrated that phosphorylation of Ser6, -15, -20, -37, -46 and -392 are dispensable for p53-dependent apoptosis. However, it should be noted that Nutlin-3 is not a potent inducer of apoptosis as compared to other p53-activating stimuli, and that many cell types fail to undergo apoptosis even after prolonged treatments with the drug (Thompson et al., 2004; Tovar et al., 2006). Overall, it is likely that p53 modifications modulate the p53 transcriptional program in certain scenarios, but a universal requirement for Ser46 phosphorylation in apoptosis can be discarded.
The Selective Context Model: life (and death) after DNA binding
In an alternative to The Selective Binding Model, The Selective Context Model proposes that p53 binds to all accessible genomic binding sites, yet only a fraction of p53 bound genes are transactivated (or transrepressed) in specific scenarios. In this model, selectivity is achieved not at the level of binding, but rather at ulterior regulatory steps such as histone modifications, recruitment of coactivators and/or stimulation of RNAP II activity at initiation and/or elongation steps. Thus, selectivity is determined by the context in which each p53 target gene exists, which in turn can be defined by the chromatin landscape or the presence of additional cis-regulatory elements and trans-acting factors.
Lessons from genome-wide studies
The Selective Context Model gathers strong support from recent genome-wide analyses of p53 occupancy and p53-dependent gene expression programs. Using ChIP–PET technology, Wei et al. (2006) created a global map of p53 occupancy in colorectal cancer cells stimulated with 5-FU. Their analysis led to the identification of ~1700 p53 genomic-binding sites. Based on their location within 100 kb of known transcription units, 474 of these p53REs were associated to genes. Interestingly, only 122 of said genes were induced (65) or repressed (57) upon p53 activation, indicating the mere act of binding is not a good predictor of p53-dependent regulation. Using the data generated by Wei et al., the Simon group created a ‘p53-focused’ microarray to monitor global p53 occupancy using ChIP-on-CHIP assays in different cell lines utilizing several distinct p53-activating stimuli. They found that the pattern of p53 occupancy did not change between different stimuli although different biological outcomes were achieved. In agreement with Wei et al., they found that regulation of specific subsets of p53 target genes cannot be attributed to specific association of p53 to said genes (I Simon, personal communication). These observations generate the question: If target gene regulation is not ultimately defined by p53 binding, how is selectivity achieved?
Stimulus-specific regulation of p21waf1
Recent work from our lab and others has provided support for the Selective Context Model. In brief, our work on stimulus-specific regulation of the p21waf1 gene demonstrates that the decision whether or not to activate its transcription is made at regulatory steps after p53 binding to chromatin and recruitment of several p53 cofactors (for example, histone acetyl-transferases, specific Mediator subunits). We observe that the levels of p21waf1 transcription vary greatly in response to distinct p53-activating stimuli. Upon p53 activation by UV-mediated DNA damage, p21waf1 is transcribed modestly and transiently, p21 protein does not accumulate and cells undergo apoptosis (Donner et al., 2007b). In contrast, robust transcription is observed in response to p53 activation by doxorubicin, 5-FU, ionizing radiation or by the non-genotoxic inhibitor of MDM2, Nutlin-3. Of note, gene-specific events are also evident, as other p53 target genes, such as Btg2 and Gadd45a, do not display such stimulus-specific regulation. ChIP analysis of the p21waf1 locus demonstrates that the amount of chromatin-bound p53 in different scenarios merely reflects the intracellular concentration of p53 and that it is not a good predictor of p21waf1 transcription (Espinosa et al., 2003; Donner et al., 2007b). Further ChIP analysis revealed the assembly of stimulus-specific transcriptional complexes acting on the p21waf1 promoter. A number of coregulators, including histone H3 and H4 acetyl-transferases, the core Mediator complex and the general transcription factor TFIIA, are recruited to the p21waf1 promoter regardless of the p53-activating agent utilized. In contrast, other coregulators, such as the CDK-module of Mediator and the general transcription factors TFIIB and TFIIF, are recruited only during conditions of sustained p21waf1 activation (Espinosa et al., 2003; Donner et al., 2007a, b). Thus, specific configurations of the transcriptional apparatus acting at the p21waf1 locus seem to determine the ultimate activation status of RNAP II at this gene. Identical stimulus-specific recruitment of transcriptional coregulators is observed at the Mdm2 promoter, but not at the Fas promoter, revealing the existence of both stimulus- and promoter-specific events. Work from the Prives lab has generated similar observations (Mattia et al., 2007). In this case, Mattia et al. analysed the p21waf1 promoter in cells treated with daunorubicin (high p21waf1 transcription) versus hydroxyurea (HU) (low p21waf1 transcription). Strikingly, ChIP analysis in these two scenarios revealed that RNAP II activity was differentially regulated at post-initiation steps, with a blockage of RNAP II elongation being evident only in HU-treated cells (Mattia et al., 2007). Once again, p53 binding and p53-mediated histone acetylation was identical in both scenarios. Importantly, Pig3 did not show this mode of stimulus-specific regulation (Mattia et al., 2007).
One interpretation of these results is that the signaling scenarios created by distinct p53-activating agents may enable and/or incapacitate different subsets of transcriptional coregulators working at p53 target loci. We should bear in mind that the process of transcriptional activation (or repression) involves the orchestrated action of hundreds of polypeptides modulating RNAP II activity at multiple levels, from chromatin architecture to cotranscriptional RNA processing. To assume that this plethora of coregulators is constitutively available to p53 and does not provide any flexibility to the p53 transcriptional program would be an oversimplification. In the past few years, it has become evident that the so-called ‘general transcription machinery’ is in fact a huge repository of regulatory diversity. We now know that subunits of quasi-universal coregulators, such as TFIID or Mediator, play promoter-, signaling- and cell type-specific functions in gene expression (Chen et al., 1994; Albright and Tjian, 2000; Lemon et al., 2001; Brunkhorst et al., 2004; Mo et al., 2004; van de Peppel et al., 2005; Wang et al., 2005; Zhang et al., 2005; Loncle et al., 2007). In a revised model of transcriptional regulation control, specificity is not only provided by sequence-specific DNA-binding proteins, but also by non-DNA-binding coregulators once thought to be ‘generic’. For those of us who have been forced to accept that the p53 transcriptional network is regulated at post-DNA-binding steps, there are many unanswered questions. How are transcriptional coregulators affected by different p53-activating stimuli? For example, why is the CDK module of Mediator not recruited to the p21waf1 promoter in UVC-treated cells? What positive regulators of RNAP II elongation are incapacitated in HU-treated cells? Despite the great advances in our understanding of the signaling cascades triggered by DNA damage and other p53-activating stimuli, little is known about how they impact on the transcriptional apparatus. We could envision that DNA damage-activated protein kinases (for example, ATM, ATR, Chk1, Chk2) could modulate the activity of transcriptional coregulators and thus impact directly on the quality of the p53 transcriptional program. Clearly, much additional work is needed to answer these questions.
Gene-specific regulatory forces within the p53 network
It is evident that p53 target genes are different in many ways, not just in the sequence, location and topology of their p53REs. For example, p53 target genes display a massive diversity in the use of core promoter elements (CPEs) (for example, TATA box, Initiator (Inr), downstream promoter elements, GC-rich stretch) (Butler and Kadonaga, 2002). These important cis-elements are recognized by components of the ‘general transcription machinery’ such as subunits of TFIID (for example, TBP, TAF1). Studies from the Kadonaga lab indicate that CPEs dictate the responsiveness of a given promoter to upstream enhancers (Butler and Kadonaga, 2001). Thus, CPEs could easily define how p53 target genes react to increased p53 binding or other DNA-binding regulators modulating the network. In addition to CPEs, other cis-elements may affect the p53 transcriptional program in a gene-specific manner. Any given human gene is targeted by a myriad of transcription factors obligate to diverse signaling pathways. For example, a survey of the literature indicates that no less than a dozen different transcription factors are known to regulate p21waf1 transcription in human cells. The impact of these third parties is clearly demonstrated in a report by the Massagué group studying the role of MYC in the cell fate choice to p53 activation (Seoane et al., 2002). It has been well established that, in addition to the positive effects of MYC on cell proliferation, increased MYC activity sensitizes cells to apoptotic stimuli. Seoane et al. demonstrated that MYC is a strong repressor of p21waf1 transcription via binding to and repression of MIZ1, a zinc-finger transcription factor that binds to the p21waf1 promoter. In this way, MYC prevents p21waf1 activation by p53 and other transactivators. Importantly, MYC action did not affect p53 binding to the p21waf1 promoter. Furthermore, MYC did not significantly repress expression of Puma, Pig3 and Bax. Therefore, by functioning as a gene-specific repressor within the p53 transcriptional network, MYC favors the p53 apoptotic response.
The effects of uneven epigenetic landscapes
In addition to their differences in cis-elements, p53 target genes are likely to exist in different epigenetic landscapes, as defined by chromatin architecture, histone modifications, DNA methylation status, even perhaps their localization within the nucleus. An example of the impact of the epigenetic landscape on gene-specific regulation within the p53 network was recently provided by the Prives lab (Tanaka et al., 2007). Using sucrose gradients to fractionate cross-linked p53-associated chromatin, Tanaka et al. (2007) identified distinct macromolecular complexes containing different subsets of p53 target genes. In particular, they could segregate a complex containing the p21waf1 and Mdm2 loci from a complex containing the Pig3 and p53aip1 loci. Using mass spectrometry, they identified human cellular apoptosis susceptibility protein (hCAS) as a differential subunit of these macromolecular entities. ChIP analysis indicated that hCAS associates with the chromatin of Pig3, p53aip1 and p53R2, but not with the p21waf1 and Puma loci. Importantly, association of hCAS with select p53 target genes is independent of p53. Furthermore, hCAS does not affect p53 binding to any of the p53REs analysed. Although hCAS is not an exhaustive discriminator of the apoptotic (Pig3, p53aip1 and Puma) versus the non-apoptotic (p21waf1, 14-3-3σ, p53R2) genes analysed, knockdown of hCAS produces nonetheless an imbalance in the program and, consequently, attenuates p53-dependent apoptosis. How does hCAS achieve its gene-selective effects? Using ChIP assays, Tanaka et al. analysed several histone modifications at hCAS-bound p53 target genes and discovered that both proteins collaborate to reduce the methylation levels of histone H3 lysine 27 (H3-K27), a modification associated with transcriptional repression and heterochromatin formation. Thus, hCAS collaborates with p53 to generate a permissive chromatin landscape without affecting p53 binding to chromatin per se. Interestingly, levels of H3-K27 methylation are very dissimilar among p53 target genes before activation, suggesting that K27 methylases may generate an ‘uneven playing field’ within the program, thus creating a differential need for K27 demethylases among p53 target genes.
Different modes of RNAP II activation
Another illustration of ‘uneven playing fields’ is provided by our analysis of RNAP II behavior on different p53 target genes. ChIP analysis reveals unequal levels of RNAP II preloading among p53 target genes before activation. Overall, non-apoptotic genes, such as p21waf1, Gadd45a, 14-3-3σ, Pcna and Mdm2, show higher levels of poised RNAP II than apoptotic genes, such as Fas, Killer, Puma, p53aip1 and Noxa (Espinosa et al., 2003). Once considered to be a rare phenomenon, RNAP II preloading and pausing is now known to be a widespread mechanism for regulation of developmentally regulated and inducible genes (O’Brien and Lis, 1991; Guenther et al., 2007). Recent genome-wide studies reveal that up to 74% of genes in certain human cell types carry paused RNAP II (Guenther et al., 2007). Whether or not quantitative differences in RNAP II preloading results in qualitatively different modes of regulation remains to be determined. We could envision that preloaded promoters may require a distinct subset of coregulators mediating promoter escape and/or elongation as compared to unloaded promoters. While testing this possibility, we discovered that there is a gene-specific requirement for positive transcription elongation factor b (P-TEFb) among p53 target genes (Gomes et al., 2006). P-TEFb is thought to be universally required for transcriptional elongation. However, we find that some p53 target genes (p21waf1, Puma) dispense with P-TEFb for activation. In contrast, genes such as 14-3-3σ, Killer and Pig3 are fully dependent on this coregulator. Although differential requirement for P-TEFb does not fully correlate with the degree of RNAP II preloading or the grouping of p53 target genes into functional categories, it reveals nonetheless that there are fundamental differences in the mechanism of RNAP II activation on different p53 target loci. Interestingly, gene-specific requirement for P-TEFb has also been demonstrated for other transcriptional networks (Luecke and Yamamoto, 2005).
Coregulators as filters for p53 target gene expression
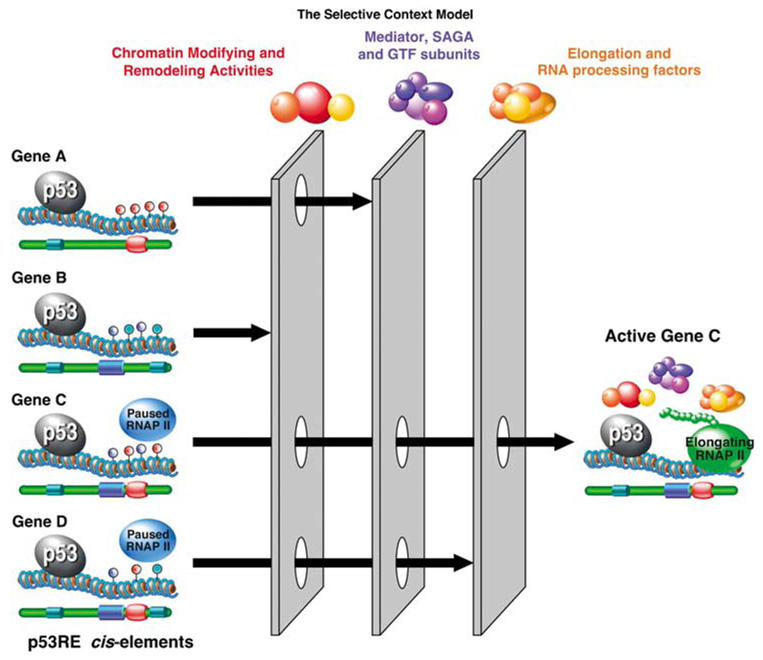
Taking all of the above into consideration, we realize that the mere act of p53 binding to a p53RE is something of a drop in a bucket, and that p53 target genes are subjected to many other regulatory forces. As depicted in Figure 2, the signal generated by p53 binding is ‘filtered’ by the availability of chromatin modifying/remodeling complexes and other coregulators of RNAP II activity at a given locus. By imposing filters at different stages of the transcription cycle, these coregulators act in a combinatorial fashion to allow expression of only a small subset of p53 target genes in a given scenario. Is there a way for chromatin-bound p53 to directly govern these forces in a gene-selective manner? In a variation of The Barcode Hypothesis, it is possible that p53 post-translational modifications provide the means to achieve selectivity at post-DNA-binding steps.
Figure 2.
The Selective Context Model. In this model, p53 binding to most response elements does not result in altered gene expression, as only a minor fraction of p53-bound loci display changes in their transcriptional status. Before p53 activation, p53 target loci display many differences, as manifested by different concurring cis-regulatory elements, distinct combinations of histone modifications and DNA methylation (blue, red and turquoise lollipops), even different amounts of preloaded paused RNAP II. These differences impose gene-specific requirements for transcriptional coregulators, such as chromatin modifying and remodeling activities, variants of the Mediator and SAGA complexes, specific subunits of general transcription factors (GTFs), even defined sets of elongation and RNA processing factors. For example, the presence of a repressive histone methylation mark at a given locus would impose the requirement for a specific histone demethylase, whereas the presence of fixed nucleosomes at the promoter would impose a requirement for a chromatin remodeling complex. Alternatively, promoters not carrying preloaded RNAP II will require the recruitment of GTFs and assembly of a pre-initiation complex (PIC) before they could be readily activated. In turn, the requirement for specific subunits of the PIC may be dictated by the core promoter elements (CPEs) present at a given locus (for example, TATA box, Initiator, downstream promoter element) (blue, red and turquoise barrels). Finally, effective gene activation will require conversion of paused RNAP II into an elongating form, as well as correct processing of the nascent mRNA (that is, 5′ capping, splicing, 3′ cleavage and polyadenylation), all of which may be also regulated in a gene-specific manner. Importantly, the quality of these combinatorial ‘filters’ could vary with the signaling scenario (that is, cell type or type of p53 activating stimuli), as the levels and activity of coregulators in each category could be modulated, thus allowing for great flexibility within the p53 transcriptional program.
K120: a signal after binding?
Two recent reports indicate that acetylation of p53 lysine 120 (K120), located within the DBD, favors selective activation of apoptotic genes at post-DNA-binding steps (Sykes et al., 2006; Tang et al., 2006). K120 is acetylated by TIP60 and MOF, two related acetyl-transferases. In ectopic expression experiments using adenoviral constructs or ER-p53 fusion constructs, a K120 mutant showed diminished transactivating potential on the Puma and Bax loci, whereas its ability to activate p21waf1 and Mdm2 remained essentially unaltered. Interestingly, the gene-specific effects of the K120 mutation took place at post-DNA-binding steps, as both groups agree that K120 mutants bind efficiently to the p53REs in Puma and Bax. However, the reports differ somewhat in the proposed mechanism of gene selectivity. Yang et al. propose that the impairment in Puma activation is due to a defect in p53-mediated acetylation of histone H4 at this locus. Although TIP60, a known H4 acetyl-transferase, was efficiently recruited by p53 K120 mutants to the Puma locus, H4 acetylation was decreased at this region as compared with cells expressing wild-type p53. It is unclear how acetylation of K120 may affect TIP60 action in a gene-specific manner. TIP60 interacts with the N-terminal domain of p53, not the DBD, where K120 resides. Some mode of allosteric regulation of TIP60 activity by the p53 DBD could be invoked. Alternatively, acetylated K120 may serve as the docking site for a bromodomain-containing H4 acetyl-transferase different than TIP60 (for example, p300, CBP). It would be interesting to identify other p53 target genes differentially affected by K120 acetylation beyond Puma and Bax and determine if the effects on H4 acetylation are conserved. Intriguingly, Sykes et al. propose that K120 acetylation occurs only at those genes which require it (Puma, Bax), as they could not detect K120 acetylation on p53 bound to the p21waf1 and Mdm2 promoters. This conflicts somewhat with the observation by Yang et al. showing that TIP60 is efficiently recruited by p53 to the p21waf1 promoter. To reconcile these findings, we could imagine that K120 is somehow refractory to acetylation by TIP60 at the p21waf1 locus. Again, further mechanistic studies are required to illuminate these issues. As discussed before for Ser46 phosphorylation, studies with Nutlin-3 represent an obstacle for The Barcode Hypothesis. It would be useful to determine the acetylation status of p53 upon Nutlin-3 treatment. Since MDM2 and some acetyl-transferases may compete with each other for the same contact surface in the p53 N terminus, it is possible that mere MDM2 inhibition leads to p53 acetylation without stress-induced signaling.
Disclaimer: there is post-transcriptional regulation as well
Throughout this review, I have purposely focused on transcriptional mechanisms providing regulatory diversity to the p53 network. However, an efficient way to alter gene expression is by the regulation of already transcribed mRNAs. In analogous fashion to transcriptional control, specific cis-acting elements, located mostly in the 3′-untranslated regions (UTRs) of mRNAs, modulate the stability and/or translation of a given mRNA species through the binding of trans-acting factors, such as RNA-binding proteins and microRNAs. During post-transcriptional repression, target mRNAs can be decapped, deadenylated and degraded by specialized cytosolic machineries (Garneau et al., 2007). Alternatively, translation of the target mRNA can be inhibited by preventing ribosome binding or elongation (Garneau et al., 2007). Despite the numerous advances in our understanding of post-transcriptional regulatory mechanisms, little is known about their impact on the p53 network. Reports from outside the p53 field reveal the importance of post-transcriptional regulation of the p21waf1 gene in several biological processes such as tissue differentiation. A number of cis-regulatory elements present in the 3′-UTR of the p21waf1 mRNA have been identified, as well as RNA-binding proteins recognizing these sequences (Johannessen et al., 1999; Figueroa et al., 2003; Lal et al., 2004; Yang et al., 2004; Shi et al., 2005). Recently, the Chen group identified a novel p53 target gene, named RNPC1, which codes for an RNA-binding protein capable of stabilizing p21waf1 mRNA (Shu et al., 2006). Thus, RNPC1 could be required for proper cell cycle arrest response. Furthermore, the discovery of p53-inducible microRNAs (p53-miRs) reinforces the idea that post-transcriptional regulation plays a significant role within the p53 tumor suppressor network (Hermeking, 2007). p53-miRs can potentially modulate the expression of hundreds of mRNAs within and outside the p53 network, which could affect the type of cellular response provoked by increased p53 activity.
Finally, the ultimate impact of p53 target gene expression can be affected by regulating the activity of the corresponding protein products. For example, the p21 protein can be inactivated via AKT-dependent phosphorylation and consequent cytoplasmic retention (Zhou et al., 2001). Alternatively, the apoptotic effects of p53 target genes coding for BH3-only proteins (for example, Puma, Noxa) could be determined by the abundance and activity of the corresponding Bcl-2-like proteins they antagonize (Jiang and Milner, 2003; Chen et al., 2005; Alves et al., 2006). Thus, the two models discussed in this review can only partially explain the regulatory diversity found within the p53 network and should be eventually integrated with post-transcriptional regulatory mechanisms.
Final remarks: the ephemeral nature of ideas
Obviously, my effort to cluster a wealth of data into two alternative models may be ill-fated from the beginning. By sharing my vision of two conceptually different mechanisms, I hope to provoke future work that will support or refute these models, with the idea that this will advance our understanding rather than generating unproductive controversy. I will not be the least surprised to soon find myself having a completely different view of the problem or acknowledging that both (or neither) of these two models are correct to some extent. My favorite scientist put it very well, ‘Science consists in grouping facts so that general laws or conclusions may be drawn from them’. ‘How odd it is that anyone should not see that all observation must be for or against some view if it is to be of any service!’. ‘I have steadily endeavored to keep my mind free so as to give up any hypothesis, however much beloved’.
Acknowledgements
I thank Aaron Donner, Nathan Gomes, Gerard Evan and Carol Prives for reading the paper and providing useful critiques. I also thank Dr Jim Bridwell for providing support and inspiration. Work in my lab is currently supported by grants from NIH (RO1-CA117907) and March of Dimes (5-FY05-1217).
References
- Albright SR, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- Alves NL, Derks IA, Berk E, Spijker R, van Lier RA, Eldering E. The Noxa/Mcl-1 axis regulates susceptibility to apoptosis under glucose limitation in dividing T cells. Immunity. 2006;24:703–716. doi: 10.1016/j.immuni.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, Jin B, Duraisingham S, Crook T, Lu X. ASPP1 and ASPP2: common activators of p53 family members. Mol Cell Biol. 2004;24:1341–1350. doi: 10.1128/MCB.24.3.1341-1350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet. 2006;38:1133–1141. doi: 10.1038/ng1879. [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkhorst A, Neuman T, Hall A, Arenas E, Bartfai T, Hermanson O, et al. Novel isoforms of the TFIID subunit TAF4 modulate nuclear receptor-mediated transcriptional activity. Biochem Biophys Res Commun. 2004;325:574–579. doi: 10.1016/j.bbrc.2004.10.078. [DOI] [PubMed] [Google Scholar]
- Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Cain C, Miller S, Ahn J, Prives C. The N terminus of p53 regulates its dissociation from DNA. J Biol Chem. 2000;275:39944–39953. doi: 10.1074/jbc.M002509200. [DOI] [PubMed] [Google Scholar]
- Chen JL, Attardi LD, Verrijzer CP, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, et al. Hzf determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–637. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison TS, Vagner C, Kaghad M, Ayed A, Caput D, Arrowsmith CH. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J Biol Chem. 1999;274:18709–18714. doi: 10.1074/jbc.274.26.18709. [DOI] [PubMed] [Google Scholar]
- Donner AJ, Hoover JM, Szostek SA, Espinosa JM. Stimulus-specific transcriptional regulation within the p53 network. Cell ycle. 2007a;6:2594–2598. doi: 10.4161/cc.6.21.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007b;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Ortega-Molina A, Velasco-Miguel S, Herranz D, Vassilev LT, Serrano M. Induction of p53-dependent senescence by the MDM2 antagonist nutlin-3a in mouse cells of fibroblast origin. Cancer Res. 2007;67:7350–7357. doi: 10.1158/0008-5472.CAN-07-0200. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Fei P, Bernhard EJ, El-Deiry WS. Tissue-specific induction of p53 targets in vivo. Cancer Res. 2002;62:7316–7327. [PubMed] [Google Scholar]
- Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, et al. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Gohler T, Reimann M, Cherny D, Walter K, Warnecke G, Kim E, et al. Specific interaction of p53 with target binding sites is determined by DNA conformation and is regulated by the C-terminal domain. J Biol Chem. 2002;277:41192–41203. doi: 10.1074/jbc.M202344200. [DOI] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorina S, Pavletich NP. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet. 2007;3:e127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jett SD, Cherny DI, Subramaniam V, Jovin TM. Scanning force microscopy of the complexes of p53 core domain with supercoiled DNA. J Mol Biol. 2000;299:585–592. doi: 10.1006/jmbi.2000.3759. [DOI] [PubMed] [Google Scholar]
- Jiang M, Milner J. Bcl-2 constitutively suppresses p53-dependent apoptosis in colorectal cancer cells. Genes Dev. 2003;17:832–837. doi: 10.1101/gad.252603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen LE, Knardal SL, Madshus IH. Epidermal growth factor increases the level of the cyclin-dependent kinase (CDK) inhibitor p21/CIP1 (CDK-interacting protein 1) in A431 cells by increasing the half-lives of the p21/CIP1 transcript and the p21/CIP1 protein. Biochem J. 1999;337(Part 3):599–606. [PMC free article] [PubMed] [Google Scholar]
- Kaeser MD, Iggo RD. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc Natl Acad Sci USA. 2002;99:95–100. doi: 10.1073/pnas.012283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Konopleva M, Samudio IJ, Shikami M, Cabreira-Hansen M, McQueen T, et al. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood. 2005;106:3150–3159. doi: 10.1182/blood-2005-02-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- Loncle N, Boube M, Joulia L, Boschiero C, Werner M, Cribbs DL, et al. Distinct roles for mediator Cdk8 module subunits in Drosophila development. EMBO J. 2007;26:1045–1054. doi: 10.1038/sj.emboj.7601566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig RL, Bates S, Vousden KH. Differential activation of target cellular promoters by p53 mutants with impaired apoptotic function. Mol Cell Biol. 1996;16:4952–4960. doi: 10.1128/mcb.16.9.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev. 2005;19:1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattia M, Gottifredi V, McKinney K, Prives C. p53-dependent p21 mRNA elongation is impaired when DNA replication is stalled. Mol Cell Biol. 2007;27:1309–1320. doi: 10.1128/MCB.01520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras induces mediator complex exchange on C/EBP beta. Mol Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Mol Cancer Res. 2004;2:371–386. [PubMed] [Google Scholar]
- O’Brien T, Lis JT. RNA polymerase II pauses at the 5′ end of the transcriptionally induced Drosophila hsp70 gene. Mol Cell Biol. 1991;11:5285–5290. doi: 10.1128/mcb.11.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- Perez CA, Ott J, Mays DJ, Pietenpol JA. p63 consensus DNA-binding site: identification, analysis and application into a p63MH algorithm. Oncogene. 2007;26:7363–7370. doi: 10.1038/sj.onc.1210561. [DOI] [PubMed] [Google Scholar]
- Resnick-Silverman L, St Clair S, Maurer M, Zhao K, Manfredi JJ. Identification of a novel class of genomic DNA-binding sites suggests a mechanism for selectivity in target gene activation by the tumor suppressor protein p53. Genes Dev. 1998;12:2102–2107. doi: 10.1101/gad.12.14.2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- Senoo M, Manis JP, Alt FW, McKeon F. p63 and p73 are not required for the development and p53-dependent apoptosis of T cells. Cancer Cell. 2004;6:85–89. doi: 10.1016/j.ccr.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, et al. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J Biol Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 2006;20:2961–2972. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee EA, Lu X. The ASPP family: deciding between life and death after DNA damage. Toxicol Lett. 2003;139:81–87. doi: 10.1016/s0378-4274(02)00421-6. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szak ST, Mays D, Pietenpol JA. Kinetics of p53 binding to promoter sites in vivo. Mol Cell Biol. 2001;21:3375–3386. doi: 10.1128/MCB.21.10.3375-3386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Thompson T, Tovar C, Yang H, Carvajal D, Vu BT, Xu Q, et al. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J Biol Chem. 2004;279:53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn TT, van Leenen D, Holstege FC. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vives V, Slee EA, Lu X. ASPP2: a gene that controls life and death in vivo. Cell Cycle. 2006a;5:2187–2190. doi: 10.4161/cc.5.19.3266. [DOI] [PubMed] [Google Scholar]
- Vives V, Su J, Zhong S, Ratnayaka I, Slee E, Goldin R, et al. ASPP2 is a haploinsufficient tumor suppressor that cooperates with p53 to suppress tumor growth. Genes Dev. 2006b;20:1262–1267. doi: 10.1101/gad.374006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl GM, Carr AM. The evolution of diverse biological responses to DNA damage: insights from yeast and p53. Nat Cell Biol. 2001;3:E277–E286. doi: 10.1038/ncb1201-e277. [DOI] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Yang X, Jones RA, Wiester MJ. A nanoscale slipped sandwich of Tb(10)-stabilization of a benzaldehyde methyl hemiacetyl. Dalton Trans. 2004 June;21:1787–1788. doi: 10.1039/B405981F. [DOI] [PubMed] [Google Scholar]
- Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, et al. MED1/TRAP220 exists predominantly in a TRAP/mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]