Abstract
Chaperone-mediated autophagy (CMA) is a selective lysosomal pathway for the degradation of cytosolic proteins. We review in this work some of the recent findings on this pathway regarding the molecular mechanisms that contribute to substrate targeting, binding and translocation across the lysosomal membrane. We have placed particular emphasis on the critical role that changes in the lipid composition of the lysosomal membrane play in the regulation of CMA, as well as the modulatory effect of other novel CMA components. In the second part of this review, we describe the physiological relevance of CMA and its role as one of the cellular mechanisms involved in the response to stress. Changes with age in CMA activity and the contribution of failure of CMA to the phenotype of aging and to the pathogenesis of several age-related pathologies are also described.
Keywords: chaperones, lysosomes, membrane proteins, protein translocation, proteases, proteolysis
1. Introduction: Intracellular protein degradation and the lysosomal system
Maintaining a balance between protein synthesis and degradation is absolutely essential for proper cellular functioning, cellular homeostasis, and cell survival in a changing extracellular environment [1, 2]. Protein degradation thus wears several hats in cells: First, as a recycling system, it mediates the breakdown of proteins that are no longer needed into constitutive amino acid components, which can then be used in the synthesis of new proteins [1]. Second, protein degradation serves also as a quality control mechanism, ensuring that proteins damaged or incorrectly synthesized are removed from the cells by degradation, preventing thus the devastating cellular consequences associated with accumulation of malfunctioning proteins inside cells [3]. Third, recent studies have identified the important role of protein degradation in cellular defense, as it contributes to the proteolytic breakdown of components of invading pathogens and other types of biological cell aggressors [4]. Fourth, protein degradation acts as a cautious controller of cellular homeostasis, because continuous degradation of most intracellular proteins limits the amount of time a given protein is exposed to the sometimes harsh and potentially altering cellular environment, thereby offering a preventative strategy against possible protein damage and dysfunction [3]. Finally, the cellular ability to rapidly change the rate of a particular protein’s degradation allows fast modulation of individual components of the proteome in response to stress or the changing extracellular millieu, making protein degradation a key player in cellular adaptation [2, 4].
Two major systems, present in almost all cell types, mediate complete degradation of intracellular proteins into their constitutive amino acids. The ubiquitin-proteasome system is a multi-subunit protease complex in the cytosol which permits entry and subsequent degradation of proteins tagged with one or more covalently bound ubiquitin molecules [5]. Subunits of the regulatory complex of the proteasome recognize the ubiquitin tag, remove it, and mediate the unfolding of the substrates required to gain access to the catalytic region of the proteasome barrel. With certain exceptions, most proteasome substrates are short-half life proteins such as newly synthesized, misfolded, and critical regulatory proteins involved in cell division, signaling, and transcription [1, 3, 5]. Postranslational modifications such as phosphorylation or oxidation also favor degradation through this cytosolic protease.
The autophagic/lysosomal system is the other major mechanism for intracellular proteolysis [6, 7]. Lysosomes are single membrane organelles dedicated to degradation of both intracellular and extracellular components. Their acidic pH and the large variety of hydrolases present in the lysosomal lumen (including proteases, lipases, glycosidases, and nucleases) mediate complete breakdown of all types of molecules and confer upon this organelle its high degradative capacity [8]. Substrates can reach lysosomes via heterophagy (including endocytosis and phagocytosis), in which the cargo to be degraded originates at the plasma membrane or extracellularly, or via autophagy, for cargo located in the cytosol, which is the main focus of this special issue of SCDB. Autophagy or “self eating” refers to the complete degradation of intracellular macromolecules (long-lived proteins and organelles) in lysosomes. Autophagy serves a variety of important cellular purposes, many related to the functions of intracellular proteolysis described above, which include, among others, constitutive protein and organelle recycling under basal conditions, generation of essential components (amino acids, free fatty acids, sugars) during conditions of nutritional stress or starvation, tissue remodeling during development and embryogenesis, antigen presentation and pathogen destruction as part of the acquisition of innate immunity, removal of altered or damaged cellular components, and it even participates in a type of cell death and in the removal of residual apoptotic bodies [6, 7].
There are three main types of autophagy in mammals. Macroautophagy (extensively reviewed in other articles of this special issue), involves the formation of a double-membrane structure (termed the phagophore or limiting membrane) which sequesters portions of the cytoplasm, including entire organelles and proteins, and seals to form a double-membrane vesicle or autophagosome. More than 30 gene products—generically known as autophagy-related, or Atg proteins participate in the formation of the autophagosome, which acquires the necessary hydrolytic enzymes to degrade its cargo upon fusion with lysosomes [6]. Like macroautophagy, microautophagy also involves nonspecific engulfment of cytoplasm, although in this case via direct invaginations of the lysosomal membrane itself to form intralysosomal vesicles which “pinch off” into the lumen and are degraded there by the lysosomal hydrolases [9]. A third type of autophagy, described so far only in mammalian cells, is chaperone-mediated autophagy (CMA), which constitutes the main focus of this review.
2. Chaperone-mediated autophagy: General properties
CMA is a uniquely selective form of autophagy by which specific cytosolic proteins are transported one-by-one across the lysosomal membrane for degradation [10, 11]. Unlike the other forms of autophagy, in which portions of the cytoplasm are typically engulfed in bulk (although there are specific types of micro- and macroautophagy), CMA is extremely selective for a subset of cytosolic soluble proteins, whereas this pathway cannot degrade organelles. CMA is constitutively active in many cell types, but, like macroautophagy, CMA is maximally activated under stress conditions (inducible CMA) such as nutritional stress or starvation and cellular stresses leading to protein damage [11]. Very often, both macroautophagy and CMA act in a synchronized or sequential manner. For example, during starvation, macroautophagy is first activated, and then, as starvation persists, cells switch from this bulk degradation to CMA, which mediates selective targeting of non-essential proteins for degradation to obtain the amino acids required for the synthesis of essential proteins [12, 13]. The intrinsic selectivity of CMA is also well suited for the removal of proteins damaged during stress without perturbing nearby normally functioning forms of the same protein. This selectivity is achieved by making the CMA-tag accessible to the chaperone in the altered protein but inaccessible when it is properly folded. Furthermore, during stress the selective removal by CMA of endogenous inhibitors of transcription factors known to contain the KFERQ-related motif, such as IκBα and c-Fos, also favors transcriptional activation of stress-related proteins, acting thus as a modulator of the severity of the cellular response to stress.
In addition to the involvement of CMA in basal cellular homeostasis and in the stress response, this autophagic pathway also bears specialized functions such as the recently proposed participation of CMA in antigen presentation [14]. Experimental reduction of CMA activity in cultured cells compromises their viability when exposed to different stressors and it is probably responsible, at least in part, for the accumulation of damaged proteins in their cytosol, highlighting the physiological importance of this pathway. In addition, improper CMA function has been identified in several nephropathies, some neurodegenerative disorders, and certain lysosomal storage diseases, and it is also a common characteristic of most cell types and tissues in aging organisms [11].
3. Molecular dissection of CMA
The distinctive characteristic of CMA—the selectivity for the degradation of a subset of soluble cytosolic proteins—is directly determined by two factors: the presence of a recognition-targeting motif in the amino acid sequence of the substrate proteins, and the fact that proteins access the lysosomal lumen one-by-one after unfolding [10]. The cytosolic and lysosomal molecular machineries that mediate this process are organized on the basis of those important features. They include the recognition of the substrate protein, mediated by the targeting signal and the complex of cytosolic chaperones that recognize this signal, and the translocation complex at the lysosomal membrane, which takes care of substrate binding, unfolding and translocation across the membrane.
3.1. The CMA targeting motif
All CMA substrates contain in their amino acid sequence a pentapeptide motif biochemically related to KFERQ, known as the CMA targeting motif [15]. The sequence KFERQ was found in the first identified CMA substrate, ribonuclease A (RNase A), where this motif was shown to be necessary and sufficient for its targeting for degradation to lysosomes. All CMA substrates have similar targeting motifs which adhere to the following characteristics: one amino acid in the sequence must be basic, another must be acidic, a third must be hydrophobic, and the fourth one can be either basic or hydrophobic, but never acidic. These residues can arrange in any order (e.g., sequences such as QREFK or VDKFQ is also recognized) but should be flanked on one of the sides by a glutamine. In particular contexts, this glutamine can be replaced by an asparagine [15]. It has been estimated through amino acid sequence analysis and affinity isolation experiments with specific antibodies that approximately 30% of cytosolic proteins bear a KFERQ-like CMA targeting motif sequence [16]. However, it is possible, at least theoretically, that the number of actual substrate proteins in a given cell becomes higher through post-translational modification of proteins bearing incomplete four-amino acid motifs. The presence of a CMA motif in the amino acid sequence of a protein is the first requirement to consider it a possible CMA substrate.
3.2. Cytosolic and lysosomal chaperones
The KFERQ sequence in the substrate proteins is recognized in the cytosol by a group of chaperones and co-chaperones, the predominant member of which is the heat shock cognate protein of 70-kDa (hsc70) [17] (Fig. 1). Hsc70 not only targets the CMA substrate to the lysosomal membrane, where it can interact with the CMA receptor, but it likely facilitates substrate unfolding [18], which is crucial for the protein’s translocation across the lysosomal membrane. Furthermore, as described in more detail below, hsc70 also regulates the dynamics of assembly and disassembly of the translocation complex.
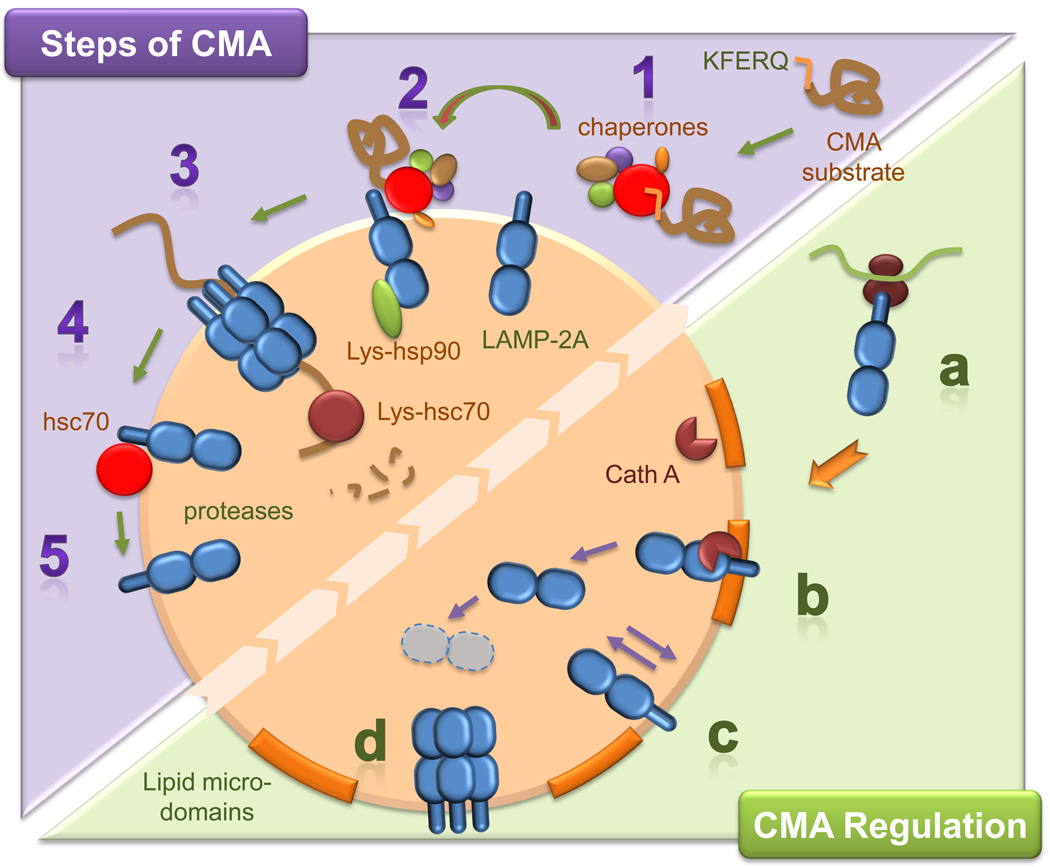
Figure 1. Schematic representation of the steps and regulatory mechanisms of CMA.
Top: Steps of CMA: Substrate proteins for CMA bear a targeting motif that, when recognized by a cytosolic chaperone complex (1), delivers them to lysosomes. At the membrane, substrate proteins bind to monomeric forms of LAMP-2A (2) and drive their multimerization into a higher order complex required for substrate translocation (3). Chaperones in the lumenal side of the membrane help stabilize LAMP-2A and mediate substrate translocation. Right after the substrate has crossed the lysosomal membrane, the same chaperone disassembles LAMP-2A from the translocation complex (4), allowing for new cycles of binding and uptake (5). Bottom: CMA Regulation: Levels of LAMP-2A at the lysosomal membrane are regulated in part by de novo synthesis (a), by changes in its regulated degradation in discrete lipid microdomains by cathepsin A (Cath A) (b), and by its insertion/deinsertion from the lysosomal lumen into the membrane (c). Besides the increase in LAMP-2A levels, multimerization of LAMP-2A (that only occurs outside the lipid microdomains) is also required to enhance the activity of this pathway (d).
In addition to the hsc70 in the cytosol, a lumenal form of hsc70 inside lysosomes is also required to complete substrate translocation via CMA [19, 20]. This lysosomal hsc70 (lys-hsc70) is the defining feature of the subset of cellular lysosomes competent to perform CMA. In fact, both the amount of lys-hsc70 in lysosomes and the number of lysosomes that contain lys-hsc70 increase under conditions such as starvation or oxidative stress, when maximal activation of this pathway is needed. For example, in rat liver, the percentage of CMA competent lysosomes fluctuates from approximately 20–30% of total lysosomes under normal feeding conditions to 60–80% of total lysosomes after 3 days of starvation [21, 22].
A second chaperone that also localizes on both sides of the lysosomal membrane is the heat shock protein of 90-kDa, or hsp90. The fraction of hsp90 at the cytosolic side of the lysosomal membrane is proposed to participate in substrate protein unfolding [18], although further direct experimental proof is necessary. Almost half of the lysosomal hsp90 associates with the lumenal side of the lysosomal membrane where it contributes to the stabilization of essential components of the translocation complex as they organize into a multimeric structure [23]. Other co-chaperones have been detected at the cytosolic side of the lysosomal membrane but their specific involvement in translocation of CMA substrates requires further investigation. Recent studies have proposed that the carboxyl terminus of hsc70-interacting protein (CHIP) may participate in the lysosomal targeting of the protein α-synuclein for its degradation in this compartment [24]. However, the type of autophagy responsible for CHIP-mediated lysosomal degradation has not been elucidated.
3.3. The lysosomal translocation complex
The identification of a protein at the lysosomal membrane, the lysosome-associated membrane protein type 2A (LAMP-2A), as a receptor for CMA substrates was the first step in the characterization of the machinery involved in binding and uptake of substrates by lysosomes via this autophagic pathway [25] (Fig. 1). LAMP-2A is one of the three splice variants of the lamp2 gene that give rise to three single-span membrane proteins, LAMP-2A, LAMP-2B, and LAMP-2C, all with a common highly N-glycosylated lumenal region, and different transmembrane and cytosolic tail regions [26]. CMA substrate proteins only bind to the cytosolic tail of LAMP-2A and not of LAMP-2B or C [27]. Mutagenesis analysis has revealed the importance of four positive residues in the LAMP-2A cytosolic tail for binding of substrate proteins [27]. However, the precise amino acids on the CMA substrate proteins required for receptor binding remain unknown. Binding of substrate proteins to LAMP-2A is limiting for CMA, and overexpression of LAMP-2A in cells in culture, but not the other two LAMP-2 variants, increases CMA activity [25]. Recent work has demonstrated that the function of LAMP-2A is not limited to that of a conventional receptor, but that it also participates in substrate translocation [23]. In fact, the complex dynamics and intricate degradation of the LAMP-2A receptor underlie almost every aspect of CMA.
Substrate proteins bind to monomers of LAMP-2A at the lysosomal membrane and this interaction drives the organization of LAMP-2A into multimeric complexes required for substrate translocation into lysosomes [23]. Once the substrate protein has reached the lysosomal lumen, LAMP-2A disassembles from this multimeric complex in order to enable subsequent rounds of substrate binding, which only takes place at free monomers of the receptor (Fig. 1). In fact, binding of substrate proteins is restricted to monomers of LAMP-2A, whereas translocation only occurs when the multimeric complex is formed. This continuous assembly and disassembly of LAMP-2A from the multimeric translocation complex highlights the importance of the lateral mobility of this protein in the lysosomal membrane. Lysosomal chaperones at both sides of the lysosomal membrane, and the subcompartmentalization of the lysosomal membrane into lipid microdomains modulate the lateral mobility of LAMP-2A [23, 28]. In addition to the well-characterized role of hsc70 in the delivery of substrate proteins to the lysosomal membrane, this chaperone is also required for the disassembly of LAMP-2A from the multimeric complexes once the substrate has reached the lysosomal lumen [23]. This disassembly function of hsc70 is counterbalanced by a novel lysosomal membrane protein, the glial fibrilar acid protein that stabilizes LAMP-2A into the multimeric complex in a GTP-dependent manner (Bandyophaday et al. under revision). Although further investigation is needed to determine the specific conformations adopted by LAMP-2A while transitioning from monomeric to multimeric complexes, the presence of hsp90 at the lumenal side of the lysosomal membrane is required to preserve the stability of LAMP-2A during this transition [23]. In addition, the organization of LAMP-2A at the lysosomal membrane is regulated by its dynamic association with discrete membrane lipid microdomains (Fig. 1). Under basal conditions, a large percentage of LAMP-2A at the lysosomal membrane localizes in lipid microdomains, where it undergoes a regulated turnover [28]. Upon CMA activation, LAMP-2A moves outside these regions into areas of higher fluidity, where multimerization into the translocation complex is possible.
4. Regulation of CMA
The signaling mechanisms involved in the activation of CMA and in the modulation of the activity of this autophagic pathway remain, for the most part, poorly characterized. In contrast, the local regulation of CMA at the lysosomal compartment has been extensively analyzed and has revealed the presence of precise, fine-tuned mechanisms that control the levels and accessibility to substrates of LAMP-2A at the lysosomal membrane (Fig. 1). CMA activity is directly proportional to the number of LAMP-2A molecules present at the lysosomal membrane at a given time [25], and changes in the membrane content of LAMP-2A are used by the cell to rapidly upregulate or downregulate the activity of this pathway. De novo synthesis of LAMP-2A, rates of turnover of this protein at the lysosomal membrane, and redistribution of LAMP-2A from the lysosomal lumen into the membrane, all contribute to modulate its lysosomal levels (Fig. 1). Under conditions of low CMA activity, LAMP-2A mobilizes into the lipid microdomains, where it is degraded in a two-step fashion, which first involves cleavage of the cytosolic/transmembrane region by a yet unidentified metalloprotease at the membrane, followed by cleavage by a serine protease, cathepsin A, that associates dynamically with the lumenal side of the lysosomal membrane [29]. This cleavage releases a truncated form of LAMP-2A, encompassing the bulk of this protein, into the lysosomal lumen, where it is rapidly degraded by the lysosomal hydrolases [29, 30]. Interestingly, when CMA substrates are present or upregulation of CMA activity is needed, mobilization of LAMP-2A and cathepsin A into the lipid microdomains decreases, thus protecting the CMA receptor from degradation. This blockage in degradation rather than increase of the synthesis of this receptor in order to increase the number of LAMP-2A molecules at the lysosomal membrane is particularly advantageous to the cells in conditions with limited access to amino acids such as when nutrients are scarce. Membrane levels of LAMP-2A can be further increased at the lysosomal membrane, without requiring de novo synthesis, through the mobilization of the pool of LAMP-2A normally resident in the lysosomal lumen back into the lysosomal membrane [30]. The exact nature of this luminal pool of LAMP-2A remains unknown, but previous studies have shown the presence of intact molecules of the protein inside lysosomes, and a gradual decrease in the percentage of LAMP-2 in this compartment as activation of CMA persists beyond 24 h. Although the lumenal LAMP-2A is not present inside visible vesicles, fractionation studies have revealed its association with lipids, supporting the existence of lumenal LAMP-2A–containing micelles [30]. By mechanisms still unclear, these micelles fuse or integrate into the lysosomal membrane under specific conditions, resulting in the incorporation of LAMP-2A in the membrane and exposure of its C terminus to the cytosol. Membrane chaperones and an intact membrane potential are also needed for this mobilization [30].
The other lysosomal component limiting for CMA activity is lys-hsc70. As indicated in the previous section, the presence of this chaperone at the lumenal side of the membrane is necessary to attain complete translocation of the substrates [19, 20]. Levels of lys-hsc70 increase gradually with the increase in CMA activity, although the mechanisms modulating this increase are still poorly understood [19, 20]. Lys-hsc70 is not delivered to lysosomes via macroautophagy or by CMA, because blockage of each of these pathways does not reduce the lysosomal content of this chaperone [13, 31]. It is possible that this chaperone reaches lysosomes through maturation of late endosomes, a compartment in which high levels of lumenal hsc70 have also been detected, highlighting thus a possible relationship between CMA and endocytosis.
In addition to the self-contained regulation of CMA in lysosomes by changes in levels of LAMP-2A and lys-hsc70, the activity of this autophagic pathway is also directly modulated by changes in other autophagic and proteolytic systems inside the cell. Cells in culture respond to CMA blockage by upregulating macroautophagy [13]. Similarly, blockage of macroautophagy results in constitutive activation of CMA [31]. These pathways are clearly not redundant, as CMA is, for example, unable to degrade organelles normally turned over by macroautophagy, and macroautophagy lacks the selectivity of CMA in the degradation of individual soluble cytosolic proteins. Nevertheless, the compensatory activation of one form of autophagy when the other is compromised is enough to preserve cellular homeostasis, at least under basal conditions. Additionally, blockage of either form of autophagy also has a direct impact on proteasomal activity. During the acute stages of CMA blockage there is an accumulation of polyubiquitinated proteins, often in the form of protein aggregates, attributable to the observed reduction in their removal through the proteasome system [32]. Similar inhibitory effects on proteasome activity have been detected upon blockage of macroautophagy, which contrasts with the upregulation in macroautophagy observed when the proteasome is inhibited [33]. The molecular mechanisms that regulate crosstalk among different pathways are currently under investigation. In the case of the interrelationship between macroautophagy and CMA, it has been shown that continuous fusion of autophagosomes to lysosomes when macroautophagy is upregulated results in transient dissipation of the lumenal lysosomal pH, which negatively affects lys-hsc70 stability. In fact, although lys-hsc70 is normally stable at pH ranges of 5.2–5.4, changes in pH values above 5.6 in the lysosomal lumen result in its rapid degradation in this compartment [19]. The reduced levels of lys-hsc70 in those lysosomes decreases their capability to perform CMA activity [31].
One topic that is gaining interest these days regarding CMA is the possible difference between basal and inducible CMA. For macroautophagy, growing evidence supports the existence of different regulatory mechanisms for the basal and inducible forms of this autophagic process and even the involvement of different effectors. Most cell types analyzed to date display some level of continuous CMA activity detectable in the absence of typical CMA inducing conditions. Basal CMA requires participation of the same effectors at the lysosomal membrane—the membrane chaperones and the protein translocation complex. However, whether the regulation of basal and inducible CMA takes place through different signaling mechanisms is unknown.
5. Physiological relevance of CMA
5.1. Activation of CMA during starvation
The first stimulus shown to activate CMA was nutritional starvation (Fig. 2A). Removal of serum in cultured cells for more than 10 hours, or prolonged starvation in animals (for up to 3 days), increase CMA activity and reduce the cellular content of KFERQ-containing proteins [21, 34]. Starvation-induced activation of CMA coincides with the decay in macroautophagy [12, 13]. Cells might benefit from switching to a more selective degradation, such that particular essential proteins could be maintained in the cytosol, while other, less critical ones, could be specifically targeted for degradation. For example, many glycolytic enzymes – unnecessary in the cells in absence of nutrients- contain in their sequence KFERQ-like targeting motifs and have been shown to undergo degradation through CMA during prolonged starvation. The specific changes that make the CMA-targeting motifs in these proteins accessible to the cytosolic hsc70 under these conditions require further investigation, but it is plausible that the lack of substrate or their disassembling from functional complexes could facilitate the exposure of the motif. CMA activation during nutrient deprivation is both tissue- and cell type-specific. Starvation is a potent CMA stimulus in liver, spleen, kidney, and heart, whereas other cells, such as particular neuronal populations, display higher basal levels of CMA activity, but starvation does not have a considerable impact on CMA-dependent degradation [35].
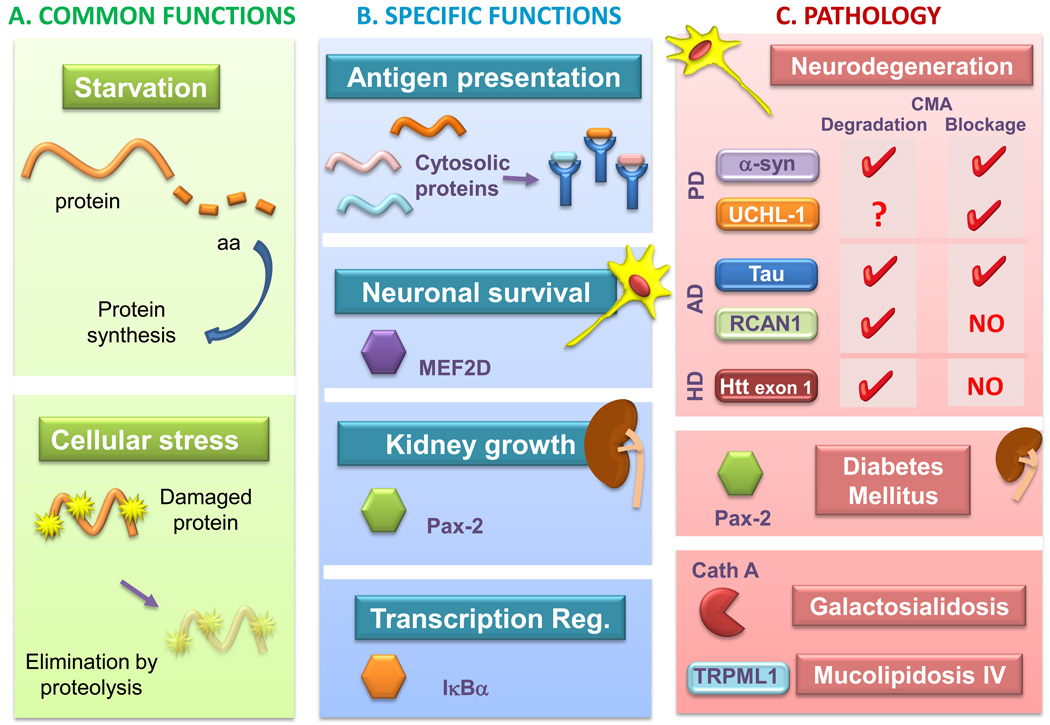
Figure 2. Pathophysiology of CMA.
This model depicts: A. Common functions of CMA in all cells. CMA is activated as part of the cellular response to stress to eliminate damaged proteins or during prolonged starvation, when it contributes amino acids for the synthesis of essential proteins. B. Specific functions of CMA. These functions are often linked to the degradation of particular proteins, such as endogenous proteins for antigen presentation, degradation of the neuronal survival factor MEF2D for neuronal homeostasis, of Pax-2 for kidney growth control, or of IκBα for transcriptional modulation in response to stress. C. Pathologies linked to CMA malfunctioning and the pathogenic proteins degraded by CMA and, where indicated, those responsible for CMA blockage. Abbreviations: aa, amino acids; MEF2D, myocyte enhancer factor 2D; Pax-2, paired box-related transcription factor; IκBα, inhibitor of kappa B; α-syn, α-synuclein; UCHL-1, ubiquitin carboxyl-terminal esterase L1; RCAN1, regulator of calcineurin 1; Htt, huntingtin; Cath A, cathepsin A; TRPML1, transient receptor potential mucolipin-1.
5.2. CMA and the cellular response to oxidative stress
The first connection between CMA and oxidative stress originated during the identification of IκBα as a substrate for this pathway. The increase in degradation of this transcriptional inhibitor by CMA in response to starvation is partially prevented upon treatment with antioxidants, suggesting that oxidation of this molecule somehow accelerates its degradation through CMA [36]. A more direct study comparing lysosomal binding and uptake of different CMA substrates, either unmodified or upon exposure to pro-oxidizing conditions, confirms that the oxidized proteins are more readily internalized in lysosomes than their unmodified counterparts [22] (Fig. 2A). Direct evidence for the upregulation of CMA during oxidative stress comes from the analysis of lysosomes isolated from cultured cells and livers of rodents treated with pro-oxidant compounds. In addition to detection of endogenous cytosolic oxidized proteins in the lumen of these lysosomes, they also show higher CMA ability, even for non-modified substrates [22]. A higher content of LAMP-2A in these lysosomes, due to transcriptional upregulation of this gene, seems behind the increased CMA activity [22]. Levels of other key CMA components, such as the lysosomal hsc70 and hsp90, are also augmented under these conditions, but the mechanisms that mediate their increase remain unknown.
Further studies in cultured cells in which CMA activity was reduced through knockdown of the lysosomal receptor for this pathway reveal that activation of CMA is required to preserve cell viability in response to diverse cellular stressors [13]. Thus, although cells respond to CMA blockage by upregulating macroautophagy, exposure of CMA-incompetent cells to oxidant and pro-oxidant factors or to ultraviolet light manifests a more severe compromise in cell viability than those with preserved CMA function. Lastly, the importance of CMA during oxidative stress has also been recently confirmed in whole animals. As described in more detail in the following sections, defective functioning of CMA with age in whole organs may contribute to the accumulation of oxidized proteins characteristic of most tissues in old organisms, because manipulations to prevent the age-dependent decrease of CMA in liver reduces the amount of oxidized proteins in this organ, even in mice of advanced age [37].
5.3. CMA and the immune response
Professional antigen presenting cells use the endosomal/lysosomal systems to internalize exogenous antigens, which can then be presented on the major histocompatibility complex (MHC) to CD4+ T cells. The proteasome and macroautophagy participate in processing and MHC loading with endogenous and exogenous antigens [4]. Involvement of CMA in antigen processing/presentation has also been recently proposed (Fig. 2B). Cells with reduced levels of LAMP-2A or hsc70 exhibit decreased display of cytoplasmic epitopes on class II molecules. Conversely, an increase in cytoplasmic autoantigen presentation is observed upon either overexpression of LAMP-2A or hsc70 [14]. The exact mechanisms that regulate this process and whether reduced CMA activity with age could contribute to the reduced immune response in old organisms require further investigation.
6. CMA and aging
Total rates of protein degradation decline with age in almost all tissues and organisms analyzed (from senescent fibroblasts in culture to fruit flies, nematodes, and mammals) (reviewed in [38]). In fact, reduced proteolytic capacity with age has been proposed to be responsible for the higher content of altered proteins and damaged organelles in old organisms. Quantitative and qualitative changes with age have been described for both the ubiquitin-proteasome system and lysosomes, and CMA is no exception [38]. CMA degradation of cytosolic proteins in senescent cultured fibroblasts is slower than in early passage cells, and this degradation is no longer upregulated upon serum starvation [39]. This decrease in CMA activity was later confirmed by comparing the ability of lysosomes isolated from livers of young and old rats to take up substrates [40]. A detailed dissection of the different steps involved in this pathway revealed that CMA substrate proteins are still recognized by hsc70 and targeted to the membrane of old lysosomes, and that if substrates are presented directly to the lysosomal hydrolases, their degradation is comparable to that observed when presented to lysosomal enzymes from young organisms [40]. However, CMA substrates exhibit a progressive reduction in their binding and uptake into lysosomes isolated from animals of increasing age. This decline in the key selective autophagy pathway is a direct result of a decrease in the levels of the CMA receptor, LAMP-2A, at the lysosomal membrane [40]. Although cells compensate initially for the reduced levels of the receptor by increasing the cellular pool of CMA-active lysosomes, with time a reduction in net CMA activity becomes apparent [40].
The decline in LAMP-2A levels with age is not due to transcriptional downregulation or changes in rates of synthesis and delivery of this receptor to the lysosomal compartment. Those parameters are comparable in young and old cells, whereas its dynamics and stability at the lysosomal membrane are severely compromised with age [41]. As previously mentioned, CMA is in part regulated by the controlled degradation of LAMP-2A at the lysosomal membrane and its dynamic distribution between the lysosomal membrane and lumen. Lysosomes isolated from livers of old rodents exhibit a pronounced decrease in LAMP-2A molecules at their membranes, due in part to inefficient reinsertion of the lumenal LAMP-2A into the lysosomal membrane and also to alterations in the regulated degradation of this receptor [41]. In fact, the regulated degradation that normally occurs at the lipid microdomains becomes inefficient—likely due to changes in the composition of these domains that diminish association of LAMP-2A and cathepsin A with these regions. A large portion of LAMP-2A is instead rapidly degraded in the lysosomal lumen contributing to the lower levels of this protein in lysosomes from old organisms [28].
Although probably not the only defect responsible for the decline of CMA with age, the decrease in the levels of LAMP-2A is the most important reason for the lower autophagic activity in old organisms. Using a double-transgenic mouse model in which LAMP-2A expression could be specifically modulated in the liver, our group found that it is possible to preserve normal CMA activity in the livers of these old transgenic mice until advanced ages [37]. The livers of these transgenic mice exhibited reduced levels of damaging oxidized and aggregated proteins, improved cellular homeostasis, preserved organelle structure and function, higher resistance to hepatotoxic compounds, and improved overall liver function as compared to old wild-type controls [37]. This work highlights that preventing the age-dependent defect in CMA is enough to preserve cellular homeostasis and may have important therapeutic applications as an anti-aging intervention.
7. CMA and disease
Conditions resulting in a primary defect in the lysosomal system may secondarily affect CMA activity. However, there are also a series of disorders in which dysfunction of CMA seems to directly contribute to their pathogenesis.
The potential role of CMA dysfunction in neurodegenerative diseases is currently a subject of increasing interest. Many of these diseases involve the aberrant accumulation of protein inclusions or aggregates in the cytosol of the affected neurons [42]. The intrinsic characteristics of CMA—translocation into lysosomes of single protein molecules upon unfolding—means that protein aggregates are unable to undergo degradation through this pathway. However, the fact that a growing number of these pathogenic proteins contain in their amino acid sequence one or several CMA-targeting motifs has opened the possibility that CMA may normally contribute to the degradation of the soluble forms of these proteins [42]. Direct connections with CMA have already been established for several of these pathogenic proteins (Fig. 2C).
The first pathogenic protein related to CMA was α-synuclein, the main component of the neuronal protein inclusions (Lewy bodies) in patients affected with Parkinson’s disease (PD). Although mutations in α-synuclein have only been described in a small percentage of PD patients, this protein aggregates in the form of Lewy bodies in all forms of PD. Wild-type α-synuclein is a CMA substrate, whereas pathogenic mutant forms of this protein are targeted to lysosomes by the CMA chaperones but fail to translocate inside the lysosomal lumen [43]. The presence at the lysosomal membrane of the tightly bound forms of mutant α-synuclein interferes with the degradation of other cytosolic proteins through this pathway [43] (Fig. 2C). Similar failure to translocate, and lysosomal blockage, was subsequently described for some post-translationally modified forms of wild-type α-synuclein, in particular as a result of the reaction of this protein with dopamine [44], supporting the idea that CMA impairment may contribute to both genetic and idiopathic forms of PD. These original findings using an in vitro system with isolated lysosomes and cortical and dopaminergic neurons in culture, have been recently confirmed by other groups [45]. In all experimental systems, cellular viability improves if lysosomal targeting of the pathogenic forms of α-synuclein is prevented by eliminating the CMA-targeting motif [43, 45]. Organization of the pathogenic forms of α-synuclein into irreversible oligomeric structures at the lysosomal membrane has been described [44], but the exact mechanisms by which the pathogenic forms of α-synuclein block translocation of CMA substrates is currently being investigated.
A second connection between CMA and PD has been recently established in studies showing that mutant forms of the ubiquitin carboxyl-terminal esterase L1 (UCHL-1) described in some familial forms of PD interact abnormally with CMA components [46] (Fig. 2C). Although no direct evidence for CMA of wild-type UCHL-1 has been found, this protein interacts with three of the main components of this pathway, namely hsc70, hsp90, and LAMP-2A. Mutant forms of the protein show an abnormally enhanced binding to LAMP-2A that was proposed to interfere with CMA activity and contribute to the higher levels of α-synuclein observed in cells expressing these mutant UCHL-1 variants [46]. Also, recent studies have revealed that changes in the degradation of the neuronal survival factor myocyte enhancer factor 2D (MEF2D) via CMA could also contribute to pathogenesis of PD [47] (Fig. 2B).
An unexpected role for CMA in Alzheimer’s disease (AD) has been proposed based also on the abnormal interaction of components of the lysosomal CMA translocation machinery with particular mutant forms of tau, a cytoskeleton-associated protein that contributes to the pathogenesis of AD (Fig. 2C). These tau mutants undergo proteolytic cleavage, which generates highly amyloidogenic fragments. In a recent study it was found that lysosomal enzymes are directly responsible for the two last cleavages, but that the tau mutants that generate amyloidogenic products do not reach lysosomes through this pathway; instead, they are targeted from the cytosol via CMA [48]. In contrast to regular CMA substrates, the truncated forms of tau undergo only partial insertion into the CMA translocon complex, enough to present their C terminus to the lysosomal hydrolases that mediate their cleavage into amyloidogenic peptides. As in the case of α-synuclein, the pathogenic forms of tau also organize into irreversible oligomeric structures at the lysosomal membrane that interfere with normal CMA, and in this case, also often result in destabilization of the lysosomal membrane and lysosomal enzyme leakage into the cytosol [48]. A second gene implicated in AD, the regulator of calcineurin 1 (RCAN1), also undergoes degradation by CMA [49] (Fig. 2C). This finding opens additional putative connections between AD and CMA such as the possible consequences to disease progression of changes in levels of RCAN1 as a result of the decline in CMA activity with age.
Analysis of the amino acid sequence of huntingtin, the protein that accumulates as nuclear and cytosolic aggregates in Huntington’s disease (HD) patients, has revealed the presence of three putative CMA-targeting motifs. None of these motifs is present inside the exon 1 of the protein, where the abnormally extended stretch of glutamines localize in the mutant pathogenic form. However, recent studies show that regulated phosphorylation by the inflammatory kinase IKK of exon 1 enhances the normal clearance of this product by both the proteasome and the lysosomes [50] (Fig. 2C). Interestingly, the lysosomal clearance is dependent on LAMP-2A and hsc70, supporting the idea that CMA may contribute to the removal of the pathogenic protein before aggregation, and that decline in activity of this pathway with age may be one of the aggravating factors in the progression of this disease.
Other diseases for which connections with CMA have already been established are different types of kidney disorders. Abnormally high levels of lys-hsc70 were originally described in different nephropathies, but the relevance of these changes was unclear [11]. Subsequent studies reveal that CMA is upregulated in tubular kidney cells in response to toxic-modification in lipid-binding proteins abundant in kidney, such as α-2-microglobulin [51]. Enhanced CMA activity favors the elimination of these toxic forms of the protein before they accumulate inside the cells. However, persistence of the toxic stimuli makes CMA degradation insufficient to deal with the protein load that accumulates in the form of hyaline droplets. Changes in the CMA degradation of the paired box-related transcription factor Pax2 in kidney has been proposed to be responsible for the renal hypertrophy associated with certain pathological conditions such as acute diabetes mellitus [52] (Fig. 2C). The decrease in CMA activity observed in the kidney of diabetic rats seems due to reduced levels of LAMP-2A and hsc70 in the diabetic kidney, although the mechanism behind the changes in these CMA components remains unknown.
As indicated above, all conditions resulting in primary alteration of the lysosomal compartment can indirectly affect CMA activity. That is the case in a group of disorders generically known as lysosomal storage diseases (LSD) in which a deficit in a particular lysosomal enzymatic activity leads to massive accumulation of undegraded substrate inside lysosomes and the consequent lysosomal dysfunction. However, there are two LSD for which alterations in CMA are not merely a consequence of the lysosomal alteration, but rather are due to the possible functional association of the mutant protein with CMA (Fig. 2C). The first one is galactosialydosis, a LSD caused by mutation or loss of protective protein/cathepsin A. This protein, in addition to functioning as a molecular chaperone for different lysosomal enzymes, contributes to their lysosomal targeting and stabilization, and is also involved in the regulated degradation of LAMP-2A through its carboxylpeptidase activity [29]. Loss of this enzymatic activity results in slower LAMP-2A degradation and the consequent maintained increase in CMA activity, even under normal basal conditions, which long term may contribute to the emaciation characteristic of these patients. The other LSD for which a direct connection with CMA has been recently described is mucolipidosis type IV [53]. In this case, the mutated protein, the transient receptor potential mucolipin-1 (TRPML1), interacts with the chaperones that participate in CMA, and cells from affected patients show decreased rates of CMA in response to serum removal. This inability to upregulate CMA could contribute to defective protein quality control of CMA substrate proteins under conditions of stress. A decrease in the levels of LAMP-2A seems behind the reduced rates of CMA, but how mutations in TRPML1 are connected to the observed changes in LAMP-2A requires future investigation.
Studies on the possible involvement of CMA in other common human disorders such as cancer, metabolic disorders, liver disease, etc. are currently ongoing, but in light of the importance of CMA in cellular homeostasis and in the cellular response to stress, it is anticipated that the list of CMA-related pathologies will keep growing. Furthermore, the possible aggravating effect of the generalized impairment in CMA activity with age in the large group of age-related disorders has just started to be unveiled.
8. Concluding remarks and pending questions
The unique characteristics of CMA—selectivity and direct translocation of substrate proteins across the lysosomal membrane—dictate the nature of the substrates for this autophagic pathway and determine the contribution of CMA to different aspects of cell physiology.
Despite the considerable advances in the molecular dissection of this pathway in recent years, there are still many pieces missing from the CMA “puzzle”. For instance, it remains unclear whether other organisms (besides mammals) perform CMA or if this selective pathway evolved from a more primitive system or as result of some specific needs of higher eukaryotes. Further characterization of the translocation machinery as well as the particular function of each of the chaperones known to associate with the hsc70-substrate complex is also needed.
Although the possibility of genetically modulated levels of LAMP-2A and subsequently CMA activity, both in cultured cells and animals, has been pivotal in our current understanding of the functional relevance of this pathway, small molecules or pharmacological compounds able to activate or inhibit CMA in an acute manner are still unavailable. These compounds would help to further elucidate key aspects of this pathway, while at the same time providing a key tool capable of separating CMA from other forms of autophagy.
Of particular interest are the interactions of CMA with other autophagic pathways. Identification of the molecular effectors of this autophagic crosstalk could provide new targets for upregulation and downregulation of autophagy in physiological and pathological conditions. This interdependence of the autophagic pathways may be of relevance in certain pathologies in which a primary defect in an autophagic pathway could be compensated for by another autophagic pathway for years. Currently, major emphasis has been placed on the analysis of the involvement of CMA in neurodegenerative diseases, but it is anticipated that there may be more diseases for which CMA dysregulation could play a key role.
Acknowledgements
Work in our laboratory is supported by NIH grants from NIA (AG021904, AG031782), NIDKK (DK041918), NINDS (NS038370), a Glenn Foundation Award and a Hirsch/Weill-Caulier Career Scientist Award. S.O. is supported by a NIH/NIA training grant T32 AG023475.
Abbreviations
- AD
Alzheimer’s disease
- CMA
chaperone-mediated autophagy
- HD
Huntington’s disease
- hsc70
heat shock cognate protein of 70 kDa
- hsp
heat shock protein
- LAMP
lysosome-associated membrane protein
- LSD
lysosomal storage disorders
- PD
Parkinson’s disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ciechanover A. Intracellular protein degradation: from a vague idea through the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Hematology Am Soc Hematol Educ Program. 2006;1:505–506. doi: 10.1182/asheducation-2006.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Knecht E, Aguado C, Carcel J, Esteban I, Esteve JM, Ghislat G, et al. Intracellular protein degradation in mammalian cells: recent developments. Cell Mol Life Sci. 2009;66:2427–2443. doi: 10.1007/s00018-009-0030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;18:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 4.Deretic V. Links between autophagy, innate immunity, inflammation and Crohn's disease. Dig Dis. 2009;27:246–251. doi: 10.1159/000228557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dice J. Lysosomal Pathways of Protein Degradation. Molecular Biology Intelligence Unit. Austin, TX: Landes Bioscience; 2000. [Google Scholar]
- 9.Ahlberg J, Glaumann H. Uptake--microautophagy--and degradation of exogenous proteins by isolated rat liver lysosomes. Effects of pH, ATP, and inhibitors of proteolysis. Exp Mol Pathol. 1985;42:78–88. doi: 10.1016/0014-4800(85)90020-6. [DOI] [PubMed] [Google Scholar]
- 10.Dice J. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 11.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab. 2009 doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuertes G, Martin De Llano J, Villarroya A, Rivett A, Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem. J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006;103:5905–5910. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou D, Li P, Lin Y, Lott JM, Hislop AD, Canaday DH, et al. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Dice J. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 16.Chiang H, Dice J. Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J Biol Chem. 1988;263:6797–6803. [PubMed] [Google Scholar]
- 17.Chiang H, Terlecky S, Plant C, Dice J. A role for a 70 kDa heat shock protein in lysosomal degradation of intracellular protein. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 18.Agarraberes F, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–2499. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- 19.Cuervo A, Dice J, Knecht E. A lysosomal population responsible for the hsc73-mediated degradation of cytosolic proteins in lysosomes. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 20.Agarraberes F, Terlecky S, Dice J. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuervo A, Knecht E, Terlecky S, Dice J. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 22.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 25.Cuervo AM, Dice J. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 26.Konecki D, Foetisch K, Zimmer K, Schlotter M, Lichter-Konecki U. An alternatively spliced form of the human lysosome-associated membrane protein-2 gene is expressed in a tissue-specific manner. Biochem Biophys Res Comm. 1995;215:757–767. doi: 10.1006/bbrc.1995.2528. [DOI] [PubMed] [Google Scholar]
- 27.Cuervo A, Dice J. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 28.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuervo AM, Mann L, Bonten E, d'Azzo A, Dice J. Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 2003;22:12–19. doi: 10.1093/emboj/cdg002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuervo A, Dice J. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 31.Kaushik S, Massey A, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massey AC, Follenzi A, Kiffin R, Zhang C, Cuervo AM. Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy. 2008;4:442–456. doi: 10.4161/auto.5654. [DOI] [PubMed] [Google Scholar]
- 33.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wing S, Chiang H, Goldberg A, Dice J. Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem J. 1991;275:165–169. doi: 10.1042/bj2750165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuervo AM. Autophagy: many pathways to the same end. Mol Cell Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- 36.Cuervo AM, Hu W, Lim B, Dice JF. IκB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell. 1998;9:1995–2010. doi: 10.1091/mbc.9.8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genetic. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dice J. Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem. 1982;257:14624–14627. [PubMed] [Google Scholar]
- 40.Cuervo AM, Dice J. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 41.Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC, et al. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 43.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Vicente M, Talloczy Z, Kaushik S, Massey A, Mazzulli J, Mosharov E, et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xilouri M, Vogiatzi T, Vekrellis K, Park D, Stefanis L. Abberant α-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS One. 2009;4:e5515. doi: 10.1371/journal.pone.0005515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabuta T, Wada K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008;283 doi: 10.1074/jbc.M801918200. 23731-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Q, She H, Gearing M, Colla E, Lee M, Shacka J, et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H, Wang P, Song W, Sun X. Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J. 2009;23:3383–3392. doi: 10.1096/fj.09-134296. [DOI] [PubMed] [Google Scholar]
- 50.Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuervo A, Hildebrand H, Bomhard E, Dice J. Direct lysosomal uptake of α2-microglobulin contributes to chemically induced nephropathy. Kidney Int. 1999;55:529–545. doi: 10.1046/j.1523-1755.1999.00268.x. [DOI] [PubMed] [Google Scholar]
- 52.Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 53.Venugopal B, Mesires N, Kennedy J, Curcio-Morelli C, Laplante JM, Dice JF, et al. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol. 2009;219:344–353. doi: 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]