Abstract
The sulfhydryl oxidase augmenter of liver regeneration (ALR) binds FAD in a helix-rich domain that presents a CxxC disulfide proximal to the isoalloxazine ring of the flavin. Head-to-tail interchain disulfide bonds link subunits within the homodimer of both the short, cytokine-like, form of ALR (sfALR), and a longer form (lfALR) which resides in the mitochondrial intermembrane space (IMS). lfALR has an 80-residue N-terminal extension with an additional CxxC motif required for the reoxidation of reduced Mia40 during oxidative protein folding within the IMS. Recently Di Fonzo et al. (Di Fonzo, A., Ronchi, D., Lodi, T., Fassone, E., Tigano, M., Lamperti, C., Corti, S., Bordoni, A., Fortunato, F., Nizzardo, M., Napoli, L., Donadoni, C., Salani, S., Saladino, F., Moggio, M., Bresolin, N., Ferrero, I., and Comi, G. P. (2009) Am. J. Hum. Genet. 84, 594–604) described an R194H mutation of human ALR that led to cataract, progressive muscle hypotonia, and hearing loss in three children. The current work presents a structural and enzymological characterization of the human R194H mutant in lf- and sfALR. A crystal structure of human sfALR was determined by molecular replacement using the rat sfALR structure. R194 is located at the subunit interface of sfALR, close to the intersubunit disulfide bridges. The R194 guanidino moiety participates in three H-bonds: two main-chain carbonyl oxygen atoms (from R194 itself, and from C95 of the intersubunit disulfide of the other protomer) and with the 2' OH of the FAD ribose. The R194H mutation has minimal effect on the enzyme activity using model and physiological substrates of short and long ALR forms. However the mutation adversely affects the stability of both ALR forms: e.g. by decreasing the melting temperature by about 10 °C, by increasing the rate of dissociation of FAD from the holoenzyme by about 45-fold, and by strongly enhancing the susceptibility of sfALR to partial proteolysis and to reduction of its intersubunit disulfide bridges by glutathione. Finally, a comparison of the TROSY-HSQC 2D NMR spectra of wild type sfALR and its R194H mutant reveal a significant increase in conformational flexibility in the mutant protein. In sum, these in vitro data document the major impact of the seemingly conservative R194H mutation on the stability of dimeric ALR and complement the in vivo observations of Di Fonzo et al.
Augmenter of liver regeneration (ALR1; also abbreviated as GFER, HPO and HSS) is a representative of a group of small sulfhydryl oxidases whose founding member, Erv1p, was first recognized as a protein essential for respiration and vegetative growth in yeast (1, 2). Since that time a number of Erv/ALR family members have been characterized including yeast Erv2p and ALR analogs from certain double-stranded DNA viruses, and from plants and animals (3). These proteins share a common flavin binding fold in which the isoalloxazine ring is inserted into the mouth of a bundle of four helices (3). This novel flavin binding mode, first recognized for yeast Erv2p (4), and then for rat ALR (5) and yeast Ero1p (6), is found in all of the well-characterized sulfhydryl oxidases including the larger multidomain Quiescin-sulfhydryl oxidase (QSOX) enzymes (7–10).
Mammalian ALR is a covalent homodimer and is found in two splice variants (11–16). The short form (sfALR, 15 kDa; starting at M81 of the human long form, lfALR, sequence depicted in Figure 1A) is a circulating growth factor (11, 12, 15, 17, 18) and interacts with specific receptors on the cell surface (19, 20). Receptors for sfALR stimulate the mitogen-activated protein kinase cascade leading to enhanced liver regeneration (19, 21) and recovery of renal tubular cells from ischemic/reperfusion injury (22). sfALR is also found in the cytosol and in the nucleus and interacts with Bcl-2/adenovirus E1B 19kDa interacting protein 2-like (23) that confers protection against the effects of viral infection and proapoptotic stimuli (24). sfALR also binds to Jun activation domain-binding protein 1 of the mammalian COP9 signalosome (25). While interest in sfALR is increasing, specific details concerning its enzymatic activity, locale, and mode of action remain unclear.
FIGURE 1.
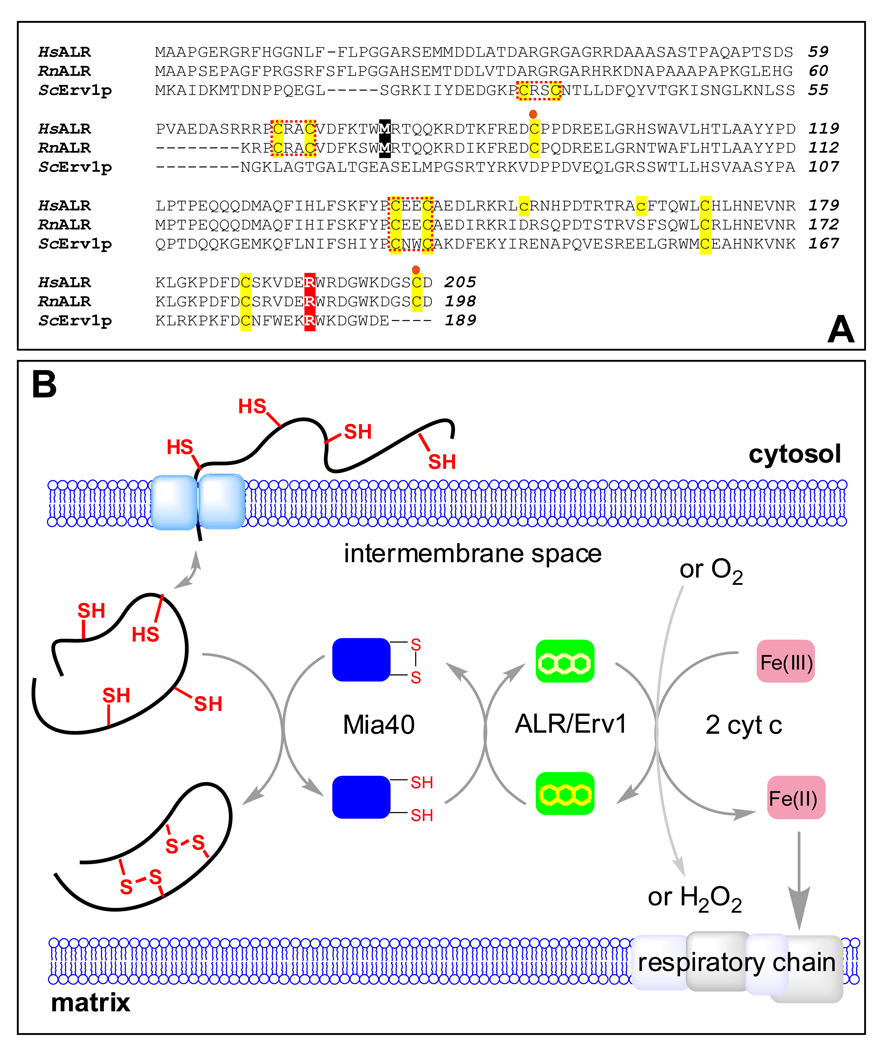
Comparison of sequences of human and rat ALR with yeast Erv1p and a depiction of the role of ALR in oxidative folding in the mitochondrial intermembrane space. Human, rat and yeast sequences are indicated by Hs Rn and Sc prefixes respectively in the alignment shown in Panel A. All cysteines are highlighted in yellow. The N-terminal (distal) and C-terminal (proximal) redox-active CxxC motifs are enclosed within red dotted boxes. The cysteine residues forming intersubunit disulfide bonds are denoted by orange circles. The two cysteine residues in human ALR that are mutated to prevent aggregation are shown in lower case. The methionine residue highlighted in black represents the N-terminus of the short form of mammalian ALR. The position of the R194 mutant in human ALR is highlighted in red. Panel B: Proteins undergoing oxidative folding in the IMS transfer electrons to Mia40 which then transmits them to long form ALR (or Erv1p in yeast). The terminal electron acceptor is either cytochrome c or molecular oxygen.
Long form ALR (23 kDa), like its yeast counterpart, Erv1p, populates the intermembrane space (IMS) of the mitochondrion, where it participates in a chain of disulfide exchange reactions that generate disulfide bonds in a number of resident proteins with twin Cx3C and Cx9C motifs (26–37) (Figure 1B). ALR is currently classified as a flavoprotein oxidase catalyzing the generation of disulfides with the following stoichiometry:
However, cytochrome c is a much better electron acceptor than oxygen for both sf- and lfALR in vitro (38, 39) This finding led us to suggest that reduction of cytochrome c could minimize the generation of reactive oxygen species associated with generation of disulfides in the IMS (Figure 1B, (38)). Subsequent in vivo studies of the yeast mitochondrial IMS have confirmed that Erv1p and cytochrome c can interact productively during oxidative folding (37, 40). Alternatively, any hydrogen peroxide liberated from these flavoprotein sulfhydryl oxidases could reoxidize cytochrome c catalyzed by cytochrome c peroxidase within the IMS (41).
In an interesting recent communication, Comi and colleagues (42) describe the first human ALR mutation to be recognized. Three children from consanguineous parents developed cataract, progressive muscle hypotonia with developmental delay, and hearing loss. Di Fonzo et al. identified the missense mutation which results in the substitution of an arginine for a histidine at position 194 (42). While this R194H mutation did not affect the levels of mRNA, it appeared to impact the import and/or stability of the mutant protein in the mitochondrial IMS. Primary myoblasts from an affected sibling showed an approximately 2-fold increase in doubling time, again consistent with a significant impact of the mutant on cell function (42). Di Fonzo et al. further sought to characterize the mutant by generating the analogous mutation (R182H) in yeast Erv1p. In this background, the mutation significantly impacted cytochrome oxidase biosynthesis because Erv1p is involved in the folding of the copper chaperones required for metal insertion into this respiratory chain component (43). However the focus of their study was neither an enzymological characterization of the human mutant nor a discussion of the molecular environment of R194 and the interactions it might make with neighboring residues.
Herein we characterize the R194H mutation in the context of our continuing enzymological studies of human ALR (38, 39, 44). While the yeast Erv1p analog of ALR proved a useful starting point for evaluation of the behavior of the R194H mutation (42), there are significant differences between the yeast and mammalian orthologs. For example, the distal CxxC disulfide responsible for shuttling reducing equivalents from Mia40 to the proximal disulfide that is in redox communication with the flavin is placed some 40 residues distant in the primary structure of the two proteins (Figure 1A) (3, 39, 45). Further, an initial examination of the location of R194 using the published structure of rat ALR (PDB: 1OQC, (5)) suggests that it is placed close to an interchain disulfide bond in the mammalian proteins and is involved in H-bonding interactions across the subunit interface (see later). Not only does Erv1p lack these interchain disulfides, but there are further significant differences between the sequences of yeast and mammalian orthologs in the vicinity of R194 (Figure 1A). It is therefore important to assess the impact of the R194H mutation with the cognate human protein.
Since a structure of human ALR was unavailable, we have determined a high resolution crystal structure of the short form ALR, intending to pair this with a companion structure of the R194H mutant. Although we have been unsuccessful in obtaining crystals of the R194H mutant, we have utilized 2D NMR methods to document a marked increase in protein mobility compared to the corresponding wild type protein. While the mutant flavoprotein remains fully active in a variety of assays, the resulting protein appears less stable, prone to loss of FAD cofactor, susceptible to reduction of its intersubunit disulfide bonds, and sensitive to partial proteolysis. Our findings suggest that R194 is an important participant in a web of molecular interactions that link the binding of flavin to the stability of the intersubunit disulfide bonds. These results support the observations of Di Fonzo et al. (42) that the pathology of this ALR mutant reflects, in part, the stability of the protein in vivo.
EXPERIMENTAL PROCEDURES
Materials
Reagents were obtained as described previously (39). Polyethylene glycol 8000 was from Thermo Fisher. Zinc acetate was from Allied Chemical and sodium cacodylate from SPI-Chemical.
Mutation, Expression, and Purification of ALR and Arg194 Mutants
The numbering of amino acid residues for sfALR followed that of lfALR. The full sequence of the His-tagged constructs of lf- and sfALR are shown in Figure S1. Primers for site directed mutagenesis (obtained from IDT) are listed in Supporting Information. Mutagenesis was performed as described previously (38, 39) and confirmed by sequence analysis (Genewiz Inc). As before, the two non-conserved and non-essential cysteines (C154 and C165) were mutated to alanine residues to avoid pronounced oxidative aggregation that was encountered in the early stages of work on human ALR (38). While two earlier papers (38, 39) designated these double mutants as ALR', we adopt the simpler abbreviation “ALR” here, and refer to the former C154A/C165A protein as “wild type”.
The Mia40 construct used in this work had a shorter N-terminal His-tag/linker than used in the earlier work (39). The amino acid sequence of this shorter construct is shown in Supporting Information (Figure S1). Expression and purification of Mia40, lfALR, and the corresponding R194H mutant followed the procedures described earlier (39). Similarly, wild type and mutant forms of sfALR were obtained as outlined previously (38), except that riboflavin was not added to the culture media, and the protease inhibitor cocktail tablets were replaced with 1 mM PMSF, and 1 µM leupeptin. Expression of 15N labeled human sfALR (using 15NH4Cl, 99%, Cambridge Isotope Laboratories) followed the protocol of Marley et al. (46).
UV-VIS Spectra
Where necessary, spectra of lf- and sfALR and their R194 mutants were correcting for slight light-scattering by using a turbidity correction using software supplied with the HP8453 diode array spectrophotometers. An extinction coefficient for the bound FAD of the R194H mutants of lfALR were determined to be 11.4 mM−1 cm−1 at 456 nm, after release of flavin using 0.1 % w/v SDS as described earlier (39). The comparable value for wild type lfALR is 11.7 mM−1 cm−1 (39).
Sulfhydryl Oxidase Assays
Assays of ALR, monitored by the consumption of oxygen in a Clarke-type oxygen electrode, by reduction of cytochrome c, and by the oxidation of reduced human Mia40, were conducted as described previously (38, 39).
Crystallization and Data Collection
Protein crystallization conditions were screened by hanging-drop vapor diffusion using Hampton Research crystal screening kits. Drops were generated at 25 °C by mixing 1 µL of the protein stock solution (8 mg/ml in 50 mM phosphate buffer, pH 7.5) with 1 µL of crystallization well solution. Promising conditions were refined to optimize crystal quality. The crystals used here (Figure S2) were grown using a solution of 18% w/v PEG 8000, 210 mM Zn-acetate dihydrate, and 100 mM sodium cacodylate, pH 6.5, that had been filtered through a 0.2 micron disposable filter unit (Nalgene). Crystals were dipped in a cryoprotectant mixture of the well solution containing 20% xylitol before flash-cooling in liquid nitrogen. Diffraction data were collected using in-house equipment (Rigaku RUH3R and R-AXIS IV). The crystal to detector distance was 100 mm and the Cu-radiation X-ray wavelength was 1.541 Å. Crystals were maintained at −180 °C, and data were collected for 15 min per 1° oscillation from a single crystal yielding a total of 180 diffraction images. The data was indexed, integrated and scaled with the program HKL2000 (47).
Crystal Structure Solution and Refinement
The human sfALR structure was solved by molecular replacement using the coordinates for the rat sfALR dimer (PDB 1OQC). Molecular replacement was carried out using the program MOLREP from the CCP4 suite of programs (48). Automated model building of human ALR employed ARP/wARP (48) and 30 cycles of refinements utilized REFMAC5 and COOT (49). Water molecules were placed during successive cycles of model building and refinement. The final human sfALR model (residues 81–205) had 3 subunits comprising of one covalent dimer and half of another covalent dimer whose dimer interface resided on a crystallographic symmetry axis. The N-terminus of each subunit was disordered, with the final model including residues 94–205 from each subunit. A final 2Fo-Fc electron density difference map (Figure S3) confirmed the quality of the final model. The final Rworking and Rfree values were 0.189 and 0.235, respectively.
In Silico Modeling
The R194H mutations of sfALR was modeled using the human sfALR structure using the program MODELLER (50–52), optimized with variable target function and molecular dynamics (51), and further minimized using the CNS program (53, 54). A total of 50 models were generated for the mutant R194H and the structure with the lowest discrete optimized protein energy (DOPE) (55) was further minimized with CNS. DOPE values for the R194H model and the wild type structure were comparable. The minimized model has RMSD of 0.013 Å and 1.74 ° pre and post CNS minimization, indicating good convergence. Solvent accessibilities for amino acid side chains and atoms were calculated using the NACCESS program (56).
Flavin Dissociation
lf- and sfALR were diluted to a concentration of 10 µM in 900 µL of 50 mM phosphate buffer, pH 7.5, 25 °C, containing 0.3 mM EDTA and 4.44 M guanidine hydrochloride. Flavin release was followed in an HP8453 diode array spectrophotometer by the decrease in absorbance at 496 nm.
FAD Association Rates
Apoproteins for sfALR wild type and the R194H mutant were prepared by incubating the sfALR holoprotein while bound to a Ni-NTA column (Invitrogen ProBond) with 10 mL of 50 mM phosphate buffer, pH 7.5, containing 6 M guanidine hydrochloride for 2 h followed by 10 mL of the same buffer without denaturant. The column was then immediately re-equilibrated with denaturant and the treatment repeated three more times to ensure complete release of FAD. The column was developed using 5 mL aliquots of 50, 200 and 500 mM imidazole and apo-ALR was desalted on a PD10 column equilibrated with 50 mM phosphate buffer, pH 7.5, containing 0.3 mM EDTA. Absorbance changes were monitored by stopped-flow (SF-61 DX2 double mixing instrument; Hi-Tech) and analyzed using KinetAsyst 3 software.
Thermal Stability
The thermal stabilities of wild type and R194H mutants of lf- and sfALR were assessed using a JASCO 810 CD spectrophotometer. Far UV (260–205 nm) CD spectra were collected at 2 °C increments every 10 min from 2 – 94 °C, in 1 mm path length cells using 10 µM ALR proteins. Mean residue ellipticities (deg cm2 dmol−1) were plotted as a function of temperature and used to calculate midpoint values.
NMR Data
The 1H-15N TROSY-HSQC NMR spectra were recorded at 25 °C using a Bruker AV600 MHz spectrometer equipped with a CryoProbe operating at 600.13 and 60.81 MHz for 1H and 15N, respectively. 15N labeled wild type and R194H sfALR proteins were expressed and purified as described above. NMR samples were prepared in 5 mm tubes containing 130 µM protein in 10 mM potassium phosphate buffer, pH 6.9, in 90% H2O/10% D2O. Acquisition parameters are listed in supporting information and raw NMR data were processed using the NMRpipe program (57).
Partial Proteolysis
Wild type and R194H mutant sfALR (100 µM in 100 µL of 50 mM phosphate buffer, pH 7.5, 0.3 mM EDTA, 25 °C) were mixed with 1% w/w chymotrypsin. Aliquots (5 µL) were periodically removed, mixed with an equal volume of 2X non-reducing Laemmli buffer to arrest proteolysis, and then analyzed using 12% crosslinked SDS-PAGE gels.
Reduction of Intersubunit Disulfide Bonds with Glutathione
Wild type and R194H sfALR (30 µM) were incubated in glass tubes with 10 mM GSH in 50 mM phosphate buffer, 0.3 mM EDTA, adjusted to pH 7.5. The tubes were sealed with serum caps and the gas-space flushed with nitrogen through entry and exit needles. A narrow gauge needle could be threaded through the exit needle to withdraw 50 µL aliquots prior to quenching samples to a final concentration of 20 mM N-ethylmaleimide. Samples were then analyzed using 12% non-reducing SDS-PAGE gels and stained with Coomassie Brilliant Blue G-250. Bands for monomeric and dimeric ALR were quantitated using ImageJ (58).
RESULTS AND DISCUSSION
Location of R194 in Human ALR
Our previous studies of human ALR (38, 39, 44) have used a homology model for the short form enzyme that was based on the rat sfALR dimer structure of Wu et al. (5). This model suggests that the R194 side chain of ALR would participate in several contacts, both with the ribose moiety of the FAD, and with main chain atoms at the subunit interface (see later). This multiplicity of interactions suggested that an evaluation of the effect of the R194H mutation would be best undertaken with the cognate wild type protein: human ALR. While we have yet to be successful in crystallizing human lfALR, with its 80-residue N-terminal extension (Figure 1A), crystals of the short form enzyme were readily obtained (Figure S2, Supporting Information) and used for the structural determination described below. Although it is possible that some effects of the R194H mutant are unique to the long form of human ALR, none of the data presented here shows a selective impact between long and short versions of this flavoenzyme.
Crystal Structure of Human sfALR
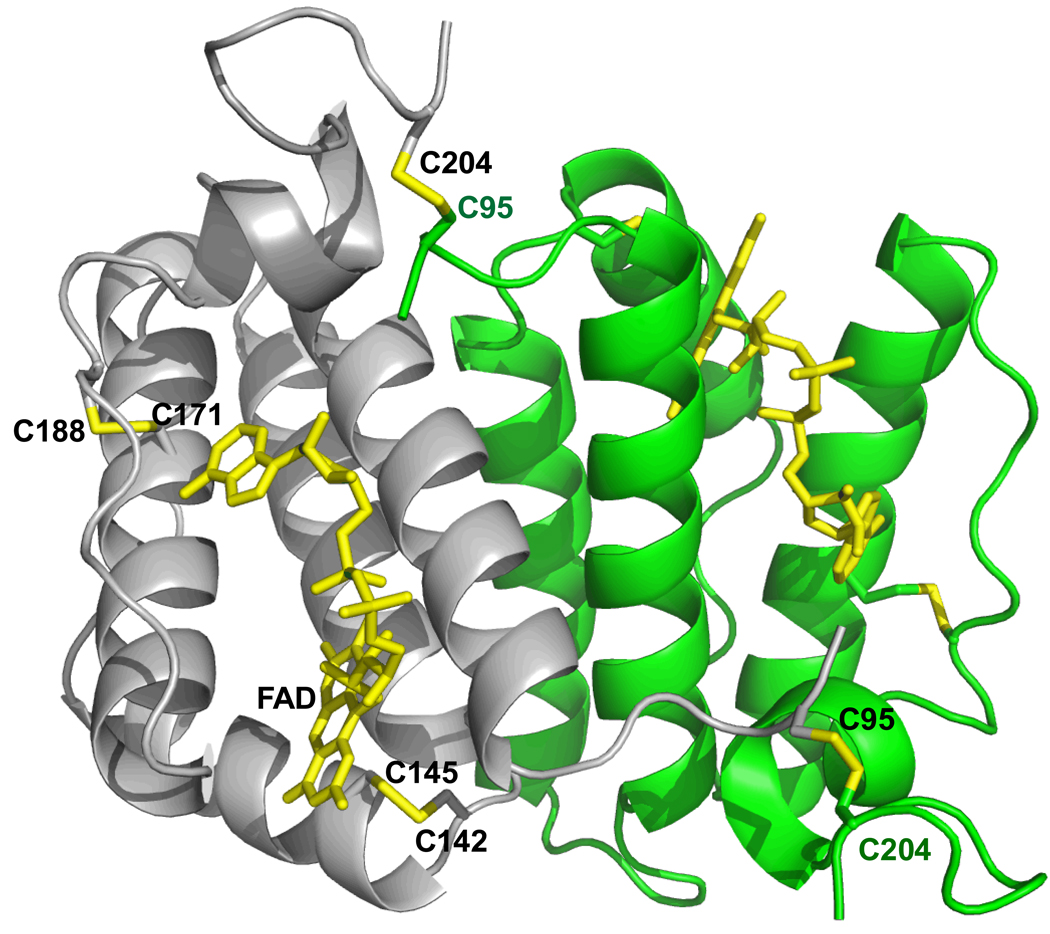
Human sfALR crystallized rapidly without removal of the N-terminal His tag. The initial structure was solved using the rat sfALR (PDB 1OQC) as a molecular replacement search model. The asymmetric unit consisted of three subunits: a disulfide linked homodimer and half of an adjacent dimer. In all subunits, the first 27 N-terminal residues of the sfALR construct (Figure S1; comprising 14 residues of the tag and linker, followed by 13 residues from the N-terminus of the cytokine form of ALR) were disordered. A summary of the data collection and refinement statistics for the crystal structure, encompassing residues D94-D205, is presented in Table 1. The overall fold of the covalent human sfALR dimer is shown in Figure 2. Figure S3 shows a difference electron density map surrounding the bound FAD prosthetic group (2Fo-Fc) of this 1.85 Å structure. As would be expected from the level of protein sequence identity between rat and human short form proteins (85%), both dimers overlay very well (with an RMSD of 0.531 Å; Figure S4 in Supporting Information).
Table 1.
Data Collection and Refinement Statistics of Human sfALRa
| Data Collection | |
| Space group | C2 |
| Unit cell dimensions a, b, c (Å), α, β, γ (°) |
112.719, 65.145, 63.767 90.0, 89.973, 90.0 |
| Resolution (Å) | 50.0–1.85 (1.92–1.85)b |
| Completeness (%) | 99.9 (100.0) |
| Redundancy | 3.6 (3.6) |
| I / σI | 36.54 (3.13) |
| Rmerge linear c | 0.044 (0.366) |
| Refinement | |
| Resolution (Å) | 63.76–1.85 |
| Rwork/Rfreed | 0.189/0.235 |
| Number of atoms (non-hydrogen) |
3429 |
| Mean B value | 31.9 |
| RMSD bond lengths (Å) | 0.017 |
| RMSD bond angles (°) | 1.94 |
| Ramachandran plot (most-favored) |
95.2% |
| Ramachandran plot (additionally and generously allowed region) |
4.8% |
Protein Data Bank accession 3MBG
Values in parentheses are for the highest resolution shell.
Rmerge = Σ|Io-Ia|/Σ(Ia), where Io is the observed intensity and Ia is the average intensity, the sums being taken over all symmetry related reflections.
Rworking = Σ|Fo-Fc|/Σ(Fo), where Fo is the observed amplitude and Fc is the calculated amplitude. Rfree is the equivalent of Rworking, except it is calculated for a randomly chosen set of reflections that were omitted (5%) from the refinement process.
FIGURE 2.
Overall chain fold of dimeric human sfALR. The two C95-C204 disulfides that join the gray and green subunits of the sfALR homodimer are shown in yellow, together with the redox-active proximal disulfide (C142–C145) and the structural disulfide (C171–C188) in the gray subunit. The FAD is depicted in yellow.
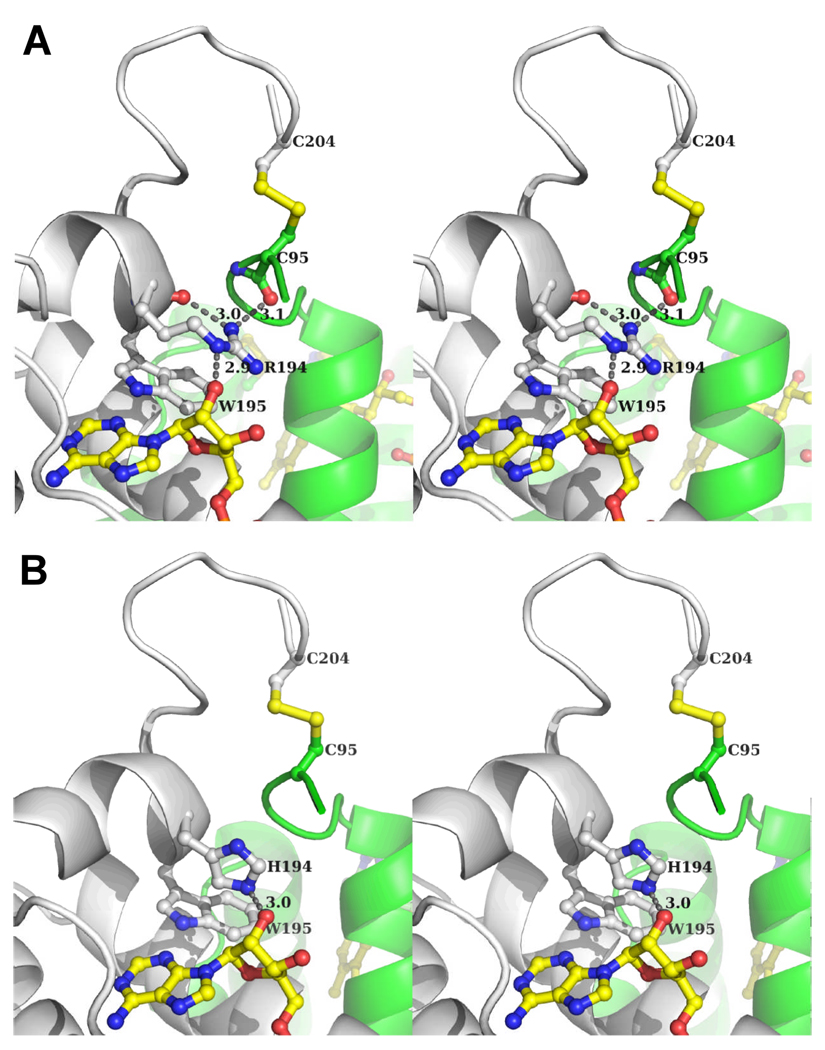
Figure 3A shows a stereoview of the human sfALR structure centered around R194. The residue is nearly coplanar with W195 suggestive of a cation-pi interaction. One of the terminal guanidino amino groups of R194 forms H-bonds with two main chain carbonyl oxygen atoms: one from R194 itself, and the other contributed by C95 of the other subunit. This particular cysteine residue participates in the two interchain disulfides (C95-C204 and its counterpart, C204-C95) that maintain ALR as a covalent head-to-tail dimer (Figure 2). A third H-bond links the δ-nitrogen of R194 to the 2'-OH of the ribose moiety of the bound FAD. This chain of interactions might reasonably be expected to link FAD binding with the conformational stabilization of the dimer interface in the vicinity of the interchain disulfide bridges (see later).
FIGURE 3.
A stereoview surrounding R194 in human sfALR and a minimized model of the R194H mutant. Panel A : R194 forms H-bonds with the 2’ OH of the ribose moiety of FAD, with its own main chain peptide carbonyl, and with the main chain carbonyl oxygen contributed by C95 of the other (green) subunit. Panel B represents a minimized model (see Experimental Procedures) of the R194H sfALR mutant.
Despite the expectation that the R194H mutation would prove structurally conservative, numerous crystallization screening attempts with the mutant protein proved unsuccessful. Indeed the mutation has an unexpectedly large effect on protein stability and flavin binding (see below) and this probably contributes to our inability to crystallize the protein over a wide range of conditions. Hence, for comparison with the native protein, we prepared an energy minimized model of R194H (Figure 3B; see Experimental Procedures). This model introduced very small changes to the backbone (with a Cα RMSD of 0.244 Å) but generated significant perturbations in the region of residue 194. Here the imidazole ring of H194 is oriented at approximately right angles to the indole ring of W195. The model shows that H194 is now 3 Å from the 2'-OH of the FAD-ribose in an orientation unfavorable for strong H-bond formation. While the importance of this potential H-bond in the R194H protein cannot be evaluated based on a modeled structure alone, the other two H-bonding interactions identified for the wild type protein appear to be absent in the mutant (Figure 3B).
Comparison of UV-VIS Spectra of Wild Type and R194H Mutant
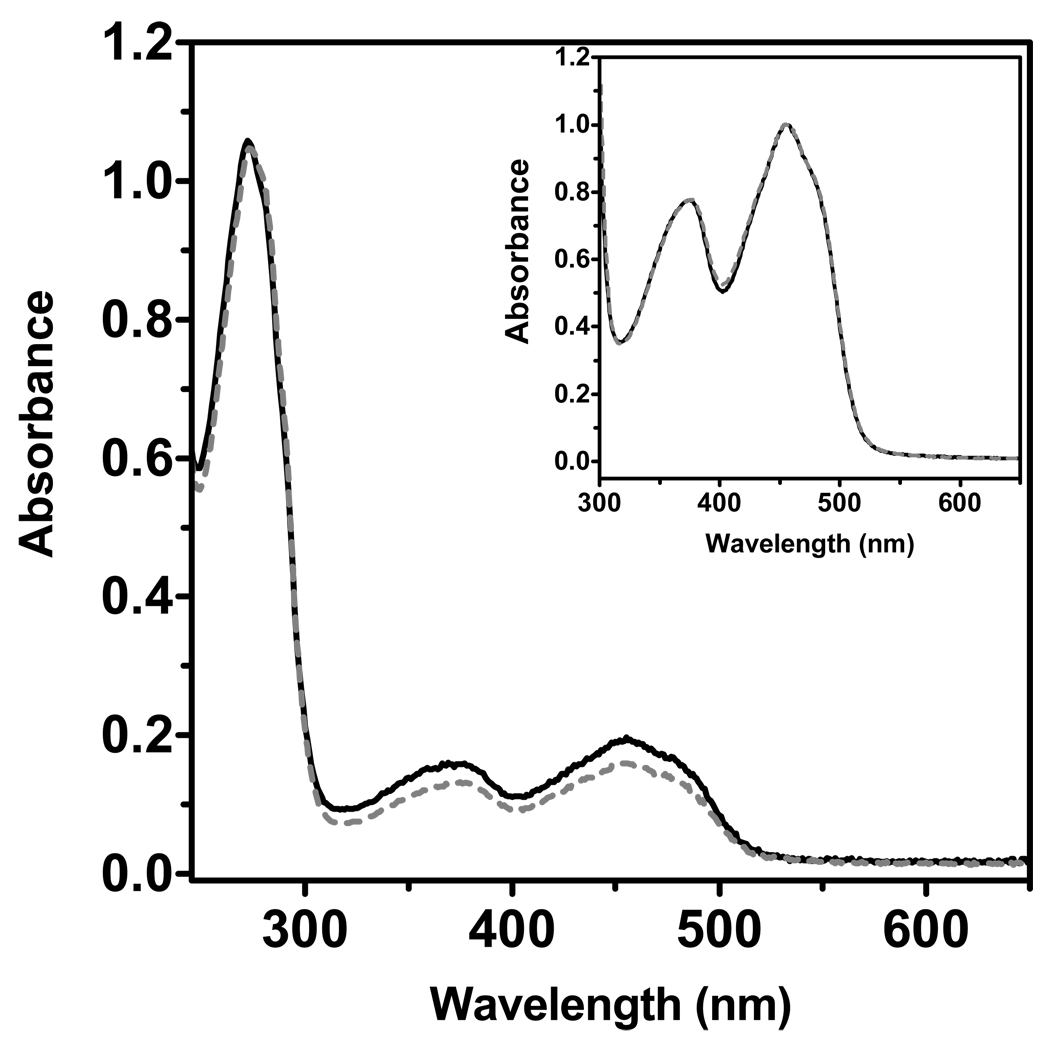
Since R194 makes H-bond contact with the 2'-OH ribose group of the FAD, and participates in a network of interactions around that region of the flavin prosthetic group, we examined whether the mutation produced significant differences in the spectrum of the bound isoalloxazine ring. First, it should be noted that there is approximately 20% less FAD bound to the R194H lfALR mutant than for the wild type protein (for comparison, the main panel in Figure 4 normalizes these spectra at 280 nm). The inset shows that spectra of the bound flavin for wild type and mutant lfALR are very similar (with comparable extinction coefficients for the bound flavin; see Experimental Procedures). Comparable measurements for sfALR show that about 15% less FAD is bound to the mutant protein; again the normalized flavin spectra are comparable (Figure S5).
FIGURE 4.
Comparison of UV-VIS spectra of wild type and R194 lfALR. Spectra were recorded in 50 mM phosphate buffer, pH 7.5, containing 0.3 mM EDTA and normalized either at 280 nm (main panel) or at 456 nm (inset). Spectra of wild type and the R194H mutant are shown by solid and dashed lines respectively.
Influence of R194H Mutation on the Enzymatic Activity of Human lf- and sfALR
Di Fonzo et al. made no direct assessment of the catalytic impact of the R194H mutant in human ALR or of the effects of the corresponding yeast Erv1p R182H mutant (42). Here we complement their studies using a range of in vitro assays of the long and short human ALR forms. Table 2 collects the steady state catalytic parameters for these experiments (see Experimental Procedures). In all cases ALR concentrations of wild type and mutant proteins were expressed in terms of flavin content (see Experimental Procedures) to normalize for the small differences in flavin loading mentioned above. Assays for lfALR utilized reduced Mia40 as the electron donor and either molecular oxygen or cytochrome c as oxidants (as described previously (39)). In terms of oxygen as an electron acceptor (Table 2; B), small compensating changes in kcat and Km terms generate essentially the same catalytic efficiency of approximately 104 M−1s−1. For cytochrome c there is again no major difference between the kcat/Km terms (Table 2; C). Using the model substrate DTT in the oxygen electrode, lfALR R194H mutant shows a catalytic efficiency some 1.7-fold higher than the native protein (Table 2; A). In contrast, sfALR gives an approximate 30% decrease in kcat/Km values, again with minor changes in turnover and Km parameters.
Table 2.
Enzyme activity of wild type and R194H mutants of lf- and sfALRa
| Donor/Acceptor | kcat (min−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| A) DTT / O2 | |||
| lfALR | 61 ± 1.5 | 3 ± 0.3 | 330 |
| lfALR R194H | 72 ± 1 | 2.2 ± 0.1 | 550 |
| sfALR | 108 ± 7 | 1.6 ± 0.4 | 1100 |
| sfALR R194H | 73 ± 3 | 1.6 ± 0.25 | 760 |
| B) Mia40b / O2 | |||
| lfALR | 42 ± 8 | 0.068 ± 0.021 | 10200 |
| lfALR R194H | 24 ± 4 | 0.042 ± 0.014 | 9600 |
| C) Mia40b / Cyt c | |||
| lfALR | 48 ± 6 | 0.045 ± 0.012 | 17600 |
| lfALR R194H | 48 ± 9 | 0.039 ± 0.015 | 20700 |
Three assay systems were used: A) oxygen consumption in the oxygen electrode using DTT as a model substrate, B) the oxygen-dependent reoxidation of reduced Mia40 followed discontinuously with DTNB, and C) the reduction of cytochrome c followed continuously in the presence of reduced Mia40.
Kinetics determined with a shorter N-terminal linker for Mia40 than used previously (see Experimental Procedures (39)).
Overall, these data show that the kinetic impact of the R194H mutation in either human lf- or sfALR forms is minor. Di Fonzo et al. speculated that, in addition to the instability of lfALR induced by the R194H mutation, the mutant protein might be less efficient at transferring electrons to cytochrome c (42). Our data suggest that this latter possibility is unlikely to be a major factor in the R194H phenotype. The impaired assembly of cytochrome c oxidase observed earlier in vivo (42) probably reflects compromised folding of copper chaperones in the IMS deriving from a lowered concentration of active lfALR. We now document the impact of R194H on the stability and flavin binding of human ALR.
Thermal Stability
Human sfALR shows considerable thermal stability with CD parameters only changing at temperatures above 70 °C (59). Thermal denaturation experiments following wild type and mutant lf- and sfALR in the CD shows that the mutants have a Tm about 10 °C lower than wild type proteins (Table 3; see Experimental Procedures).
Table 3.
Melting temperature of wild type and R194H mutant of lf- and sfALRa
| Construct | Tm (°C) |
|---|---|
| lfALR | 86 |
| lfALR R194H | 75 |
| sfALR | 86 |
| sfALR R194H | 78 |
Measured by circular dichroism at 222 nm in 50 mM phosphate buffer, pH 7.5, containing 0.3 mM EDTA (see Experimental Procedures).
Flavin Dissociation
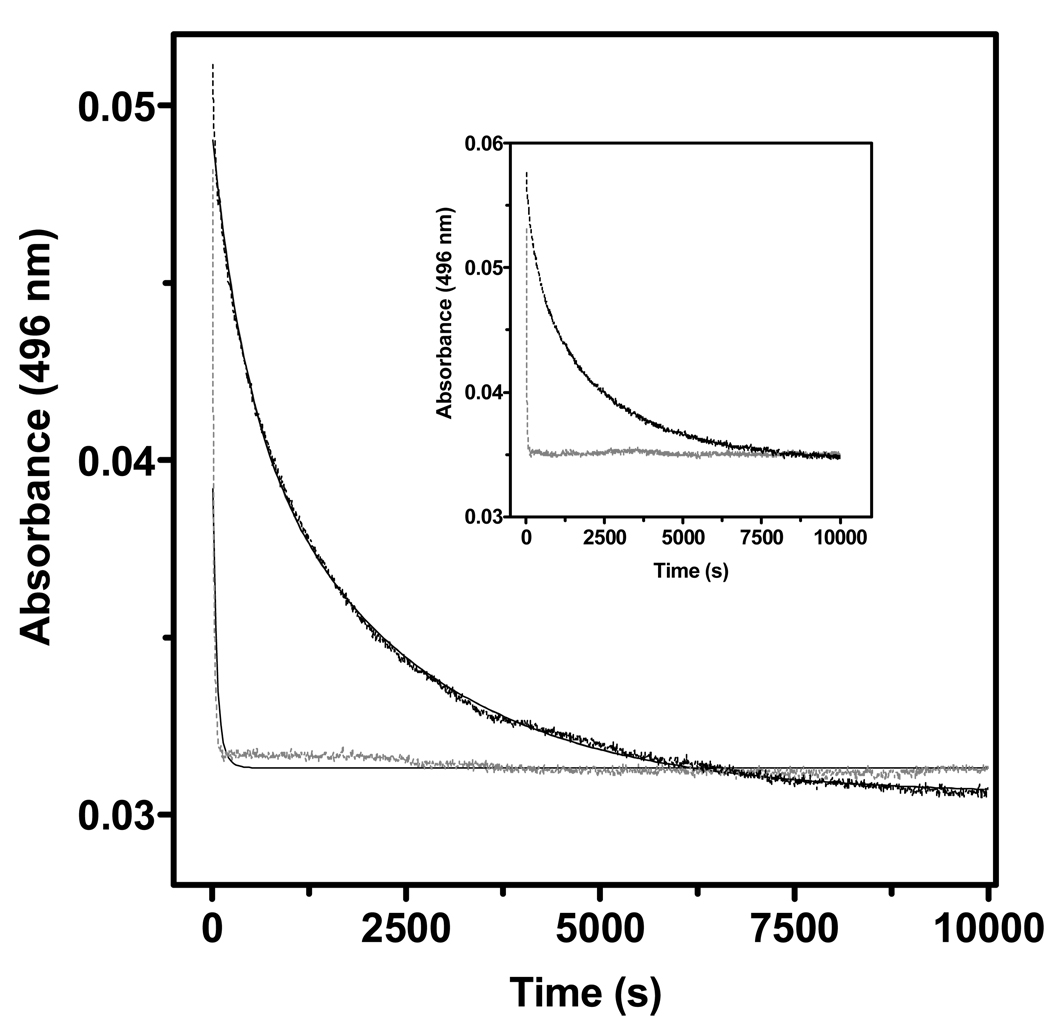
A striking demonstration of the impact of the R194H mutation is provided in Figure 5. Both wild type and the R194H mutants of lf- and sfALR were challenged with 4.4 M guanidine hydrochloride and the release of FAD was evaluated by the decrease in absorbance at 496 nm. For wild type lfALR, the half-time required for release of FAD from the wild type protein is 900 s compared to about 20 s for the mutant. The same relative susceptibility is observed for sfALR (inset Figure 5) again suggesting that the impact of the R194H mutant can be appropriately assessed in the context of the shorter ALR construct.
FIGURE 5.
Comparison of rate of flavin release from wild type and R194H mutants of ALR. The main panel monitors the absorbance at 496 nm after the addition of 4.44 M guanidine hydrochloride in 50 mM phosphate buffer, pH 7.5, 25 °C (black line). The gray line represents the rapid release of FAD from the R194H lfALR mutant. The inset presents the corresponding experiments with the short form of ALR.
FAD binding to Wild Type and R194H ALR
Since all the studies mentioned above show comparable effects of the R194H mutant on the long- and short forms of ALR, we have utilized the more readily accessible sfALR in the following sections. When FAD binding to apo-sfALR was evaluated in the stopped flow spectrophotometer by following the absorbance increase at 490 nm, both wild type and mutant behaved comparably (with closely superimposable kinetics; Figure S6 in Supporting Information).
In complementary experiments, we examined the return of ALR activity using DTT as a substrate in the oxygen electrode following the addition of FAD to the apo-forms of lf- and sfALR. In both mutant and wild type instances the oxygen traces observed were the same as those observed with the corresponding concentration of holo-sfALR (data not shown). While equilibration of the oxygen electrode requires several seconds, these results suggest that flavin binding and activity return for wild type and mutant ALR are comparably rapid. Hence the weaker FAD binding encountered with the mutant seems to largely reflect the dissociation rate constant from the holoprotein.
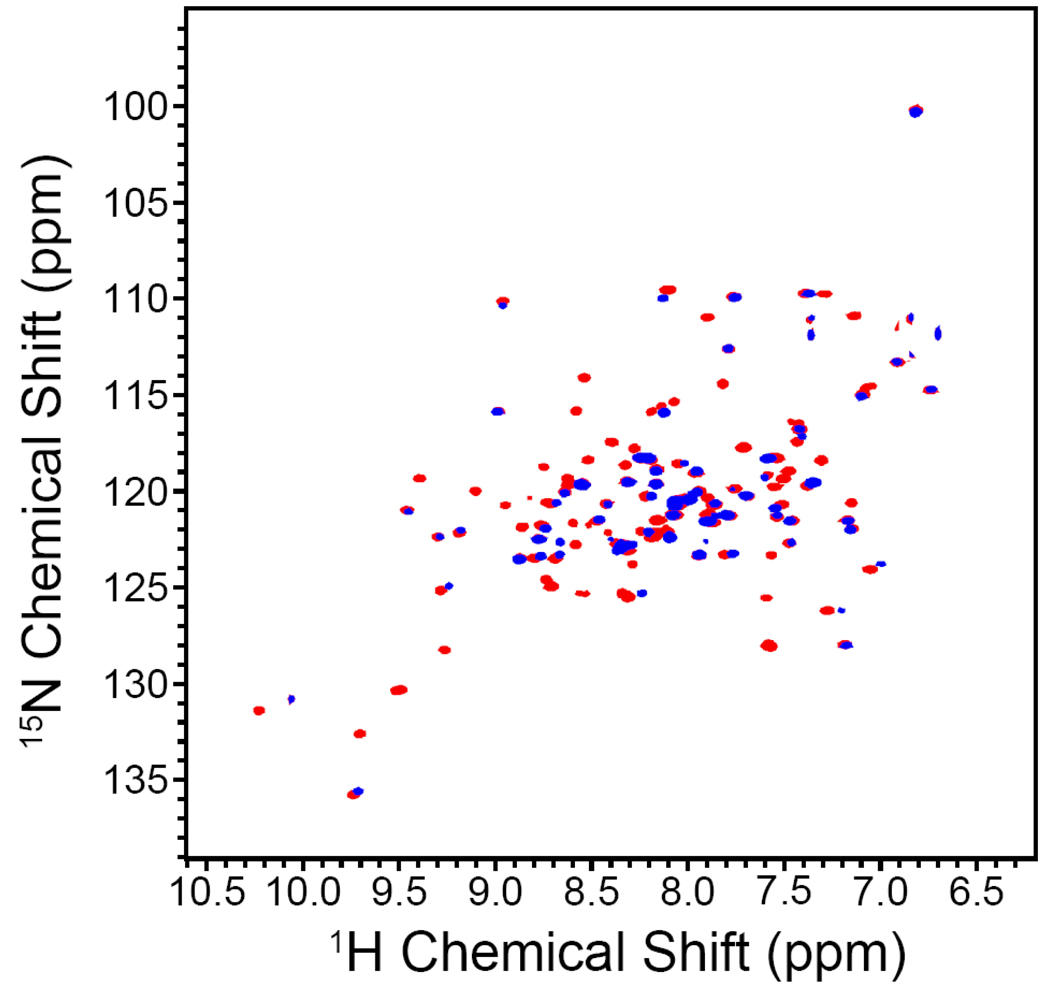
Comparison Between Wild Type and R194H Mutants by TROSY-HSQC NMR
Figure 6 presents an overlay of 2D NMR spectra of 15N-labeled wild type (red) and R194H sfALR (blue) proteins acquired under identical conditions (see Experimental Procedures). The dispersion of chemical shifts show that both proteins are well ordered, although the mutant shows considerably fewer resonances over the range depicted in Figure 6 (approximately 40% less). These absences are consistent with enhanced selective flexibility of the R194H mutant compared to the native protein.
FIGURE 6.
2D 1H-15N TROSY-HSQC spectra of wild type and R194H mutant of sfALR. Wild type (red) and R194H (blue) spectra were acquired in 10 mM phosphate buffer, pH 6.9 at 25 °C (see Experimental Procedures).
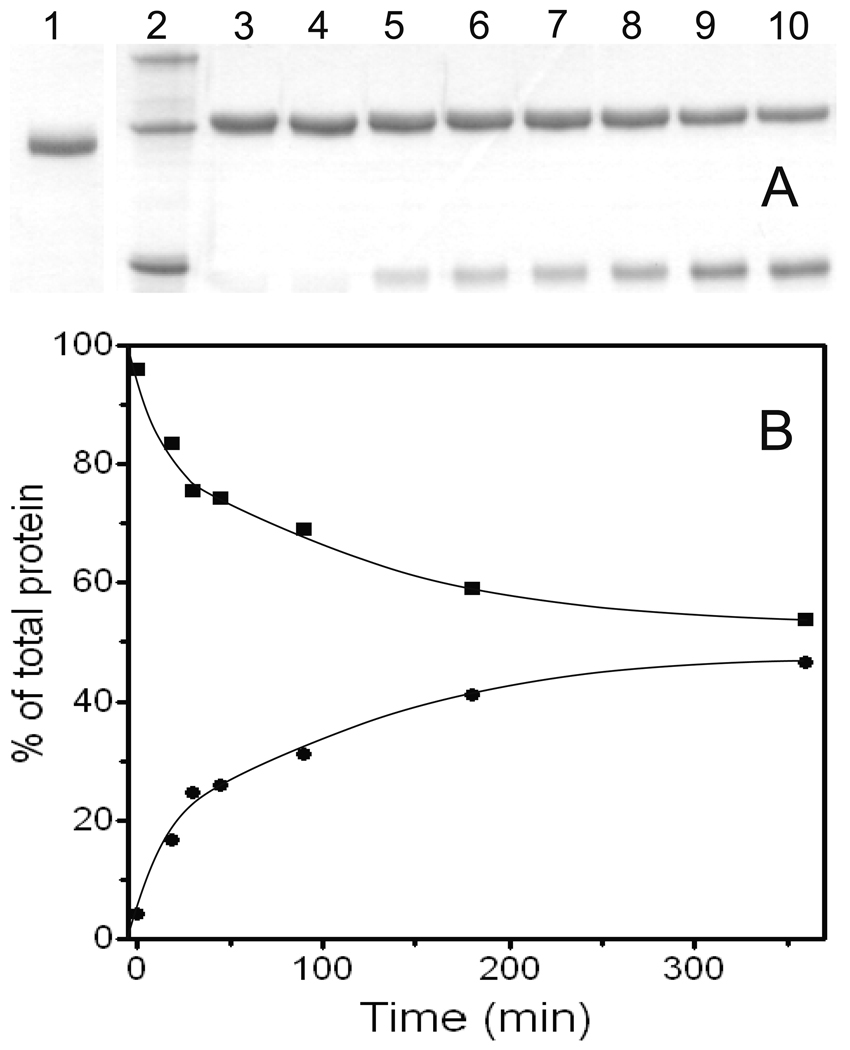
The R194H Mutant of Human ALR Acquires Protease Sensitivity
One consequence of the increased protein mobility noted above may be a corresponding increase in susceptibility to intracellular proteases. Figure 7 shows that wild type sfALR was resistant to incubation with 1% w/w chymotrypsin over 2 h at 25 °C in potassium phosphate buffer pH 7.5. This stability extended to 5 h without significant change in the appearance of bands in these non-reducing gels (data not shown). In contrast, the mutant protein showed significant proteolysis after 1 min (with the appearance of a band running slightly faster than the original; Figure 7). Further incubation with chymotrypsin generated additional truncation of R194H sfALR and disappearance of the band corresponding to the original protein after 5 min.
FIGURE 7.
The R194H mutant of sfALR gains sensitivity to partial proteolysis using chymotrypsin. Non-reducing SDS-PAGE gels were run with protein standards (35, 25 and 15 kDa from the top of panel A, lane 1; and panel B lane 2). R194H (lane 1 panel B) reproducibly runs slightly slower than wild type protein (lane 2, panel A) on SDS-PAGE. Proteins were treated with 1% w/w chymotrypsin in 50 mM phosphate buffer, pH 7.5, 25 °C, containing 0.3 mM EDTA, and samples were quenched in non-reducing Laemmli buffer (for panel A, lanes 2–10 were quenched 0, 1, 3, 5, 10, 15, 30, 60 and 120 min after protease treatment of sfALR). For the mutant in panel B, lane 1 is at time zero, lane 2 is the protein ladder and lanes 3–10 reflect the same time intervals as in panel A. For clarity, the dotted lines on the gels define the limits of band migration.
R194H Mutant is Susceptible to Reduction by Glutathione
The sensitivity of the mutant protein to limited proteolysis using chymotrypsin, and the pronounced flexibility apparent from the TROSY-HSQC experiments, suggests that the region surrounding R194 has a significant effect on both local and global structure of human ALR. Hence we tested whether the mutation might destabilize the unusual interchain disulfides between the N-terminus of one subunit and the C-terminus of the other (Figure 2). Specifically we investigated whether these cross-links in the mutant protein are prone to reduction by realistic cellular concentrations of GSH. The reductive monomerization of wild type sfALR is very slow using 10 mM GSH at 25 °C; under anaerobic conditions no monomer was evident over 6 h (Figure 8). Samples were analyzed in this experiment by non-reducing SDS-PAGE after quenching with excess NEM (see Experimental Procedures). Reductive incubations were performed anaerobically because, while GSH is an extremely poor substrate of ALR (38, 39), GSSG might slowly accumulate over prolonged aerobic incubation using micromolar concentrations of the oxidase. In contrast to wild type sfALR, reduction of the C95-C204 disulfide bonds is clearly evident after 18 min incubation with the mutant protein (Figure 8A, lane 5). Figure 8B shows a time-course of the reduction of these interchain disulfide bridges in the R194H sfALR mutant. Finally, the sensitivity of the apoprotein forms of wild type and mutant sfALR to the same reductive conditions was investigated (see Experimental Procedures). The wild type apoprotein again remains resistant to 10 mM GSH (with 90 % dimer retained after 5 h), compared to only 16% with the mutant protein (data not shown).
FIGURE 8.
The R194H sfALR mutant is sensitive to reduction of intersubunit disulfide bonds. Panel A: after 6 h anaerobic incubation of 30 µM wild type sfALR with 10 mM GSH followed by quenching with 20 mM NEM (see Experimental Procedures), the protein runs exclusively as a dimer on non reducing SDS-PAGE (lane 1). Lane 2 represents molecular weight markers (from the top: 35, 25 and 15 kDa). Lane 3 shows the R194H mutation in the absence of GSH and lanes 4–10 are 0, 18, 30, 45, 90, 180 and 360 min after the addition of 10 mM GSH. Panel B: time-dependence of the integrated intensities of the stained bands (square and circles represent dimer and monomer bands, respectively; see Experimental Procedures).
The sluggish reduction of the wild type protein by GSH is of interest because the interchain C95–204 disulfides appear significantly solvent-accessible in the holoprotein (the side chains of C95 and C204 are 45 and 8% accessible respectively; see Experimental Procedures (56)). Nevertheless in vitro these disulfides in the wild type protein remain intact for hours under anaerobic conditions in the presence of 10 mM GSH. While the unusual topology of these disulfides, securing the N- and C-termini within a homodimer, is suggestive of functional importance, their role is currently obscure. While such disulfides are found in many eukaryotic ALR analogs (from Aspergillus to humans) they are absent in both budding and non-budding yeast.
Conclusions
These data show the unexpectedly severe impact of R194H on the conformational stability of both long and short forms of human ALR. They complement the studies of Comi and coworkers (42) and provide strong in vitro corroboration of their suggestions that the R194H mutant is significantly destabilizing. Our studies provide a molecular rationale for this marked loss of stability - evident by an increased rate of dissociation of FAD from the mutant ALR, an enhanced susceptibility to partial proteolysis and to reduction of the interchain disulfide bonds, and a major loss of order as measured by 2D NMR. The unanticipated severity of these effects points to a strong influence of the interchain disulfide bonds, the neighboring amino acid residues, and the bound FAD on the stability of dimeric ALR. In contrast the catalytic activity of the mutant protein is not compromised by the replacement of R194 by a histidine residue.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mr. Shangjin Sun, and Drs. Steve Bai and Tatyana Polenova for help with the NMR measurements and analysis. Dr. Vamsi Kodali is acknowledged for helpful comments.
Footnotes
This work was supported in part by National Institutes of Health Grant GM26643 (CT), 2P20RR015588 from the National Center for Research Resources (BB), and USPHS Training Grant 1-T32-GM08550 (SAS). The content of this work is solely the responsibility of the authors, and does not necessarily reflect the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Protein Data Bank entry code 3MBG represents the atomic coordinates and structure factors for the short form of human augmenter of liver regeneration.
SUPPORTING INFORMATION
Table S1 and Figures S1–S6 provide supplementary data, sequences and analysis for human sfALR and its R194H mutant. This material is available free of charge via the Internet at http://pubs.acs.org.
Abbreviations: ALR, augmenter of liver regeneration (lfALR and sfALR refer to long and short forms of the protein, respectively); DTNB, 5,5′-dithiobis(2-nitrobenzoate); DTT, dithiothreitol; GSH, reduced glutathione; GSSG, oxidized glutathione; TROSY, transverse relaxation optimized spectroscopy; HSQC, 15N-heteronuclear single quantum coherence; IMS, mitochondrial intermembrane space; IPTG, isopropyl d-thiogalactopyranoside; NEM, N-ethylmaleimide; QSOX, Quiescin-sulfhydryl oxidase; RMSD, root mean square deviation.
REFERENCES
- 1.Lee J, Hofhaus G, Lisowsky T. Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett. 2000;477(1–2):62–66. doi: 10.1016/s0014-5793(00)01767-1. [DOI] [PubMed] [Google Scholar]
- 2.Lisowsky T. ERV1 is involved in the cell-division cycle and the maintenance of mitochondrial genomes in Saccharomyces cerevisiae. Curr. Genet. 1994;26:15–20. doi: 10.1007/BF00326299. [DOI] [PubMed] [Google Scholar]
- 3.Fass D. The Erv family of sulfhydryl oxidases. Biochim Biophys Acta. 2008;1783:557–566. doi: 10.1016/j.bbamcr.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Gross E, Sevier CS, Vala A, Kaiser CA, Fass D. A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat. Struct. Biol. 2002;9:61–67. doi: 10.1038/nsb740. [DOI] [PubMed] [Google Scholar]
- 5.Wu CK, Dailey TA, Dailey HA, Wang BC, Rose JP. The crystal structure of augmenter of liver regeneration: A mammalian FAD-dependent sulfhydryl oxidase. Protein Sci. 2003;12:1109–1118. doi: 10.1110/ps.0238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross E, Kastner DB, Kaiser CA, Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell. 2004;117:601–610. doi: 10.1016/s0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 7.Hoober KL, Glynn NM, Burnside J, Coppock DL, Thorpe C. Homology between egg white sulfhydryl oxidase and quiescin Q6 defines a new class of flavin-linked sulfhydryl oxidases. J. Biol. Chem. 1999;274:31759–31762. doi: 10.1074/jbc.274.45.31759. [DOI] [PubMed] [Google Scholar]
- 8.Alon A, Heckler E, Thorpe C, Fass D. QSOX contains a pseudo-dimer of functional and degenerate sulfhydryl oxidase domains. FEBS Lett. 2010;584:1521–1525. doi: 10.1016/j.febslet.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Thorpe C, Coppock DL. Generating disulfides in multicellular organisms: Emerging roles for a new flavoprotein family. J. Biol. Chem. 2007;282:13929–13933. doi: 10.1074/jbc.R600037200. [DOI] [PubMed] [Google Scholar]
- 10.Heckler EJ, Rancy PC, Kodali VK, Thorpe C. Generating disulfides with the Quiescin-sulfhydryl oxidases. Biochim. Biophys. Acta. 2008;1783:567–577. doi: 10.1016/j.bbamcr.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Li Y, Wei K, Li L, Liu W, Zhu Y, Qiu Z, He F. The potentiation role of hepatopoietin on activator protein-1 is dependent on its sulfhydryl oxidase activity. J. Biol. Chem. 2003;278:49022–49030. doi: 10.1074/jbc.M304057200. [DOI] [PubMed] [Google Scholar]
- 12.Pawlowski R, Jura J. ALR and Liver Regeneration. Mol. Cell. Biochem. 2006;288:159–169. doi: 10.1007/s11010-006-9133-7. [DOI] [PubMed] [Google Scholar]
- 13.Lisowsky T, Lee JE, Polimeno L, Francavilla A, Hofhaus G. Mammalian augmenter of liver regeneration protein is a sulfhydryl oxidase. Dig. Liver Dis. 2001;33:173–180. doi: 10.1016/s1590-8658(01)80074-8. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Wei K, Lu C, Li M, Xing G, Wei H, Wang Q, Chen J, Wu C, Chen H, Yang S, He F. Identification of hepatopoietin dimerization, its interacting regions and alternative splicing of its transcription. Eur. J. Biochem. 2002;269:3888–3893. doi: 10.1046/j.1432-1033.2002.03054.x. [DOI] [PubMed] [Google Scholar]
- 15.Gatzidou E, Kouraklis G, Theocharis S. Insights on augmenter of liver regeneration cloning and function. World J. Gastroenterol. 2006;12:4951–4958. doi: 10.3748/wjg.v12.i31.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polimeno L, Lisowsky T, Francavilla A. From yeast to man--from mitochondria to liver regeneration: a new essential gene family. Ital. J. Gastroenterol. Hepatol. 1999;31:494–500. [PubMed] [Google Scholar]
- 17.LaBrecque DR, Pesch LA. Preparation and partial characterization of hepatic regenerative stimulator substance (SS) from rat liver. J. Physiol. 1975;248:273–284. doi: 10.1113/jphysiol.1975.sp010973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagiya M, Francavilla A, Polimeno L, Ihara I, Sakai H, Seki T, Shimonishi M, Porter KA, Starzl TE. Cloning and sequence analysis of the rat augmenter of liver regeneration (ALR) gene: expression of biologically active recombinant ALR and demonstration of tissue distribution. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8142–8146. doi: 10.1073/pnas.91.17.8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G, Yang X, Zhang Y, Wang Q, Chen H, Wei H, Xing G, Xie L, Hu Z, Zhang C, Fang D, Wu C, He F. Identification and characterization of receptor for mammalian hepatopoietin that is homologous to yeast ERV1. J. Biol. Chem. 1999;274:11469–11472. doi: 10.1074/jbc.274.17.11469. [DOI] [PubMed] [Google Scholar]
- 20.Gandhi CR, Murase N, Starzl TE. Cholera toxin-sensitive GTP-binding protein-coupled activation of augmenter of liver regeneration (ALR) receptor and its function in rat kupffer cells. J. Cell. Physiol. 2010;222:365–373. doi: 10.1002/jcp.21957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Li M, Xing G, Hu Z, Wang Q, Dong C, Wei H, Fan G, Chen J, Yang X, Zhao S, Chen H, Guan K, Wu C, Zhang C, He F. Stimulation of the mitogen-activated protein kinase cascade and tyrosine phosphorylation of the epidermal growth factor receptor by hepatopoietin. J. Biol. Chem. 2000;275:37443–37447. doi: 10.1074/jbc.M004373200. [DOI] [PubMed] [Google Scholar]
- 22.Liao XH, Zhang L, Liu Q, Sun H, Peng CM, Guo H. Augmenter of liver regeneration protects kidneys from ischaemia/reperfusion injury in rats. Nephrol. Dial. Transplant. 2010 doi: 10.1093/ndt/gfq151. (EPUB ahead of print) [DOI] [PubMed] [Google Scholar]
- 23.Shen L, Hu J, Lu H, Wu M, Qin W, Wan D, Li YY, Gu J. The apoptosis-associated protein BNIPL interacts with two cell proliferation-related proteins, MIF and GFER. FEBS Lett. 2003;540:86–90. doi: 10.1016/s0014-5793(03)00229-1. [DOI] [PubMed] [Google Scholar]
- 24.Boyd JM, Malstrom S, Subramanian T, Venkatesh LK, Schaeper U, Elangovan B, D'Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 25.Lu C, Li Y, Zhao Y, Xing G, Tang F, Wang Q, Sun Y, Wei H, Yang X, Wu C, Chen J, Guan KL, Zhang C, Chen H, He F. Intracrine hepatopoietin potentiates AP-1 activity through JAB1 independent of MAPK pathway. FASEB J. 2002;16:90–92. doi: 10.1096/fj.01-0506fje. [DOI] [PubMed] [Google Scholar]
- 26.Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Terziyska N, Grumbt B, Bien M, Neupert W, Herrmann JM, Hell K. The sulfhydryl oxidase Erv1 is a substrate of the Mia40-dependent protein translocation pathway. FEBS Lett. 2007;581:1098–1102. doi: 10.1016/j.febslet.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Gabriel K, Milenkovic D, Chacinska A, Muller J, Guiard B, Pfanner N, Meisinger C. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J. Mol. Biol. 2007;365:612–620. doi: 10.1016/j.jmb.2006.10.038. [DOI] [PubMed] [Google Scholar]
- 29.Hell K. The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim. Biophys. Acta. 2008;1783:601–609. doi: 10.1016/j.bbamcr.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Herrmann JM, Kohl R. Catch me if you can! Oxidative protein trapping in the intermembrane space of mitochondria. J. Cell Biol. 2007;176:559–563. doi: 10.1083/jcb.200611060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beverly KN, Sawaya MR, Schmid E, Koehler CM. The Tim8-Tim13 complex has multiple substrate binding sites and binds cooperatively to Tim23. J. Mol. Biol. 2008;382:1144–1156. doi: 10.1016/j.jmb.2008.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stojanovski D, Milenkovic D, Muller JM, Gabriel K, Schulze-Specking A, Baker MJ, Ryan MT, Guiard B, Pfanner N, Chacinska A. Mitochondrial protein import: precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J. Cell Biol. 2008;183:195–202. doi: 10.1083/jcb.200804095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokatlidis K. A disulfide relay system in mitochondria. Cell. 2005;121:965–967. doi: 10.1016/j.cell.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 34.Reddehase S, Grumbt B, Neupert W, Hell K. The Disulfide Relay System of Mitochondria Is Required for the Biogenesis of Mitochondrial Ccs1 and Sod1. J. Mol. Biol. 2009;385:331–338. doi: 10.1016/j.jmb.2008.10.088. [DOI] [PubMed] [Google Scholar]
- 35.Kawamata H, Manfredi G. Different regulation of wild-type and mutant Cu,Zn superoxide dismutase localization in mammalian mitochondria. Hum. Mol. Genet. 2008;17:3303–3317. doi: 10.1093/hmg/ddn226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lange H, Lisowsky T, Gerber J, Muhlenhoff U, Kispal G, Lill R. An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep. 2001;2:715–720. doi: 10.1093/embo-reports/kve161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen S, Balabanidou V, Sideris DP, Lisowsky T, Tokatlidis K. Erv1 Mediates the Mia40-dependent Protein Import Pathway and Provides a Functional Link to the Respiratory Chain by Shuttling Electrons to Cytochrome c. J. Mol. Biol. 2005;353:937–944. doi: 10.1016/j.jmb.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 38.Farrell SR, Thorpe C. Augmenter of liver regeneration: a flavin dependent sulfhydryl oxidase with cytochrome C reductase activity. Biochemistry. 2005;44:1532–1541. doi: 10.1021/bi0479555. [DOI] [PubMed] [Google Scholar]
- 39.Daithankar VN, Farrell SR, Thorpe C. Augmenter of liver regeneration: substrate specificity of a flavin-dependent oxidoreductase from the mitochondrial intermembrane space. Biochemistry. 2009;48:4828–4837. doi: 10.1021/bi900347v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell. Biol. 2007;179:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dabir DV, Leverich EP, Kim SK, Tsai FD, Hirasawa M, Knaff DB, Koehler CM. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007;26:4801–4811. doi: 10.1038/sj.emboj.7601909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Fonzo A, Ronchi D, Lodi T, Fassone E, Tigano M, Lamperti C, Corti S, Bordoni A, Fortunato F, Nizzardo M, Napoli L, Donadoni C, Salani S, Saladino F, Moggio M, Bresolin N, Ferrero I, Comi GP. The Mitochondrial Disulfide Relay System Protein GFER Is Mutated in Autosomal-Recessive Myopathy with Cataract and Combined Respiratory-Chain Deficiency. Am. J. Hum. Genet. 2009;84:594–604. doi: 10.1016/j.ajhg.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cobine PA, Pierrel F, Winge DR. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta. 2006;1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Kay CWM, Elsasser C, Bittl R, Farrell SR, Thorpe C. Determination of the distance between the two neutral flavin radicals in augmenter of liver regeneration by pulsed ELDOR. J. Am. Chem. Soc. 2006;128:76–77. doi: 10.1021/ja057308g. [DOI] [PubMed] [Google Scholar]
- 45.Ang SK, Lu H. Deciphering structural and functional roles of individual disulfide bonds of the mitochondrial sulfhydryl oxidase Erv1p. J. Biol. Chem. 2009;284:28754–28761. doi: 10.1074/jbc.M109.021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Cryst., Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti-Renom MA, Davis FP, Sali A. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2009;37:D347–D354. doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 52.Eswar N, Webb B, Marti-Renom M, Madhusudhan M, Eramian D, Shen M-y, Pieper U, Sali A. Current Protocols in Protein Science. John Wiley; 2007. Comparative Protein Structure Modeling Using MODELLER; pp. 1–31. [DOI] [PubMed] [Google Scholar]
- 53.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 54.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 55.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Prot. Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hubbard S, Thornton J. NACCESS, Computer Program. London: Department of Biochemistry and Molecular Biology, University College; 1993. [Google Scholar]
- 57.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 58.Abramoff M, Magelhaes P, Ram SJ. Image Processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 59.Yang ZC, Yang L, Zhang YX, Yu HF, An W. Effect of heat and pH denaturation on the structure and conformation of recombinant human hepatic stimulator substance. Protein J. 2007;26:303–313. doi: 10.1007/s10930-007-9072-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.