Abstract
D-eritadenine and (S)-DHPA are aliphatic adenosine analogues known to target S-adenosylhomocysteine hydrolase (SAHH) and potent antiviral compounds. In the present study, we demonstrate that these two compounds also display efficacy against recombinant SAHH enzyme of the protozoan parasite Cryptosporidium parvum, as well as inhibition of parasite growth in vitro. Our data confirm that SAHH could serve as a rational drug target in cryptosporidial infection and antiviral adenosine analogues are potential candidates for drug development against cryptosporidiosis.
Keywords: Cryptosporidium parvum, Apicomplexa, S-adenosylhomocysteine hydrolase, E.C. 3.3.1.1, D-eritadenine, (S)-DHPA, paromomycin, RT-qPCR
1. Introduction
More than three decades ago the compound (S)-9-(2,3-dihydroxypropyl)adenine [(S)-DHPA] was first identified as a broad spectrum antiviral adenosine analogue (De Clercq et al., 1978; Votruba and Holy, 1980). In the following years, (S)-DHPA (Fig. 1), D-eritadenine (Fig. 1) and several other adenosine analogues were found to target the S-adenosylhomocysteine hydrolase (SAHH) (De Clercq et al., 1984; De Clercq 2008; Holy et al., 1985; Schanche et al., 1984; Votruba and Holy, 1982). SAHH (E.C. 3.3.1.1) is an essential enzyme in all cells that hydrolyzes S-adenosylhomocysteine (SAH) into adenosine and L-homocysteine. Therefore, this enzyme is also explored as potential drug target in many bacteria and parasites (Bitonti et al., 1990; Henderson et al., 1992; Parker et al., 2003; Singh et al., 2006; Tanaka et al., 2004). The inhibition of SAHH activity could result in the accumulation of SAH and reduce the S-adenosylmethionine (SAM):SAH ratio in the cell, in which SAH further acts as a potent feedback inhibitor, blocking the SAM-dependent methylation required for the metabolism of a wide variety of biological compounds such as nucleic acids, proteins, phospholipids and other small molecules (Chiang et al., 1996; Chiang 1998; Nozaki, et al., 2005).
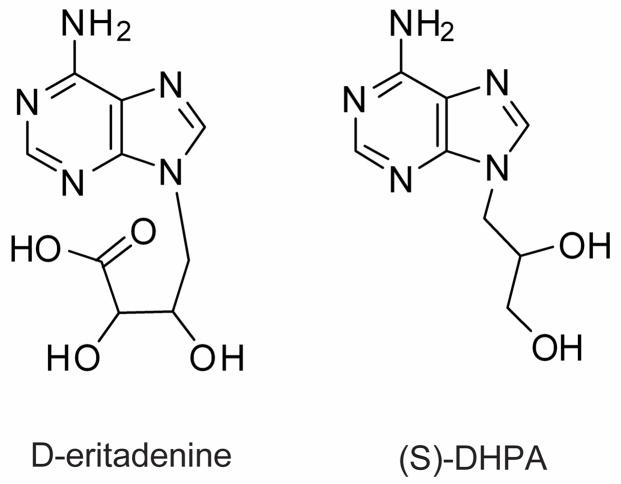
Fig. 1.
Structures of the compounds D-eritadenine and (S)-DHPA.
We have previously cloned, expressed and functionally characterized SAHH from the parasite Cryptosporidium parvum (CpSAHH), and principally observed that (S)-DHPA, D-eritadenine and Ara-A could inhibit the activity of recombinant CpSAHH, implying that CpSAHH could be explored as a potential drug target in this parasite (Ctrnacta et al., 2007). Here we extended that study by further testing detailed inhibitory kinetics of selected aliphatic adenosine analogues against recombinant CpSAHH, as well as their efficacies against the growth of C. parvum using an in vitro cryptosporidial infection model.
Cryptosporidium is a genus of unicellular parasites belonging to the Phylum Apicomplexa, of which C. parvum and C. hominis are the major species infecting humans. Their infection typically results in mild to severe, but self-limiting watery diarrhea in immunocompetent patients. However, their infection in immunocompromised individuals, such as AIDS patients, could be prolonged and life-threatening (Chen et al., 2002; Thompson et al., 2005; Tzipori and Widmer, 2008). Currently, no effective specific treatment is yet available to treat cryptosporidial infection in AIDS patients. New, specific drugs against this parasite are still urgently needed. Our discovery that aliphatic nucleoside analogs could effectively block the growth of the parasite could be another step in long search for new anticryptosporidial drug candidates.
2. Materials and methods
2.1. Recombinant CpSAHH inhibition assays
The cloning and expression of maltose-binding protein (MBP)-fused CpSAHH protein has been previously reported by us (Ctrnacta et al., 2007). Briefly, the CpSAHH gene was engineered into a pMAL-c2x expression vector and the expression and purification with an amylose-resin-based chromatography followed the manufacturer’s protocol (New England Biolabs). Purified MBP-CpSAHH fusion protein was digested with factor Xa to cleave the MBP-tag, and the tag was removed using a CHT 5-I hydroxyapatite column according to the manufacturer’s protocol (Bio-Rad). The purity of recombinant CpSAHH without the MBP-tag was analyzed using SDS-PAGE, and concentrations were determined by a Bradford protein assay. Protein aliquots were stored at −20 °C until use.
The enzymatic activity of the recombinant protein CpSAHH was spectrophotometrically assayed in the hydrolytic direction (Lozada-Ramirez et al., 2006) using 50 μM SAH as a substrate. The drugs used in this study were neutral (S)-DHPA [9-(S)-(2,3-dihydroxypropyl)adenine] and acidic derivate of (S)-DHPA, D-eritadenine [(2R,3R)-4-(6-aminopurin-9-yl)-2,3-dihydroxy-butanoic acid] (Fig. 1). Both inhibitors are adenosine analogues with sugar moieties replaced by aliphatic chains. Inhibitors were provided by Professor Antonin Holy at the Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic. Inhibition of CpSAHH was evaluated using various concentrations of D-eritadenine (0.01 μM – 1 μM) or (S)-DHPA (1 μM – 300 μM). The assay was carried out by pre-incubating 5 μL of 1 mg/ml CpSAHH with different concentrations of inhibitors for 10 min at 37 °C. The reaction started with the addition of the CpSAHH–inhibitor mixture into an enzyme reaction buffer (50 μM S-adenosylhomocysteine, 4 U Ado deaminase, 250 mM DNTB in 50 mM potassium phosphate buffer with 1 mM EDTA, pH 7.2) in a final volume of 1 ml. Enzyme activity was spectrophotometrically detected at 412 nm at 37 °C using a Shimadzu UV 1601 spectrophotometer. Controls included reactions without inhibitors, and those containing inhibitors, but no enzyme. Reactions were performed in at least four replicates.
2.2. Cultivation of parasite in vitro and drug treatments
All experiments used C. parvum oocysts (Iowa-1 strain) that were less than 3 months old, purified by Percoll gradient centrifugation and bleached as previously described (Nesterenko and Upton, 1996). HCT-8 (ATCC # CCL-244) cells (1.0 × 105 per well) were seeded into 48-well plates and allowed to grow until reaching ~80% confluence at 37 °C with 5% CO2 in RPMI 1640 medium containing 10% fetal bovine serum, 15 mM HEPES, and other supplements as previously described (Cai et al., 2005; Upton et al., 1995). For the generation of parasite standard curves, host cells were infected with 10-fold serial dilutions of oocysts (50 – 50,000). For all drug testing experiments, host cells were infected with 5,000 oocysts per well. Parasites were allowed to incubate with host cells at 37 °C for 4 h to allow for excystation and invasion into host cells. At this time, an exchange of culture medium was performed to remove parasites that failed to invade the host cells. The compounds D-eritadenine and (S)-DHPA were dissolved in water and added to the infected cell cultures at the specified final concentrations (0.01 μM – 1000 μM) during the medium exchange. Parasite-infected cultures were then incubated for 44 h at 37 °C in the presence of 5% CO2 (Cai et al., 2005). Each experimental condition was assayed in at least duplicates, and all experiments were repeated at least three times. Negative controls included cultures that received no parasites, and parasite-infected cultures that received no drug. Positive controls used various concentrations of paromomycin that is a commonly used standard inhibitor of Cryptosporidium growth in vitro (Cai et al., 2005).
Cytotoxicity of both inhibitors at 1 mM concentration on uninfected HCT-8 host cells was also examined using a standard fluorescent LIVE/DEAD viability/cytotoxicity assay for mammalian cells (Invitrogen/Molecular Probes). Cytotoxicity values for each inhibitor were also compared to cultures with and without the presence of paromomycin.
2.3. Efficacy of aliphatic adenosine analogs assayed by RT-qPCR
Total RNA was isolated from infected cultures 48 h post-infection (i.e., 44 h after drug addition) using an RNeasy isolation kit (Qiagen). The concentration and quality of RNA from each sample was determined by measuring their absorbance at 260 nm and 280 nm, and adjusted to a final concentration of 20 ng/μl. A SYBR green-based one-step real-time RT-qPCR method was performed using a QuantiTect SYBR Green RT-PCR kit (Qiagen). Each PCR reaction contained 20 ng of total RNA and proper amounts of reaction components as recommended by the manufacturer. The detection of parasite and human host cell 18S rRNA were detected using following primers: Cp18S-1011F (5′ TTG TTC CTT ACT CCT TCA GCA C 3′) and Cp18S-1185R (5′ TCC TTC CTA TGT CTG GAC CTG 3′) for C. parvum, and Hs18S-1373F (5′ CCG ATA ACG AAC GAG ACT CTG G 3′) and Hs18S-1561R (5′ TAG GGT AGG CAC ACG CTG AGC C 3′) for human host cells. Quantitative real-time RT-PCR was carried out in an iCycler iQ real-time PCR detection system (Bio-Rad). Reagents and primers were incubated at 48 °C for 30 min to synthesize cDNA, heated to 95 °C for 15 min to inactivate the reverse transcriptase. Then 40 thermal cycles were performed to amplify cDNA (95 °C for 20 s, 50 °C for 30 s, 72 °C for 30 s). At least two replicate reactions were performed for each sample in all PCR amplifications. The melting analysis showed a clear single peak without shoulder for each amplicon from each organism. Data was generated and analyzed following previously described methods (Cai et al., 2005) using GraphPad Prism v.5 software.
3. Results and discussion
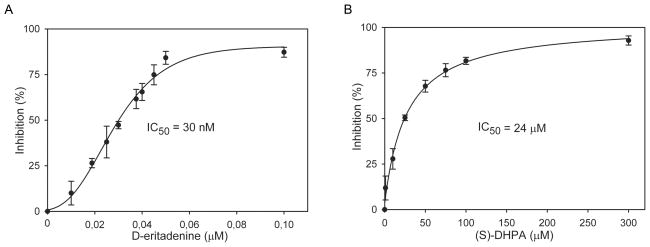
The two potent acyclic adenosine analogues, (S)-DHPA and D-eritadenine, displayed a dose-dependent inhibitory effect against the recombinant CpSAHH. The 50% inhibitory concentrations (IC50 values) calculated by nonlinear regression were at 30 nM (8.3 ng/ml) for D-eritadenine and 24 μM (5 μg/ml) for (S)-DHPA, respectively (Fig. 2). It was described previously that D-eritadenine was very potent inhibitor of isolated or recombinant SAHH from various sources (Holy et al., 1982; Huang et al, 2002; Schanche et al., 1984; Votruba and Holy, 1982). We confirmed that D-eritadenine is also an extremely good inhibitor of recombinant CpSAHH. Substrate (SAH) to inhibitor ratio of D-eritadenine was 1667:1, while substrate to inhibitor ratio for (S)-DHPA was only about 2:1. Maximum inhibitions were achieved between 0.1 μM – 1 μM for D-eritadenine (~ 91% inhibition) or at 300 μM for (S)-DHPA (~ 94% inhibition), respectively (Fig. 2).
Fig. 2.
Inhibition of D-eritadenine (A) and (S)-DHPA (B) on the activity of recombinant Cryptosporidium parvum S-adenosylhomocysteine hydrolase. The percent inhibition curves were derived by nonlinear regression using a sigmoidal model. Bars represent standard deviations derived from at least four replicates.
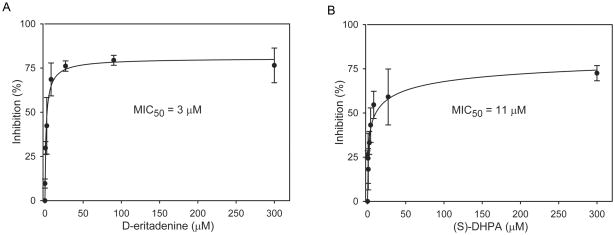
To evaluate drug effects against C. parvum in vitro, parasites were cultured by infecting human cells (HCT-8). From multiple techniques which are available for the evaluation of drug efficacy against C. parvum in vitro, an improved RT-qPCR based protocol described by Cai et al. (2005) was chosen. This method assesses the relative expression between parasite and host rRNA levels to minimize experimental and operational errors. Using RT-qPCR based protocol, (S)-DHPA and D-eritadenine also displayed dose-dependent inhibitory effects against the parasite growth in vitro (Fig. 3). Despite that the IC50 values on the recombinant CpSAHH activity for these two compounds differed by 800-fold, their minimum 50% inhibition concentrations (MIC50 values) on the parasite growth were much closer to each other, i.e., at 3 μM for D-eritadenine and 11 μM for (S)-DHPA, respectively. The notable difference between IC50 and MIC50 of D-eritadenine implied a possible very poor penetration of this polar compound into cells. The RT-qPCR assay was reliable, as the positive control compound paromomycin displayed an MIC50 value at 113 μM, which is comparable to the previously published value at 126 μM (Cai et al., 2005). At the maximal tested concentration of 0.3 mM, D-eritadenine and (S)-DHPA inhibited the parasite growth at approximately 80% and 86%, respectively (Fig. 3).
Fig. 3.
Inhibition of D-eritadenine (A) and (S)-DHPA (B) on the growth of C. parvum in vitro as determined by a RT-qPCR assay. The percent inhibition curves were derived by a nonlinear regression using a sigmoidal model. Bars represent standard deviations derived from at least six replicates.
Previous data on many cell lines have shown that neither D-eritadenine nor (S)-DHPA at effective concentrations significantly impair host cell viability (De Clercq et al., 1978; Holy, 1982; Votruba et al., 1983). In this study, we tested the cytotoxicity of these two compounds on HCT-8 host cells together with paromomycin at a concentration of 1 mM that is more than a hundred times higher than their MIC50 values (Tab. 1). At this high concentration, D-eritadenine displayed a similar level of cytotoxicity in comparison with paromomycin, while (S)-DHPA showed a slightly higher degree of cytotoxicity. Our results echo those previously published data, suggesting that both inhibitors would have very little effect on host cell growth if applied at administrable doses.
Tab. 1.
Cytotoxicity test of D-eritadenine, (S)-DHPA and paromomycin (1 mM each) on the uninfected HCT-8 cells cultured for 48 hours as determined by a fluorescent LIVE/DEAD assay (Invitrogen/Molecular Probes). The percent viability values in (S)-DHPA-treated cells differ significantly from those of paromomycin (by t-test, P = 0.017). Standard deviations were derived from at least four replicates.
| 1 mM drug | Viability of HCT-8 | SD |
|---|---|---|
| Paromomycin | 85% | 7% |
| D-eritadenine | 80% | 6% |
| (S)-DHPA | 70% | 4% |
Although nitazoxanide (NTZ) is currently available as the only drug approved by the United States Food and Drug Administration (FDA) for the treatment of cryptosporidiosis, NTZ is not completely effective, nor approved for use in AIDS patients (Anderson and Curran, 2007). Therefore, there is a need to develop new drugs against human Cryptosporidium infection, particularly for the immunocompromised patients. New drugs are also needed for the virtually untreatable cryptosporidiosis in animals. Recently published genome sequences have revealed that both C. parvum and C. hominis possess highly streamlined metabolic pathways with the absence of any de novo synthetic capacities such as the de novo synthesis of any amino acids (Abrahamsen et al., 2004; Xu et al., 2004). However, Cryptosporidium retains the ability to interconvert a number of amino acids, such as the interconversion between methionine and homocysteine that provides an essential methyl pool needed for the methylation of various nucleotides and proteins. We hypothesize that CpSAHH could serve as a drug target, as the inhibition of this third-step enzyme in this pathway would interrupt the vital methylation processes, thus killing the parasite (Ctrnacta et al., 2007).
The present study not only shows that two antiviral aliphatic adenosine analogues could inhibit the recombinant CpSAHH activity, but also confirms that these two compounds could inhibit the parasite growth in vitro, thus validating that SAHH is indeed a rational drug target in C. parvum. Even though these two compounds could only inhibit the parasite growth at micromolar concentrations, the convenience of the enzyme assay and the availability of a large number of analogues developed in the pursuit of antiviral therapy will permit the high-throughput screening of compounds against CpSAHH for highly selective inhibitors for future drug development.
Acknowledgments
We thank Dr. Janet S. Keithly at the Wadsworth Center, New York State Department of Health for co-mentoring the study and providing technical support. We thank Dr. Ivan Votruba at the Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic for the critical reading of the manuscript. We also thank Prof. Antonin Holy at the Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic for providing the inhibitors. This research was supported in part by: the New York State International Training and Research Program grant 2D43TW000233 (V.C.), NIH - Fogarty International Center, USA; the research projects MSM 0021620806 (V.C., F.S.), MSM0021620858 (I.H.) and AV0Z50520701 (V.C.) from the Ministry of Education of the Czech Republic; GAUK 4295/09 (V.C., F.S., M.S.) from the Grant Agency of the Charles University in Prague, Czech Republic; the National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health (NIH), USA (R01 AI44594) (G.Z., J.F., V.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Anderson VR, Curran MP. Nitazoxanide: a review of its use in the treatment of gastrointestinal infections. Drugs. 2007;67(13):1947–67. doi: 10.2165/00003495-200767130-00015. [DOI] [PubMed] [Google Scholar]
- Bitonti AJ, Baumann RJ, Jarvi ET, McCarthy JR, McCann PP. Antimalarial activity of a 4′,5′-unsaturated 5′-fluoroadenosine mechanism-based inhibitor of S-adenosyl-L-homocysteine hydrolase. Biochemical Pharmacology. 1990;40:601–606. doi: 10.1016/0006-2952(90)90562-y. [DOI] [PubMed] [Google Scholar]
- Cai X, Woods KM, Upton SJ, Zhu G. Application of quantitative real-time reverse transcription-PCR in assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum in vitro. Antimicrobial Agents and Chemotherapy. 2005;49:4437–4442. doi: 10.1128/AAC.49.11.4437-4442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XM, Keithly JS, Paya CV, LaRusso NF. Cryptosporidiosis. The New England Journal of Medicine. 2002;346:1723–1731. doi: 10.1056/NEJMra013170. [DOI] [PubMed] [Google Scholar]
- Chiang PK. Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacology & Therapeutics. 1998;77:115–134. doi: 10.1016/s0163-7258(97)00089-2. [DOI] [PubMed] [Google Scholar]
- Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. S-Adenosylmethionine and methylation. The FASEB Journal. 1996;10:471–480. [PubMed] [Google Scholar]
- Ctrnacta V, Stejskal F, Keithly JS, Hrdy I. Characterization of S-adenosylhomocysteine hydrolase from Cryptosporidium parvum. FEMS Microbiology Letters. 2007;273:87–95. doi: 10.1111/j.1574-6968.2007.00795.x. [DOI] [PubMed] [Google Scholar]
- De Clercq E. The discovery of antiviral agents: ten different compounds, ten different stories. Medicinal Research Reviews. 2008;28:929–953. doi: 10.1002/med.20128. [DOI] [PubMed] [Google Scholar]
- De Clercq E, Bergstrom DE, Holy A, Montgomery JA. Broad-spectrum antiviral activity of adenosine analogues. Antiviral Research. 1984;4(3):119–33. doi: 10.1016/0166-3542(84)90012-3. [DOI] [PubMed] [Google Scholar]
- De Clercq E, Descamps J, De Somer P, Holy A. (S)-9-(2,3-Dihydroxypropyl)adenine: an aliphatic nucleoside analog with broad-spectrum antiviral activity. Science. 1978;200:563–565. doi: 10.1126/science.200.4341.563. [DOI] [PubMed] [Google Scholar]
- Henderson DM, Hanson S, Allen T, Wilson K, Coulter-Karis DE, Greenberg ML, Hershfield MS, Ullman B. Cloning of the gene encoding Leishmania donovani S-adenosylhomocysteine hydrolase, a potential target for antiparasitic chemotherapy. Molecular and Biochemical Parasitology. 1992;53:169–183. doi: 10.1016/0166-6851(92)90019-g. [DOI] [PubMed] [Google Scholar]
- Holy A. Novel adenosine analogues with broad-spectrum biological activity: structure and mechanism of action. Nucleic Acids Symposium Series. 1982:199–202. [PubMed] [Google Scholar]
- Holy A, Votruba I, De Clerq E. Synthesis and antiviral activity of stereoisomeric eritadenines. Collection of Czechoslovak Chemical Communications. 1982;47:1392–1407. [Google Scholar]
- Holy A, Votruba I, De Clercq E. Structure activity studies on open chain analogues of nucleosides: inhibition of S-adenosyl-L-homocysteine hydrolase and antiviral activity I. Neutral open chain analogues. Collection of Czechoslovak Chemical Communications. 1985;50:245–261. [Google Scholar]
- Huang Y, Komoto J, Takata Y, Powell DR, Gomi T, Ogawa H, Fujioka M, Takusagawa F. Inhibition of S-adenosylhomocysteine hydrolase by acyclic sugar adenosine analogue D-eritadenine. Crystal structure of S-adenosylhomocysteine hydrolase complexed with D-eritadenine. The Journal of Biological Chemistry. 2002;277(9):7477–82. doi: 10.1074/jbc.M109187200. [DOI] [PubMed] [Google Scholar]
- Lozada-Ramirez JD, Martinez-Martinez I, Sanchez-Ferrer A, Garcia-Carmona F. A colorimetric assay for S-adenosylhomocysteine hydrolase. Journal of Biochemical and Biophysical Methods. 2006;67:131–140. doi: 10.1016/j.jbbm.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Nesterenko MV, Upton SJ. A rapid microcentrifuge procedure for purification of Cryptosporidium sporozoites. Journal of Microbiological Methods. 1996;25:87–89. [Google Scholar]
- Nozaki T, Ali V, Tokoro M. Sulfur-containing amino acid metabolism in parasitic protozoa. Advances in Parasitology. 2005;60:1–99. doi: 10.1016/S0065-308X(05)60001-2. [DOI] [PubMed] [Google Scholar]
- Parker NB, Yang X, Hanke J, Mason KA, Schowen RL, Borchardt RT, Yin DH. Trypanosoma cruzi: molecular cloning and characterization of the S-adenosylhomocysteine hydrolase. Experimental Parasitology. 2003;105:149–158. doi: 10.1016/j.exppara.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Schanche JS, Schanche T, Ueland PM, Holy A, Votruba I. The effect of aliphatic adenine analogues on S-adenosylhomocysteine and S-adenosylhomocysteine hydrolase in intact rat hepatocytes. Molecular Pharmacology. 1984;26:553–558. [PubMed] [Google Scholar]
- Singh V, Shi W, Almo SC, Evans GB, Furneaux RH, Tyler PC, Painter GF, Lenz DH, Mee S, Zheng R, Schramm VL. Structure and inhibition of a quorum sensing target from Streptococcus pneumoniae. Biochemistry. 2006;45:12929–12941. doi: 10.1021/bi061184i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Nakanishi M, Kusakabe Y, Shiraiwa K, Yabe S, Ito Y, Kitade Y, Nakamura KT. Three-dimensional structure of S-adenosyl-L-homocysteine hydrolase from Plasmodium falciparum. Nucleic Acids Symposium Series (Oxf) 2004;48:281–2. doi: 10.1093/nass/48.1.281. [DOI] [PubMed] [Google Scholar]
- Thompson RC, Olson ME, Zhu G, Enomoto S, Abrahamsen MS, Hijjawi NS. Cryptosporidium and cryptosporidiosis. Advances in Parasitology. 2005;59:77–158. doi: 10.1016/S0065-308X(05)59002-X. [DOI] [PubMed] [Google Scholar]
- Tzipori S, Widmer G. A hundred-year retrospective on cryptosporidiosis. Trends in Parasitology. 2008;24:184–189. doi: 10.1016/j.pt.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton SJ, Tilley M, Brillhart DB. Effects of select medium supplements on in vitro development of Cryptosporidium parvum in HCT-8 cells. Journal of Clinical Microbiology. 1995;33:371–375. doi: 10.1128/jcm.33.2.371-375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votruba I, Holy A. Inhibition of S-adenosyl-L-homocysteine hydrolase by the aliphatic nucleoside analogue 9-(S)-(2,3-dihydroxypropyl)adenine. Collection of Czechoslovak Chemical Communications. 1980;45:3039–3044. [Google Scholar]
- Votruba I, Holy A. Eritadenines - novel type of potent inhibitors of S-adenosyl-L-homocysteine hydrolase. Collection of Czechoslovak Chemical Communications. 1982;47:167–174. [Google Scholar]
- Votruba I, Holy A, De Clercq E. Metabolism of the broad-spectrum antiviral agent, 9-(S)-(2,3-dihydroxypropyl) adenine, in different cell lines. Acta Virologica. 1983;27:273–276. [PubMed] [Google Scholar]
- Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, Pearson WR, Dear PH, Bankier AT, Peterson DL, Abrahamsen MS, Kapur V, Tzipori S, Buck GA. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]