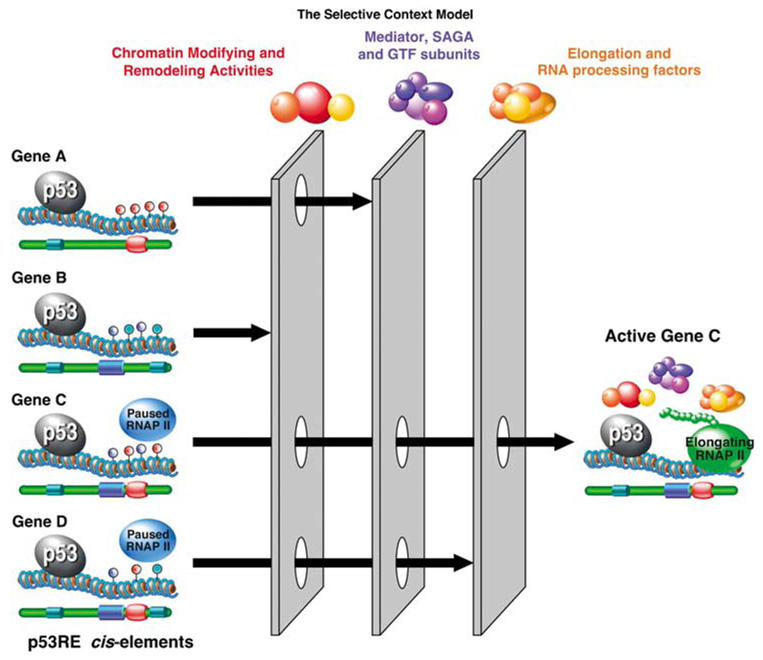
Figure 2.
The Selective Context Model. In this model, p53 binding to most response elements does not result in altered gene expression, as only a minor fraction of p53-bound loci display changes in their transcriptional status. Before p53 activation, p53 target loci display many differences, as manifested by different concurring cis-regulatory elements, distinct combinations of histone modifications and DNA methylation (blue, red and turquoise lollipops), even different amounts of preloaded paused RNAP II. These differences impose gene-specific requirements for transcriptional coregulators, such as chromatin modifying and remodeling activities, variants of the Mediator and SAGA complexes, specific subunits of general transcription factors (GTFs), even defined sets of elongation and RNA processing factors. For example, the presence of a repressive histone methylation mark at a given locus would impose the requirement for a specific histone demethylase, whereas the presence of fixed nucleosomes at the promoter would impose a requirement for a chromatin remodeling complex. Alternatively, promoters not carrying preloaded RNAP II will require the recruitment of GTFs and assembly of a pre-initiation complex (PIC) before they could be readily activated. In turn, the requirement for specific subunits of the PIC may be dictated by the core promoter elements (CPEs) present at a given locus (for example, TATA box, Initiator, downstream promoter element) (blue, red and turquoise barrels). Finally, effective gene activation will require conversion of paused RNAP II into an elongating form, as well as correct processing of the nascent mRNA (that is, 5′ capping, splicing, 3′ cleavage and polyadenylation), all of which may be also regulated in a gene-specific manner. Importantly, the quality of these combinatorial ‘filters’ could vary with the signaling scenario (that is, cell type or type of p53 activating stimuli), as the levels and activity of coregulators in each category could be modulated, thus allowing for great flexibility within the p53 transcriptional program.