Abstract
Background
Aspartic protease Cathepsin (Cath) E and D are two different proteases, but they share many common characteristics, including molecular weight, catalytic mechanism, substrate preferences, proteolytic conditions and inhibition susceptibility. To define the biological roles of these proteases, it is necessary to elucidate their substrate specificity. In the present study, we report a new peptide-substrate that is only sensitive to Cath E but not Cath D.
Methods
Substrate e, Mca-Ala-Gly-Phe-Ser-Leu-Pro-Ala-Lys(Dnp)-DArg-CONH2, designed in such way that due to the close proximity of a Mca-donor and a Dnp-acceptor, near complete intramolecular quenching effect was achieved in its intact state. After the proteolytic cleavage of the hydrophobic motif of peptide substrate, both Mca and Dnp would be further apart, resulting in bright fluorescence.
Results
Substrate e showed a 265 fold difference in the net fluorescence signals between Cath E and D. This Cath E selectivity was established by having -Leu**Pro- residues at the scissile peptide bond. The confined cleavage site of substrate e was confirmed by LC-MS. The catalytic efficiency (Kcat/KM) of Cath E for substrate e was 16.7 µM−1.S−1. No measurable catalytic efficiency was observed using Cath D and no detectable fluorescent changes when incubated with Cath S and Cath B.
Conclusions
This study demonstrated the promise of using the developed fluorogenic substrate e as a selective probe for Cath E proteolytic activity measurement.
General Significance
This study forms the foundation of Cath E specific inhibitor development in further studies.
Keywords: Cathepsin E, Cathepsin D, fluorescent, peptide, substrate, aspartic protease
1. Introduction
Cathepsin (Cath) E and Cath D, members of aspartic proteolytic enzymes family, have very similar substrate selectivity [1, 2]; hence, why Cath E was once named Cathepsin-D like proteinase [3, 4]. Unlike the relatively ubiquitous Cath D, Cath E has a limited cellular localization and tissue distribution. Cath E is contained mainly within vesicular structures associated with the endoplasmic reticulum and endosomal compartments [5–9] of the macrophage [6, 10], gastric epithelial cells [7], lymphocytes [7], microglia [11] and dendritic cells [10, 12]. Its function as a major intracellular nonlysosomal aspartic proteinase is not entirely clear; whereas, its primary localization within the cells of the immune system suggests it might play an important immunological and physiological role in the host defense process [9, 11, 13, 14]. In contrast, Cath D is known to be involved in various diseases. For example, it has been associated with cancer growth, metastasis [15–22], Alzheimer and several other diseases [23–26]. Although Cath E and Cath D have different tissue localization, cellular distribution and physiological function, they share many enzymatic characteristics including molecular weight (43 kDa), catalytic mechanism, substrate preferences, proteolytic conditions and inhibition susceptibility. Both of them are aspartic endopeptidases, which prefer hydrophobic amino acids residues at P1 and P1’ positions of the scissile bond [27, 28]. Their optimal pH of proteolysis is between pH 3.5 and 5.0 [29, 30]. A wide variety of peptiomimic inhibitors of Cath E have been reported [31–33], but none can provide satisfactory discrimination against Cath D.
In order to verify the actual localization and define the biological role of Cath E and Cath D, it is necessary to elucidate their substrate specificity profile. Although a few peptide substrates have been identified, they were unable to differentiate the proteolytic activity of Cath E from that of Cath D [13, 27, 28, 34, 35]. A selective probe, which could report only Cath E proteolytic activity, has not been developed. In this study, we aimed to identify a new peptide substrate that is not only sensitive to the proteolytic activity of aspartic peptidases but also selectively distinguishes that activity of Cath E from Cath D. A series of fluorogenic peptide substrates derived from prior reported sequences were designed with a fluorescent donor, 7-Methoxycoumarin-4-acetic acid (Mca), and an energy acceptor, dinitrophenyl (Dnp), at either end of the substrates [13, 36]. Due to the close proximity of a Mca-donor and a Dnp-acceptor, near complete intramolecular quenching effect was achieved in its intact state. After the proteolytic cleavage of the hydrophobic motif of peptide substrate, both Mca and Dnp would be further apart, resulting in bright fluorescence. Using this approach, we have identified a novel peptide substrate allowing selective detection of Cath E proteolytic activity.
2. Materials and methods
2.1. Materials
All reagents used were of analytical or HPLC grade. Dicholormethane (DCM), N-Methylpyrolidone (NMP) and Methanol (MeOH) were purchased from Fisher (Fair Lawn, NJ, USA). N,N-Dimethylformamide (DMF), Diethylether, Acetonitrile (MeCN), Diisoproylethylamine (DIPEA), Piperidine, Pepstatin A, Triisopropylsilane (TIS), 2,4,6-trinitrobenzenesulfonic acid (TNBS), Triethylamine (TEA) and 1,2-ethanedithiol (EDT) were purchased from Sigma-Aldrich (Milwaukee, WI, USA). HOBt and HBTU were purchased from Applied Biosystems (Foster City, CA, USA). Fmoc protected amino acids were purchase from AnaSpec (Fremont, CA, USA). Fmoc-Rink Amide MBHA-resin, and Fmoc-Lys(Dnp)-OH were purchased from Novabiochem (La Jolla, CA, USA). Cath E, Cath D, Cath S and Cath B were purchased from Calbiochem, EMD Bioscience (Gibbstown, NJ, USA). Anti-Cathepsin E Antibody was purchased from R&D Systems (Minneapolis, MN, USA).
2.2 Peptide Substrate Synthesis
All peptide probes were synthesized by solid-phase peptide synthesis (SPPS) using the standard Fmoc chemistry on an automatic synthesizer (ABI-433A, Applied Biosystems). Rink amide MBHA resin, 100 µmol, with substitution level of 0.7 µmol/mg was used as the support for peptide amide synthesis. Ten folds molar excess, relative to the resin loading, of each Fmoc protected amino acids were coupled sequentially to the resin using the HBTU/HOBT coupling strategy. Dnp group was attached to the ε-amino group of Lysine side chain of all peptides during solid phase synthesis using Fmoc-Lys(Dnp)-OH as the building block.
After completion of the peptide chain elongation, Coumarin-based Flurophore, 7-Methoxycoumarin-4-acetic acid (Mca), was coupled to the N-terminal amino group via in situ activation. Three equivalents of Mca (300 µmol, 70.26 mg), relative to the resin loading, were dissolved in 9 mL NMP. HBTU (270 µmol, 102.39 mg) and HOBt (300 µmol, 39.45 mg) were dissolved in 2 mL DMF. These two solutions were mixed, and six equivalents (600 µmol, 103 µL) of DIPEA were added and vortexed thoroughly for 10–15 min. This solution was added directly to Rink amide MBHA resin bound peptide in a manual SPPS reaction vessel and agitated gently for six hours under N2 at rt in the dark. The reagents were drained and washed twice with NMP. The completion of Mca coupling was confirmed by TNBS assay. A small sample of peptidyl resin beads (~10 mg) was placed in a plastic filter tube to be colorimetrically tested for free –NH2 groups. The resin was washed with THF twice for 2 min. A few drops of 10% DIPEA in NMP were added, followed by 2 drops of TNBS. If the resin does not show reddish color, the coupling is considered complete.
Thereafter, all protecting groups were removed and the peptides were cleaved from the resin using a deprotection-scavenger cocktail (TFA:H2O:TIS:EDT = 94:2.5:1.0:2.5, 10 mL/gm peptidyl resin) in a manual SPPS reaction vessel at rt in the dark with gentle agitation under N2 for three hours. The cleavage cocktail containing peptide substrates were filtered and reduced in volume to ~1 mL. Cold diethyl ether was added to precipitate peptides. Crude peptides were redissolved in MeCN:H2O (50:50 v/v, ~6 mL) and purified by reversed phase high performance liquid chromatography (RP-HPLC) using a C18 preparative Column (Nova-Pak® HR, 6 µm, 60 Å, 19 mm ID × 300 mm L; Waters, Milford, MA, USA) with a linear gradient from 10 % B to 50 % B (8 ml/min) in 60 minutes on a Varian-ProStar 210 Chromatography system (Palo Alto, CA, USA). HPLC solvent A is H2O containing 0.1% TFA, and solvent B is MeCN containing 0.1% TFA. Detection was carried out at 220 nm and 280 nm using a Varian-ProStar L-345 UV-Vis detector (Varian, Palo Alto, CA, USA). The purity of substrates was analyzed by analytical RP-HPLC using a C-18 column (5 µm, 4.6 mm ID × 150 mm L Vydac, GRACE, Deerfield, IL, USA) on a Varian 920-LC Liquid Chromatography system coupled to a UV-Vis/Fluorescence diodarray detector, and equipped with Galaxie Chromatography Data system™ (version 1.9). Fractions with the same purity were collected together and lyophilized to yield orange-yellowish powders with > 97% purity. Purity represents the percentage of the area under the UV peak of interest to the total areas of all detected UV peak in the HPLC chromatogram.
HPLC showed the aspartic peptidase substrates a, b, c, d and e with retention factors (k’) of 7.01, 6.99, 7.03, 6.64, and 6.78 respectively. The molecular weight of the purified substrate and hydrolyzed fragments was confirmed by ESI-MS (Thermo Finnigan LCQ Fleet mass spectrometer, West Palm Beach, FL, USA), and the raw data were analyzed using Xcalibur software. ESI-MS showed the molecular ions (m/z) of each aspartic peptidase substrate. The conjugate was stored at 4 °C in the dark.
2.3. Enzyme Cleavage and Substrates Specificity Assay
The catalytic selectivity of Cath E and Cath D was determined fluorometerically by FRET-based hydrolysis of intramolecularly quenched peptide substrates a–e. Fluorogenic substrates a–e (10 µL, 200 µM) were incubated with 23 pmole of Cath E or Cath D in 50 mM sodium acetate buffer, pH 4, containing 150 mM NaCl, and the total volume was brought to 100 µL using the same buffer. Cath S and Cath B were pre-activated by incubation with 100 mM sodium acetate buffer solution, pH 6.5, containing 5 mM DTT and 5 mM EDTA for 5 minutes. Cath S and Cath B (23 pmole) were incubated with 10 µL of 200 µM fluorogenic substrate e at 100 mM sodium acetate buffer solution, pH 6.5, containing 5 mM DTT and 5mM EDTA, and the total volume was brought to 100 µL using the same buffer. All assays were performed in triplicate in 96 well black walls, clear-bottom plates (Corning, NY, USA). The change in the fluorescence intensity was monitored over time using SpectraMax.M2e fluorescence spectrophotometer (Molecular Devices, Sunnyvale, CA, USA) at excitation wavelength (λex) of 340 nm and emission wavelength (λem) of 405 nm at 27 °C. Control experiments were performed simultaneously by replacing the enzyme with assay buffer.
2.4. Enzyme Digestion and Fragments Characterization
Substrate e was subjected to aspartic peptidase digestion using Cath E and Cath D. Fluorogenic substrate e (10 µL, 100 µM ) was incubated at 37°C for 3 hours with excess Cath E or Cath D (~ 119 pmole) in 100 µL of 50 mM sodium acetate buffer (pH 4) containing 150 mM NaCl. The resulting digestion fragments were analyzed by the aforementioned analytical RP-HPLC and ESI-MS.
2.5. Enzyme Inhibition and Selective Immunoprecipitation of Cathepsin E
Stock solution of pepstain A was prepared by dissolving 2 mg pepstatin A in 1 mL of 10% (v/v) acetic acid in methanol. Substrate e solution (10 µL, 200 µM) in 50 mM sodium acetate buffer (pH 4) containing 150 mM NaCl, 46 pmole of Cathepsin E or D were added followed by 1 µL of 1 mM pepstain A/ methanol. Two sets of control experiments were performed simultaneously by replacing the enzymes with assay buffer and by using methanol instead of pepstain A/methanol. All assays were performed in triplicate in 96 well assay plates and the change in the fluorescence intensity was monitored over time using λex=340 nm and λem= 405 nm at 27 °C.
In a microcentifuge tube, 5 µg Cath E enzyme was incubated with a specific Cath E-antibody in 1:1 ratio using 500 µL 1× PBS as an immunoprecipitation buffer. The mixture was gently mixed for 60 min at 4°C. Protein-G-sepharose was added and allowed to immobilize the specific Cath E antibody for 60 more min on ice. The mixture was centrifuged at 3000 rpm for 2 min, and the supernatant was collected and used for the subsequent enzyme assay as described before.
2.6. Cathepsin E Dose Response
The initial rates of substrate e hydrolysis by Cath E were monitored using a fluorescence platereader. Hydrolysis of different substrate e concentrations, 10, 20, 40 and 60 µM, was performed at pH 4 in 50 mM sodium acetate buffer containing 150 mM NaCl. Cath E with various concentrations, 2.27, 4.55, 6.82 nM, were incubated with substrate e and the changes in the fluorescence intensities were monitored for 10 minutes. The substrate e hydrolysis was determined by integrating the fluorescence intensity, and plotted by the amount of the hydrolyzed substrate e over time.
2.7. Kinetic Parameters of Substrate e
The initial velocities of the hydrolysis reactions were measured between 0 to 10 min using several concentrations of substrate e including: 5, 10, 20, 40, 60, 80, 100 µM. All of the used substrate e concentrations were much higher than the concentrations of enzyme (2.3, 4.5, 6.8 nM). To measure the initial rates of the substrate e proteolysis and to determine Michaelis-Menten kinetic parameters Vmax and Km of the Cath E and Cath D, the Michaelis-Menten equation was transformed using the Woolf kinetic transformation [S/v = (S/Vmax) + (Km/Vmax)]. The initial velocity was calculated from the slope during the linear phase of the reaction and S/v plotted versus substrate e concentration. The graph representation gave straight lines and Vmax and Km were calculated by the linear regression of the line to obtain the slope and intercept. The turnover number of Cath E enzyme (Kcat) was calculated using the equation Kcat = Vmax/[Cath E], where [Cath E] represents the Cath E concentration in µM and Vmax is the maximum velocity in µM.S−1. All the reactions and measurements were carried out in 50 mM sodium acetate buffers (pH 4.0) using the aforementioned setting.
2.8. Statistical Analysis
Statistical Package for the Social Sciences (version 13, SPSS, Chicago, Illinois) was used for examining the null hypothesis and assessing the statistical significance of the observed fluorescence intensity differences. Paired-Samples T-Test with two-tailed P-values was employed with statistical significance attributed to P ≤ 0.05.
3. Results and discussion
3.1. Substrate Design and Synthesis
Proteases are usually highly specific with respect to substrates because of the structure of their active cleft [37]. This specificity can be evidenced by the enzyme ability to distinguish between very similar molecules. However, not all proteases are so specific. Cath E and Cath D accept numbers of closely related substrates if they possess some common structural features. The bulky hydrophobic phenylalanine amino acid residues at position P1 and P1’ of the scissile bond have been described in the majority of reported substrates for both Cath E and Cath D [13, 28, 38–40]. Only few amino acid residues at P1 and P1’ positions have been reported [28].
The distinction of the catalytic activity of Cath E from that of Cath D is still problematic even with the large number of developed substrates. Lacking distinction between proteolytic activity of Cath E and Cath D might be in part due to the persistent usage of Phe as the common residues at the cleavage position [13, 17, 28, 38–40]. We hypothesized that using a hydrophobic amino acid residue other than Phe at the scissile bond and/or switching amino acids at P1 and P1’ positions might lead to a better substrate that is capable of differentiating Cath E and Cath D.
A previously reported peptide substrate described as the most sensitive sequence (substrate a, Table 1) for Cath E was used as a reference [13]. Substrates b and c, derived from the peptide substrate Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys, previously described as sensitive substrate for Cath E and Cath D [39], were also selected to compare sequence preference and examine the effect at P4’ position. Gly residue was inserted at P4’ position of substrate b based on the substrate sequence previously described as sensitive and specific for Cath D [17]. After examining the hydrophobicity of the P1 residue in the reported substrates, it is believed that a hydrophobic group is critical. According to Kyte and Doolittle hydropathy scale, the hydrophobicity index of Leu is slightly higher (3.8) than it of Phe (2.8) [41]. Thus, Leu was selected as the hydrophobic amino acid residue at P1 position in substrates d and e. The effect of the P1’ position was studied by using two conformationally distinctive amino acid residues. One is a conformationally unrestricted Gly residue and the other one is a rigid Pro residue. A charged D-Arg residue, which resists enzymatic hydrolysis, was placed at the C-termini of all substrates to increase solubility.
Table 1.
List of developed peptide substrates and their characteristics.
Due to significant spectral overlap between the emission spectrum of Mca and the absorbance spectrum of Dnp, they are widely used as a fluorophore and quencher pair. While the Mca-fluorophore moieties were attached to the N-terminal residues of the substrates, the Dnp-quencher moieties were anchored to the ε-amino group of Lysine residue’s side chain after the prime scissile position. The characteristics of the prepared peptide probes were confirmed using ESI-MS (Table 1).
3.2. Enzyme Cleavage Sensitivity and Substrates Specificity Assay
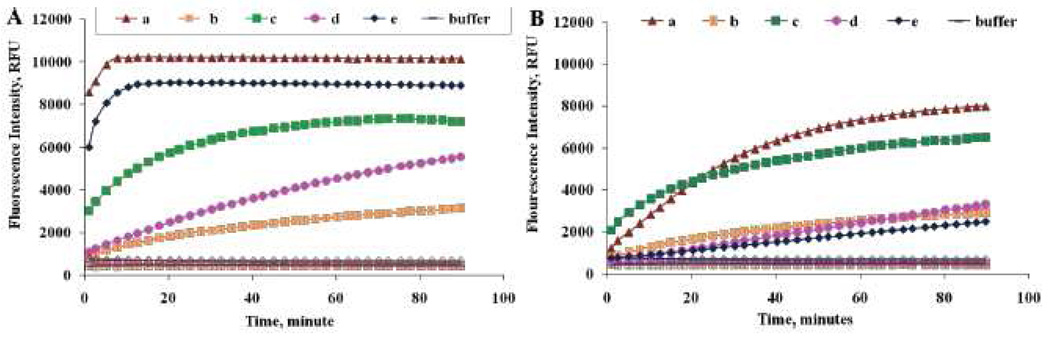
The protease mediated hydrolysis of the prepared fluorogenic substrates was followed by measuring changes in fluorescence intensity over time (Figure 1). Without enzymes, substrates a–e showed and maintained an evident quenching over the monitored period, thus the fluorescence intensity of the substrates was comparable to that of buffer solution. (Figure 1A, B).
Figure 1.
Changes in the fluorescence intensity of substrates a–e (200 µM) with 23 picomole of (A) Cath E (B) Cath D in 50 mM NaOAc buffer containing 150 mM NaCl (pH 4.0). Solid filled and unfilled markers denote the enzyme treated and untreated substrates, respectively. Without enzymes, fluorescence intensities of all tested substrates remain at the base line level. Values represent the mean of triplicate measurements.
Prominent fluorescence increase of substrate a was observed instantaneously upon incubation with Cath E (Figure 1A). Substrate a showed the fastest increase and the highest fluorescence signal among all investigated substrates. Unfortunately, such leading increase in fluorescence signal with Cath E was observed as well upon incubation with Cath D (Figure 1B). As seen with Cath E, the increase in fluorescence signals of substrate a upon incubation with Cath D was the highest among all investigated substrates. Apparently, Phe-Leu at P1 and P1’ positions in substrate a might allow constructive interaction with the binding sites of Cath D and Cath E.
Compared to substrate b, substrate c shows superior increase in the fluorescence intensity upon hydrolysis by both Cath E and Cath D (Figure 1A, B). Knowing that the scissile peptide bond of both substrates b and c share the same sequence of amino acids except at the distant P4’ position, point out to the significant role that P4’ position might play in the substrate cleavage susceptibility. Although substrate b and c exhibit distinctive susceptibility to Cath E and Cath D, they could not distinguish their proteolytic activities effectively. Most likely, having Phe-Phe at P1 and P1’position of the scissile peptide bond could not offer preferred selectivity.
Substrates d and e differ only in the amino acid residues at P1’ and P2’ positions, nevertheless their proteolytic activation by Cath E was dramatically different (Figure 1A). While substrate e was activated by Cath E quickly and reached saturation within 10 minutes, substrate d showed relatively slow hydrolysis and did not reach saturation within the monitored period. On the other hand, both substrates d and e exhibited similar slow hydrolysis by Cath D (Figure 1B). It is conceivable that placing a conformationally restricted Pro residue at P1’ position boosts the activation of the Cath E intermediate, whereas a Gly residue at the same position failed to demonstrate the same influence. While it was considered essential to have Phe in the substrate scissile bond to be susceptible by Cath E and Cath D in the literature [39], the results obtained from this study demonstrate that the susceptibility of -Leu-Pro- is critical to Cath E, but not to Cath D.
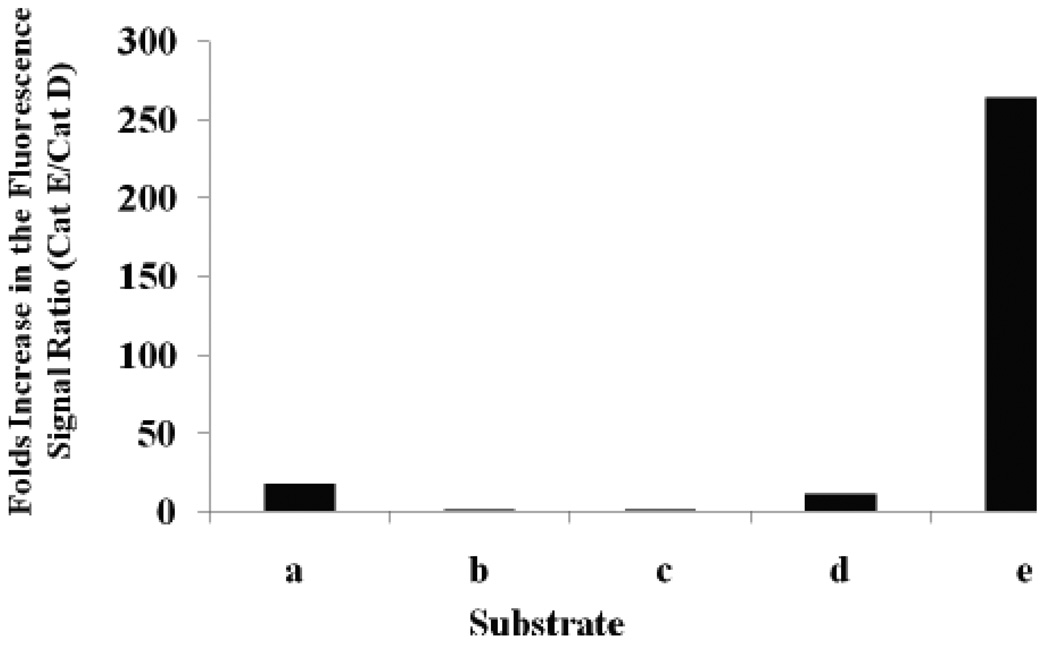
Substrate selectivity was represented by the number fold difference in fluorescence signal of Cath E over Cath D (Figure 2). Substrate e showed a striking 265-fold higher fluorescent signal ratio upon incubation with Cath E at 1 minute (Figure 2) and the ratio maintained high during the entire time period monitored. Comparatively, small differences were observed for substrates a and d, 17.9 and 12 fold, respectively, at the initial time point. The ratios continued to drop further to 4.5 and 4.0 fold at 10 minutes and to 1.6 and 2.3 fold, respectively, at the end of 45 min.
Figure 2.
Profile of net fluorescence signals ratio (Cath E/Cath D) of substrates a–e encountered at 1 minute after starting the enzymatic catalytic cleavage. Net fluorescence signals represent the signals after correction for the substrates quenched background signals.
Although the peptide sequences of substrates d and e are similar except at the prime side of the scissile bond, P1’ and P2’ positions, the difference in their fluorescence signal ratio was instantly recognizable. Placing a residue with exceptional conformational restrain, such as Pro at P1’ position of substrate e, has a constructive influence on the selectivity between Cath E and Cath D.
Negligible fluorescent signal ratios were observed for substrate b and c, 1.6 and 1.5 folds, respectively, immediately after incubation and remained low (Figure 2). This insignificant fluorescence signal ratio implicates the failure of the sequences possessing Phe-Phe, substrate b and c, at their scissile peptide bond in distinguishing between Cath E and Cath D. These fluorescence signals ratio results demonstrated the superiority of substrate e over the other investigated substrates in distinguishing Cath E and Cath D.
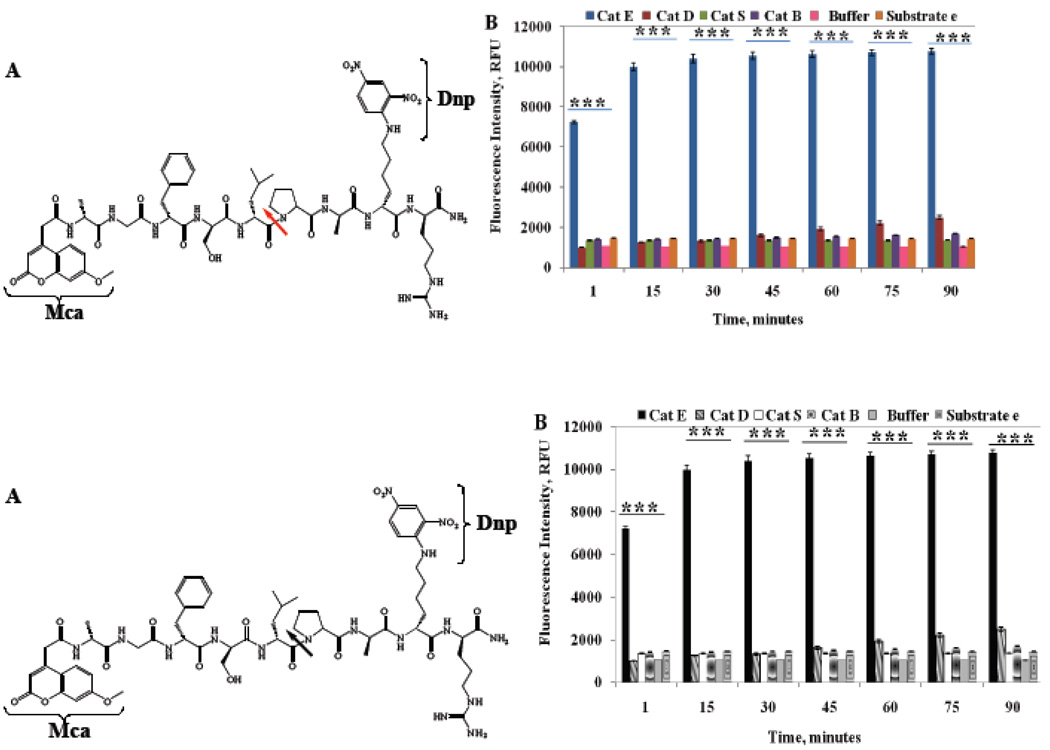
To further confirm the confined cleavage specificity to Cath E, the peptide substrate e was subjected to hydrolysis using two additional major lysosomal cathepsins, Cath S and Cath B, believed to be involved in similar biological catalytic activities (Figure 3A). As Cath E, upregulation and secretion of Cath B has been shown to occur in many types of tumors and to correlate positively with their invasive and metastatic capabilities by dissolving extracellular barriers [26]. Both Cath E and Cath S, a major lysosomal cysteine proteinase mediating degradation of class II major histocompatibility complex (MHC) in antigen presenting cells, have been reported to be involved in formation of amyloid proteins [9, 14].
Figure 3.
(A) Structure of intramolecular quenched substrate e, Mca-Ala-Gly-Phe-Ser-Leu**Pro-Ala-Lys(Dnp)-D-Arg-CONH2; (B) Change in the fluorescence intensity of substrate e (200 µM) during the incubation with 23 picomole of Cath E, Cath D, Cath S and Cath B in 50 mM NaOAc buffer of pH 4.0 for Cath E and D and 100 mM NaOAc buffer of pH 6.5 for Cath S and Cath B. Values represent the mean of at least three independent experiments. Error bars represent the upper and lower values of the Standard Error Mean (SEM). Asterisks represent the statistical significance of the two tailed P-values (***, P ≤ 0.001).
Similar to Cath D, limited increases in the fluorescence signal were observed by incubation of substrate e with Cath S or Cath B (Figure 3B). The fluorescent signal increased less than 15% of that observed with Cath E in 90 min. Excessive increases in the fluorescence signal of substrate e was observed immediately upon incubation with Cath E. The fluorescence signal continued increasing fairly quickly at 30 min of incubation with Cath E. Statistical analysis using Paired-Samples T-Test indicates the fluorescence differences are significant (two-tailed P-values of < 0.001). These results verify the pronounced specificity of substrate e to Cath E.
3.3. Cleavage Site Identification
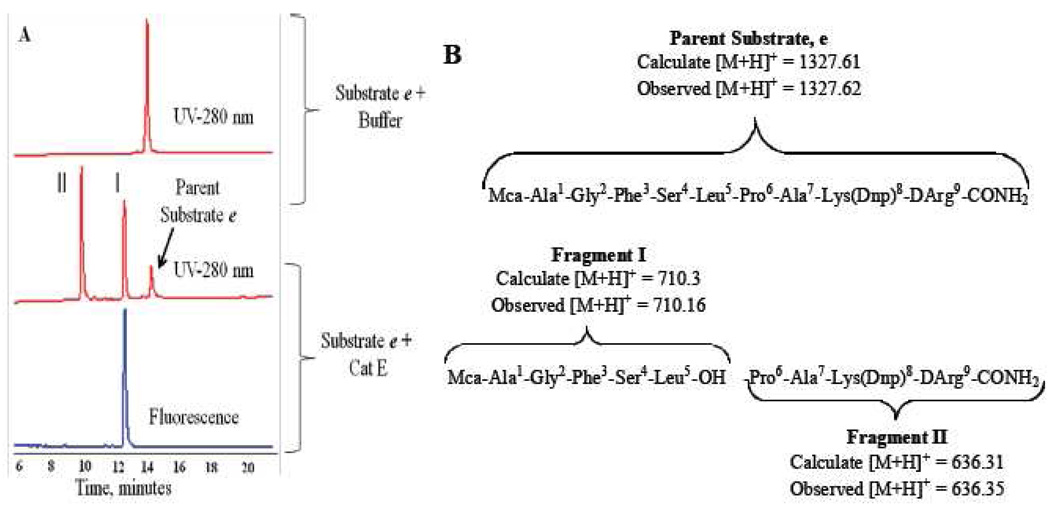
To identify the exact scission site, substrate e was incubated with five times more Cath E at 37 °C for 3 h. The incubation mixtures were analyzed by analytical HPLC (Figure 4A) and the identity of the digestion substrate fragments were characterized by ESI-MS (Figure 4B).
Figure 4.
(A) RP-HPLC Profile of peptide fragments obtained after digestion of 100 µM fluorogenic substrate e with Cath E (~119 picomole) in 50 mM sodium acetate buffer, pH 4.0, 150 mM NaCl at 37 °C for 3 h. UV absorbance detected at 280 nm. (B) The identified proteolytic fragments of substrate e and their ESI-MS characteristics.
The RP-HPLC chromatogram of the digested substrate e with Cath E showed only two major substrate fragments I and II (Figure 4A). The fluorescence-HPLC chromatogram revealed single fluorescent-peak overlaid with UV-peak of substrate fragment I. Based on LC-MS analysis, Peaks I and II resulting from Cath E digestion were corresponding to Mca-Ala1-Gly2-Phe3-Ser4-Leu5-OH m/z 709.3 [M+H+, 710.16], and -Pro6-Ala7-Lys(Dnp)8-DArg9-CONH2 m/z 635.31 [M+H+, 636.35], respectively. The LC-MS and fluorescence-HPLC results demonstrate that Cath E enzyme cleaves substrate e exclusively between Leu5 and Pro6.
When incubated with a high dose of Cath D for overnight hydrolysis, two minor cleavage sites were identified, one is between the P1 and P1’ sites (-Leu5-Pro6-) and a secondary site was found between P3 and P4 positions (-Gly2-Phe3-). (Data not shown)
3.4. Enzyme Inhibition and Selective Immunoprecipitation of Cath E
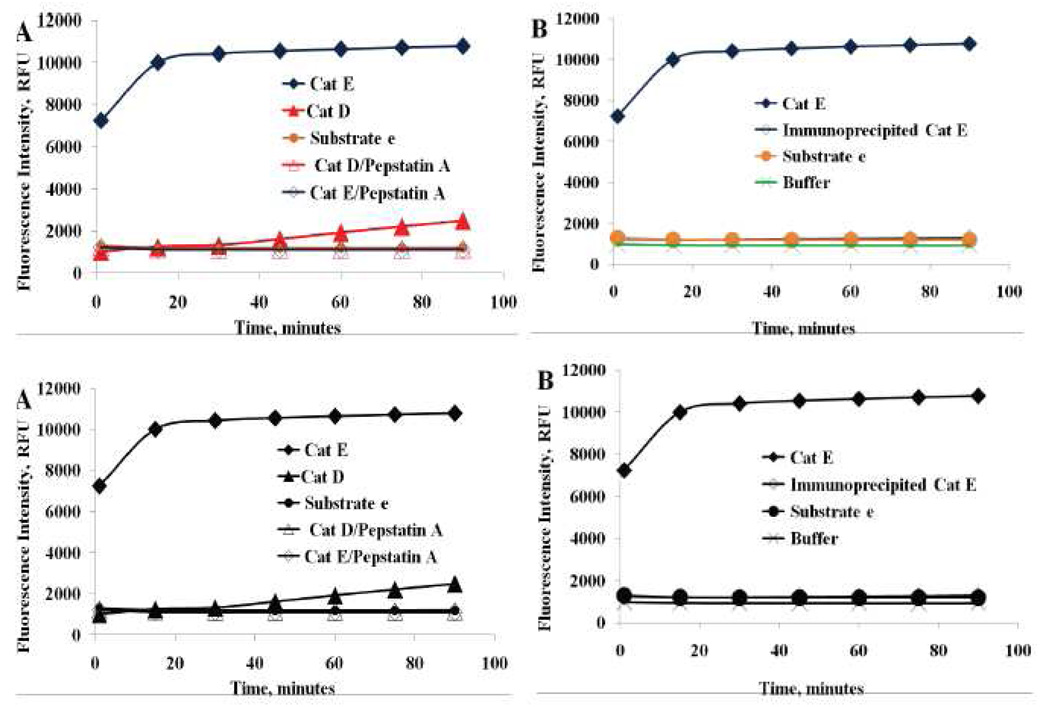
To further confirm the specificity of the substrate e activation, the inhibition effect of aspartic peptidases on substrate e hydrolysis was tested. Pepstain A was selected based on its specific universal-inhibition capabilities for acid peptidases [13, 36, 42]. It forms a complex with almost all aspartic peptidases without inhibiting cysteine or serine proteases [13, 36, 39], thus it would non-selectively inhibit the proteolytic activity of Cath E and D. In addition, selective inhibition of Cath E could be conducted using a specific anti-Cath E antibody, which recognizes both pro and mature Cath E with less than 1% cross-reactivity with Cath D according to the manufacturer data sheet. The antibody-Cath E complexes were pulled down by addition of antibody binding protein-G coupled insoluble matrix (sepharose beads). After centrifugation, the remaining supernatant was used for the enzyme specificity assay. The inhibition effect of pepstain A and anti-Cath E antibody on substrate e hydrolysis by of Cath E and Cath D was examined over the investigated time period (Figure 5).
Figure 5.
Effect of inhibition the enzymatic catalytic activity of Cath E and Cath D. Substrate e (200 µM) in 150 mM NaCl, 50 mM NaOAc buffer (pH 4.0) with 1µL of 1 mM pepstatin A/Methanol (panel A) and Selective immunoprecipitation using Cath E specific antibody in 1× PBS (panel B). All fluorescent measurements were collected with λex=340 nm and λem=405 nm.
The complete inhibition of the fluorescence signal by pepstatin A confirms that the registered fluorescence signal of substrate e is solely due to the aspartic peptidase Cath E and/or Cath D (Figure 5A). The specificity of the inhibition process was further confirmed by selective immunological precipitation of Cath E using a specific Cath E antibody. Complete absence of substrate e activation was observed after the selective immunoprecipitation of Cath E (Figure 5B). The evident inhibition by the specific Cath E antibody substantiate that the observed increase in the fluorescence signal of substrate e is exclusively due to the catalytic activation by Cath E enzyme.
3.5. Enzyme Kinetics
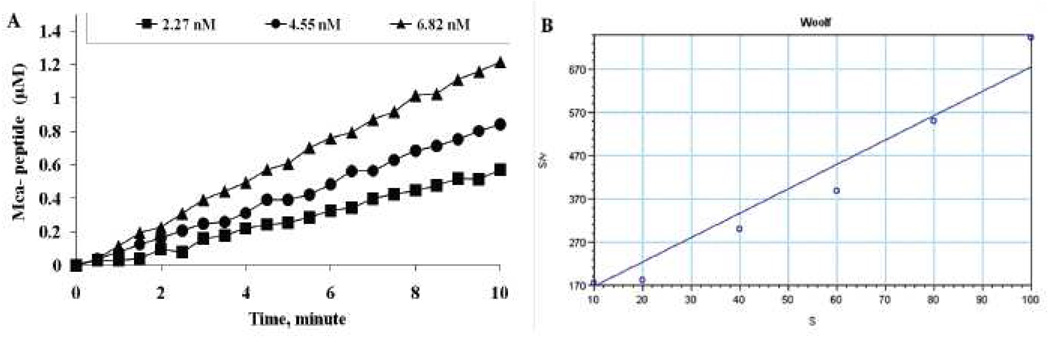
To quantify the kinetic constants of the Cath E enzyme, the proteolysis of substrate e was examined by incubating 10, 20, 40 and 60 µM of substrate e with 2.27, 4.55, 6.82 nM of Cath E for 10 minutes at 37°C. The change in the fluorescence intensity of each substrate e concentration was monitored over time using fluorescence spectrophotometer at λex= 340 nm and λem= 405 nm. Linear increases in the concentration of the fluorescently labeled digestion product were observed for all the investigated concentrations of substrate e and Cath E. Under the assay conditions, the linear phases of hydrolysis were maintained within the investigated time period between 0 and 10 minutes, even with the lowest concentration of substrate e (Figure 6A). The observed linear increase in the rate of the hydrolysis with increasing substrate e concentrations implies that the catalysis rate of Cath E is directly proportional with the concentrations of substrate e within the monitored period.
Figure 6.
(A) Dose response of proteolysis. Substrate e (10 µM) was incubated with various amount of Cathepsin E (2.27, 4.55, 6.82 nM). (B) Hanes-Woolf kinetic transformation diagrams of Cathepsin E (6.82 nM). Values represent the mean of triplicate measurements.
To estimate Michaelis-Menten Parameters (Vmax & Km), the velocities of the hydrolysis at early time points were measured at different concentrations (10, 20, 40, 60, 80, 100 µM) of substrate e. Initial rates of cleavage at varying concentrations of peptide substrate e were determined and values for Km and Kcat were calculated (Table 2). While the obtained Km value, 19.37 µM, indicates higher binding affinity of Cath E for the substrate e., the high Kcat value, 322.5 S−1, suggests a higher efficiency of Cath E in transforming substrate e to its hydrolyzed products. In contrast, no measurable cleavage was observed using up to 54 nM of Cath D under the same measurements condition.
Table 2.
Kinetic parameters for hydrolysis of fluorogenic substrate e by Cath E. All reactions and measurements were carried out in 50 mM sodium acetate buffers (pH 4.0) for 10 minutes at 37°C. Fluorescent measurements were collected with 340 nm excitation and 405 nm emissions.
| Cathepsin Enzyme |
Concentration (nM) |
Woolf Kinetic Transformation | ||||
|---|---|---|---|---|---|---|
| R2 | Vmax (µM.S−1) |
Km (µM) |
Kcat (S−1) |
Kcat/Km (µM−1.S−1) |
||
| E | 6.8 | 0.96 | 2.2 | 19.37 | 322.5 | 16.7 |
Table 3 compiles the reported specificity constant (Kcat/Km ) of Cath E and Cath D using various substrates including peptide substrate e. The calculated specificity constant value of Cath E enzyme for substrate e, 16.7 µM−1.S−1, was comparable to those reported for other substrates (Table 3). The high Kcat/Km value reveals the high specificity of Cath E in binding and transforming substrate e. Although sequences 6 and 7 show a considerably high Kcat/Km value for Cath E, 10.9 and 12.2, respectively, they also exhibit high values for Cath D, 15.6 and 16.3, respectively. Sequences 6 and 7, thus, were not able to distinguish Cath E from Cath D. Sequence 2 and sequence 3 possess almost the same succession except at P3 and P3’ positions, however, their Kcat/Km values for Cath E were vastly different. The Kcat/Km value of sequence 3 for Cath E was around 16 fold higher than the value of sequence 2. Interestingly, their Kcat/Km values for Cath D were extremely close. To the best of our knowledge, the specificity constant of Cath E for substrate e represents one of the highest reported Kcat/Km values. This is not only a reflection of the efficacy of Cath E in binding substrate e, but it also displays the effectiveness of transforming the bound substrate e into hydrolyzed fragments. Importantly, no measurable cleavage was observed using the Cath D enzyme under the same reaction conditions.
Table 3.
Specificity constant (Kcat/km) of Cath E for various substrates.
| Sequence Remarks |
Substrate Sequence | Catalytic Efficiency Kcat/Km (µM−1.S−1) |
Refs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P5 | P4 | P3 | P2 | P1 | *P1’ | P2’ | P3’ | Cath E | Cath D | ||
| 1, Substrate e | Ala | Gly | Phe | Ser | Leu | *Pro | Ala | Lys(Dnp) | 16.7 | NM§ | |
| 2 | Gly | Ser | Ser | Ala | Phe | *Leu | Ala | Phe | 0.69 | 0.35 | [13] |
| 3, Substrate a | Gly | Ser | Pro | Ala | Phe | *Leu | Arg | Lys | 11.2 | 0.92 | [13] |
| 4 | Lys | Pro | Ile | Leu | Phe | *Phe | Arg | Leu | NR¶ | 0.03 | [17] |
| 5 | Lys | Pro | Ile | Ser | Phe | *Phe | Arg | Leu | NR¶ | 0.14 | [17] |
| 6, Substrate c | Lys | Pro | Ile | Leu | Phe | *Phe | Arg | Leu | 10.9 | 15.6 | [39] |
| 7 | Lys | Pro | Ile | Ile | Phe | *Phe | Arg | Leu | 12.2 | 16.3 | [39] |
| 8 | Lys | Pro | Ile | Met | Phe | *Phe | Arg | Leu | NR¶ | 5.4 | [38] |
| 9 | Lys | Pro | Ile | Leu | Phe | *Phe | Arg | Leu | NR¶ | 7 | [38] |
| 10 | Lys | Pro | Ile | Cys | Phe | *Phe | Arg | Leu | NR¶ | 14.0 | [38] |
No measurable cleavage was observed using up to 54 nM of Cathepsin D under the same measurements condition.
Not Reported
4. Conclusions
The peptide substrate e, Mca-Ala-Gly-Phe-Ser-Leu-Pro-Ala-Lys(Dnp)-D-Arg-CONH2, was developed and characterized as a highly selective substrate for Cath E. Contrary to the previous perception that the existence of Phe at the substrates’ scissile bond is essential for Cath E hydrolysis, this study revealed the advantage of using Leu-Pro residues instead. The results obtained from this study exclusively establish that placing a conformationally restricted Pro residue at P1’ position of the substrate’s scissile bond yields selective cleavage by Cath E. This study demonstrates the promise of using the developed fluorogenic substrate e as a potential candidate for specific and sensitive detection of the proteolytic activity of Cath E while significantly distinguishing it from Cath D. Together with other substrates, it is possible to differentiate various proteases. This study also forms the foundation of Cath E specific inhibitor development in further studies.
Acknowledgments
This research was supported, in part, by National Institute of Health (NIH) Grants CA135312, and CA114149. We thank Dr. Prince V. Jeyabal for the helpful discussions on the immunoprecipitation experiment.
Abbreviations
- Cath
Cathepsin
- Mca
7-Methoxycoumarin-4-acetic acid
- Dnp
dinitrophenyl
- RP-HPLC
reversed phase high performance liquid chromatography
- MS
mass spectra
- UV
ultraviolet
- TNBS
2,4,6-trinitrobenzenesulfonic acid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Foltmann B, Szecsi PB. In: Handbook of Proteolytic Enzymes. Barrett AJ, Rawlings ND, Woessner JF, editors. San Diego: Academic Press; 1998. pp. 819–823. [Google Scholar]
- 2.Tang J. In: Handbook of Proteolytic Enzymes. Barrett AJ, Rawlings ND, Woessner JF, editors. San Diego: Academic Press; 1998. pp. 828–836. [Google Scholar]
- 3.Kageyama T, Takahashi K. J Biochem. 1980;87:725–735. doi: 10.1093/oxfordjournals.jbchem.a132801. [DOI] [PubMed] [Google Scholar]
- 4.Muto N, Arai KM, Tani S. Biochim Biophys Acta. 1983;745:61–69. doi: 10.1016/0167-4838(83)90170-x. [DOI] [PubMed] [Google Scholar]
- 5.Bennett K, Levine T, Ellis JS, Peanasky RJ, Samloff IM, Kay J, Chain BM. Eur J Immunol. 1992;22:1519–1524. doi: 10.1002/eji.1830220626. [DOI] [PubMed] [Google Scholar]
- 6.Sakai H, Saku T, Kato Y, Yamamoto K. Biochim Biophys Acta. 1989;991:367–375. doi: 10.1016/0304-4165(89)90130-x. [DOI] [PubMed] [Google Scholar]
- 7.Muto N, Yamamoto M, Tani S, Yonezawa S. J Biochem. 1988;103:629–632. doi: 10.1093/oxfordjournals.jbchem.a122318. [DOI] [PubMed] [Google Scholar]
- 8.Zaidi N, Herrmann T, Voelter W, Kalbacher H. Biochem Biophys Res Commun. 2007;360:51–55. doi: 10.1016/j.bbrc.2007.05.187. [DOI] [PubMed] [Google Scholar]
- 9.Zaidi N, Hermann C, Herrmann T, Kalbacher H. Biochem Biophys Res Commun. 2008;377:327–330. doi: 10.1016/j.bbrc.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 10.Kakehashi H, Nishioku T, Tsukuba T, Kadowaki T, Nakamura S, Yamamoto K. J Immunol. 2007;179:5728–5737. doi: 10.4049/jimmunol.179.9.5728. [DOI] [PubMed] [Google Scholar]
- 11.Sastradipura DF, Nakanishi H, Tsukuba T, Nishishita K, Sakai H, Kato Y, Gotow T, Uchiyama Y, Yamamoto K. J Neurochem. 1998;70:2045–2056. doi: 10.1046/j.1471-4159.1998.70052045.x. [DOI] [PubMed] [Google Scholar]
- 12.Chain BM, Free P, Medd P, Swetman C, Tabor AB, Terrazzini N. J Immunol. 2005;174:1791–1800. doi: 10.4049/jimmunol.174.4.1791. [DOI] [PubMed] [Google Scholar]
- 13.Yasuda Y, Kohmura K, Kadowaki T, Tsukuba T, Yamamoto K. Biol Chem. 2005;386:299–305. doi: 10.1515/BC.2005.036. [DOI] [PubMed] [Google Scholar]
- 14.Nakanishi H, Amano T, Sastradipura DF, Yoshimine Y, Tsukuba T, Tanabe K, Hirotsu I, Ohono T, Yamamoto K. J Neurochem. 1997;68:739–749. doi: 10.1046/j.1471-4159.1997.68020739.x. [DOI] [PubMed] [Google Scholar]
- 15.Rochefort H, Liaudet-Coopman E. Apmis. 1999;107:86–95. doi: 10.1111/j.1699-0463.1999.tb01530.x. [DOI] [PubMed] [Google Scholar]
- 16.Glondu M, Coopman P, Laurent-Matha V, Garcia M, Rochefort H, Liaudet-Coopman E. Oncogene. 2001;20:6920–6929. doi: 10.1038/sj.onc.1204843. [DOI] [PubMed] [Google Scholar]
- 17.Baechle D, Cansier A, Fischer R, Brandenburg J, Burster T, Driessen C, Kalbacher H. J Pept Sci. 2005;11:166–174. doi: 10.1002/psc.607. [DOI] [PubMed] [Google Scholar]
- 18.Ullmann R, Morbini P, Halbwedl I, Bongiovanni M, Gogg-Kammerer M, Papotti M, Gabor S, Renner H, Popper HH. J Pathol. 2004;203:798–807. doi: 10.1002/path.1584. [DOI] [PubMed] [Google Scholar]
- 19.Terris B, Blaveri E, Crnogorac-Jurcevic T, Jones M, Missiaglia E, Ruszniewski P, Sauvanet A, Lemoine NR. Am J Pathol. 2002;160:1745–1754. doi: 10.1016/S0002-9440(10)61121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uno K, Azuma T, Nakajima M, Yasuda K, Hayakumo T, Mukai H, Sakai T, Kawai K. J Gastroenterol Hepatol. 2000;15:1333–1338. [PubMed] [Google Scholar]
- 21.Matsuo K, Kobayashi I, Tsukuba T, Kiyoshima T, Ishibashi Y, Miyoshi A, Yamamoto K, Sakai H. Hum Pathol. 1996;27:184–190. doi: 10.1016/s0046-8177(96)90373-1. [DOI] [PubMed] [Google Scholar]
- 22.Caruso M, Moore J, Goodall GJ, Thomas M, Phillis S, Tyskin A, Cheetham G, Lerda N, Takahashi H, Ruszkiewicz A. Virchows Arch. 2009;454:291–302. doi: 10.1007/s00428-009-0731-0. [DOI] [PubMed] [Google Scholar]
- 23.Ladror US, Snyder SW, Wang GT, Holzman TF, Krafft GA. J Biol Chem. 1994;269:18422–18428. [PubMed] [Google Scholar]
- 24.Mackay EA, Ehrhard A, Moniatte M, Guenet C, Tardif C, Tarnus C, Sorokine O, Heintzelmann B, Nay C, Remy JM, Higaki J, Van Dorsselaer A, Wagner J, Danzin C, Mamont P. Eur J Biochem. 1997;244:414–425. doi: 10.1111/j.1432-1033.1997.00414.x. [DOI] [PubMed] [Google Scholar]
- 25.Dreyer RN, Bausch KM, Fracasso P, Hammond LJ, Wunderlich D, Wirak DO, Davis G, Brini CM, Buckholz TM, Konig G, et al. Eur J Biochem. 1994;224:265–271. doi: 10.1111/j.1432-1033.1994.00265.x. [DOI] [PubMed] [Google Scholar]
- 26.Nomura T, Katunuma N. J Med Invest. 2005;52:1–9. doi: 10.2152/jmi.52.1. [DOI] [PubMed] [Google Scholar]
- 27.Kageyama T. Methods Enzymol. 1995;248:120–136. doi: 10.1016/0076-6879(95)48010-2. [DOI] [PubMed] [Google Scholar]
- 28.Scarborough PE, Guruprasad K, Topham C, Richo GR, Conner GE, Blundell TL, Dunn BM. Protein Sci. 1993;2:264–276. doi: 10.1002/pro.5560020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto K, Katsuda N, Kato K. Eur J Biochem. 1978;92:499–508. doi: 10.1111/j.1432-1033.1978.tb12772.x. [DOI] [PubMed] [Google Scholar]
- 30.Jupp RA, Richards AD, Kay J, Dunn BM, Wyckoff JB, Samloff IM, Yamamoto K. Biochem J. 1988;254:895–898. doi: 10.1042/bj2540895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jupp RA, Dunn BM, Jacobs JW, Vlasuk G, Arcuri KE, Veber DF, Perlow DS, Payne LS, Boger J, de Laszlo S, et al. Biochem J. 1990;265:871–878. doi: 10.1042/bj2650871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bird JE, Waldron TL, Little DK, Asaad MM, Dorso CR, DiDonato G, Norman JA. Biochem Biophys Res Commun. 1992;182:224–231. doi: 10.1016/s0006-291x(05)80134-2. [DOI] [PubMed] [Google Scholar]
- 33.Rao CM, Scarborough PE, Kay J, Batley B, Rapundalo S, Klutchko S, Taylor MD, Lunney EA, Humblet CC, Dunn BM. J Med Chem. 1993;36:2614–2620. doi: 10.1021/jm00070a004. [DOI] [PubMed] [Google Scholar]
- 34.Kageyama T. Eur J Biochem. 1993;216:717–728. doi: 10.1111/j.1432-1033.1993.tb18191.x. [DOI] [PubMed] [Google Scholar]
- 35.Scarborough PE, Dunn BM. Protein Eng. 1994;7:495–502. doi: 10.1093/protein/7.4.495. [DOI] [PubMed] [Google Scholar]
- 36.Zaidi N, Herrmann T, Baechle D, Schleicher S, Gogel J, Driessen C, Voelter W, Kalbacher H. Febs J. 2007;274:3138–3149. doi: 10.1111/j.1742-4658.2007.05846.x. [DOI] [PubMed] [Google Scholar]
- 37.Dunn B. Nat Biotechnol. 2000;18:149–150. doi: 10.1038/72591. [DOI] [PubMed] [Google Scholar]
- 38.Gulnik SV, Suvorov LI, Majer P, Collins J, Kane BP, Johnson DG, Erickson JW. FEBS Lett. 1997;413:379–384. doi: 10.1016/s0014-5793(97)00886-7. [DOI] [PubMed] [Google Scholar]
- 39.Yasuda Y, Kageyama T, Akamine A, Shibata M, Kominami E, Uchiyama Y, Yamamoto K. J Biochem. 1999;125:1137–1143. doi: 10.1093/oxfordjournals.jbchem.a022396. [DOI] [PubMed] [Google Scholar]
- 40.Rao-Naik C, Guruprasad K, Batley B, Rapundalo S, Hill J, Blundell T, Kay J, Dunn BM. Proteins. 1995;22:168–181. doi: 10.1002/prot.340220209. [DOI] [PubMed] [Google Scholar]
- 41.Kyte J, Doolittle RF, Mol Biol J. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 42.Kitamura K, Yoshida C, Kinoshita Y, Kadowaki T, Takahashi Y, Tayama T, Kawakubo T, Naimuddin M, Salimullah M, Nemoto N, Hanada K, Husimi Y, Yamamoto K, Nishigaki K. J Mol Biol. 2009;87:1186–1198. doi: 10.1016/j.jmb.2008.12.028. [DOI] [PubMed] [Google Scholar]



 P1’ P2’ P3’ P4’
P1’ P2’ P3’ P4’