Abstract
The retinoblastoma tumour suppressor (RB) is a crucial regulator of cell-cycle progression that is invoked in response to a myriad of anti-mitogenic signals. It has been hypothesized that perturbations of the RB pathway confer a synonymous proliferative advantage to tumour cells; however, recent findings demonstrate context-specific outcomes associated with such lesions. Particularly, loss of RB function is associated with differential response to wide-ranging therapeutic agents. Thus, the status of this tumour suppressor may be particularly informative in directing treatment regimens.
In cancer it is well accepted that tumour cells invoke multiple mechanisms to bypass proliferative control. A crucial junction in the control of cellular proliferation is linked to the retinoblastoma (RB) tumour suppressor protein, whose primary function is to prevent unscheduled entry into the mitotic cell cycle1–3 (FIG. 1). RB exerts its antiproliferative effects, at least in part, through the ability to mediate the transcriptional repression of genes required for DNA replication and mitosis4–7. Through these actions, RB impinges on a sophisticated network of target genes to limit cell-cycle progression8–11. Mitogens must counteract this action of RB, and do so through signals that promote activation of cyclin-dependent kinase (CDK)–cyclin complexes, which phosphorylate RB and attenuate its capacity to induce transcriptional repression12. Typically, RB remains in this inactive state until passage through mitosis, at which point it is re-engaged through the action of a phosphatase13,14. Alternatively, RB action can be invoked during an active cycle in response to specific cellular stresses (for example, genotoxic insult) and induce cell-cycle arrest, thus protecting against continued inappropriate proliferation1,15,16. Collectively, these functions of RB are thought to be crucial in preventing tumour formation, on the basis of several criteria. First, loss of heterozygosity of RB1 contributes to tumour formation in the human retina17,18. Second, mutations that disrupt RB-mediated transcriptional regulation or genetic events that facilitate RB phosphorylation (for example, amplification of cyclin D1) are frequently observed in tumours19. Third, RB inactivation is mediated by and cooperates with oncogenes that contribute to human cancer20. These observations support a significant role for RB-mediated cell-cycle control in human tumours and predict that RB deficiency serves to confer a common proliferative advantage. However, recent studies support the hypothesis that the consequence of RB inactivation is quite complex, and can result in disparate outcomes dependent on tumour type. Moreover, it is apparent that different mechanisms used by tumour cells to disrupt the tumour-suppressive function of RB are not synonymous in consequence, suggesting that both tissue type-specific and lesion-specific variances exist with regard to cellular outcome. The findings, reviewed herein, demonstrate that RB inactivation evokes specific responses to cancer therapeutics and suggest that RB status could be developed as a metric to direct therapeutic agents.
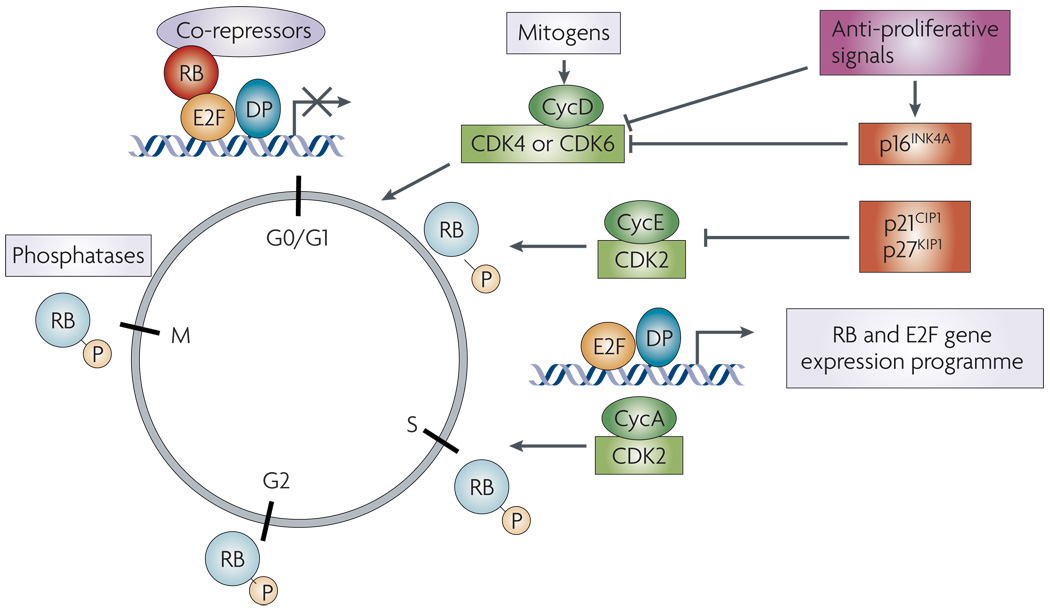
Figure 1. Schematic of RB in cell cycle control.
Mitogenic signals stimulate the expression of D-type cyclins (Cyc) and a concomitant increase in cyclin-dependent kinase 4 (CDK4) and CDK6 activity. These factors initiate RB phosphorylation, which is augmented by the activity of CDK2 complexes with cyclins A and E. The phosphorylation of RB disrupts its association with E2F. This inactivation of RB allows for the expression of a transcriptional programme that enables progression through S-phase and mitosis. At the transition from mitosis to G1, RB is dephosphorylated through the action of phosphatases. Importantly, a large number of anti-mitogenic signals function to prevent RB phosphorylation either by limiting the activity of CDK4, CDK6 and CDK2 complexes or by inducing the activity of CDK inhibitors.
RB and cell-cycle control
The contemporary model of RB function in cell-cycle control and tumour suppression is well-founded based on investigation in multiple model systems (FIG. 1). In the absence of mitogenic stimuli, RB activity is engaged to inhibit cell-cycle progression. Although this function of RB can be ascribed to multiple mechanisms, it is clear that RB serves to inhibit the transcription of multiple genes required for S-phase entry5,7,15,19,21. The best-studied of these target genes are regulated by the E2F family of transcription factors1,5–7,22. In this context, RB mediates transcriptional repression dependent on histone deacetylases, SWI/SNF chromatin-remodelling enzymes and additional chromatin modifiers1,23–25. Mitogens reverse transcriptional inhibition of E2F–dependent promoters through sequential activation of CDK–cyclin complexes, which phosphorylate RB and attenuate its transcriptional co-repressor capability26–28. The D-type cyclins (cyclins D1, D2 and D3) are considered focal points of this process, as the majority of mitogens signal for D-cyclin accumulation and concomitant formation of complexes between cyclin D and CDK4 or CDK6 (REF 19,REF 29).
Active CDK4 and CDK6 kinases initiate RB phosphorylation26,30, thus relaxing E2F target gene suppression. Unbiased gene expression analyses revealed that the RB–E2F regulatory targets consist of approximately 150–200 genes8–11,31–33, many of which are involved in S-phase and mitosis. Within this gene expression programme there is evidence for a feed-forward loop wherein E2F activity stimulates the expression of key factors to activate CDK2 and further promote RB hyperphosphorylation. Thus, mitogenic stimulation initiates an elegant cascade of events to control CDK–cyclin pairing, RB phosphorylation and cell-cycle transition. Once rendered inactive by phosphorylation, RB remains hyperphosphorylated until mitosis, when the regulatory function is restored through RB phosphatase activity13,14,34. However, RB action can also be induced during an active cycle in response to anti-mitogenic signals: these return RB to its hypophosphorylated active state through attenuation of cyclin expression, induction of CDK inhibitors or direct modulation of RB by phosphatases12,15,35–37. The combined strength and duration of these signals ultimately defines whether RB activity is engaged and whether the overall signalling milieu is permissive for cellular proliferation.
RB pathway and human tumours
From analyses of human tumours, it is evident that RB is disrupted through differential mechanisms to perturb tumour suppressor action. First, amplification and/or overexpression of D-type cyclins is observed in a subset of tumour types19,38. Cyclin D1 is a protooncogene that is overexpressed as a result of chromosomal rearrangements in parathyroid adenoma and mantle cell lymphoma19,38. Additionally, cyclin D1 is overexpressed in a large fraction of breast cancer cases, and mutations of cyclin D1 that enhance its nuclear function are found in oesophageal cancer39. Such deregulation of cyclin D1 action is suspected to enhance RB inactivation and thereby promote a proliferative advantage. Second, a similar effect on RB is expected in response to loss of the p16INK4A (CDKN2A) tumour suppressor, which is deleted or epigenetically silenced in a large number of human cancers40,41. p16INK4A is a true CDK inhibitor: it limits the activity of CDK complexes by binding to CDK4 or CDK6 moieties and disrupting their interaction with D-type cyclins42–45. In doing so, p16INK4A also triggers the release of p21CIP1 or p27KIP1 (which serve to support CDK4 activity but suppress CDK2 activity44,46) from cyclin D–CDK4 and cyclin D–CDK6 complexes. Thus, p16INK4A potently regulates RB phosphorylation both directly (through inhibition of CDK4) and indirectly (through release of CDK2-specific inhibitors). The signals that induce p16INK4A are diverse, including those that elicit cell-cycle exit (for example, catastrophic DNA damage) or paradoxically those that are associated with tumorigenesis (for example, activated oncogenic Ras). In both contexts, upregulation of p16INK4A is considered to be part of a senescence programme that limits inappropriate proliferation and tumorigenesis47–49. Consistent with a role for limiting CDK4 activity in the suppression of tumorigenesis, mutations of CDK4 that bypass the action of p16INK4A are observed in specific cancers, as is the amplification of CDK4 (REF 50–REF 52). Lastly, direct perturbations of RB action occur through deletion or mutation of RB1 itself, as is observed at high frequency in retinoblastoma or small-cell lung cancer40,53,54. Additionally, RB is the target of the HPV-E7 oncoproteins involved in the aetiology of cervical cancer55,56. Moderate frequency of RB loss has been observed in other tumour types (for example, breast cancer, bladder cancer and prostate cancer), as determined by analyses of loss of heterozygosity or immunohistochemical staining. Thus, disruption of at least one arm of the RB–p16INK4A–cyclin D1 triumvirate appears crucial to tumour formation and/or progression21,40,41,45.
Are all RB pathway lesions equal?
Based on mechanisms of action it was suspected that the differential mechanisms used by tumours to evade RB function resulted in overlapping biochemical and cellular consequence, and would therefore occur as mutually exclusive events in human disease. Evidence for the latter supposition has been documented in human tumour samples. For example, tumour cell lines or primary tumours lacking RB remain proficient for p16INK4A, and express low levels of cyclin D1 (REF 21,REF 40,REF 41,REF 45,REF 54,REF 57,REF 58). Not only are these events mutually exclusive, but there is a tumour-selective preference for ablation of any one pathway participant (TABLE 1). For example, within lung cancer, it is apparent that small-cell lung cancer is characterized by RB loss, whereas non-small-cell lung cancer is associated with a higher frequency of p16INK4A loss54,59,60. In other tumour types, differing frequencies of p16INK4A loss, RB loss and cyclin D1 amplification or overexpression are observed (TABLE 1). The impetus behind the observed selection of differential genetic or epigenetic alterations between tumour types and the influence on disease outcome is only now beginning to emerge, and is probably attributed to non-synonymous consequences of each genetic lesion. However, homogeneity in the RB pathway is the exception rather than the rule.
Table 1.
RB pathway heterogeneity in primary tumours
| Cancer type |
Loss of p16INK4A |
Overexpression of cyclin D1* |
Loss of RB | RB activity (indirect) |
Refs |
|---|---|---|---|---|---|
| SCLC | <10% ND | <10% ND | >90% ND | Not assessed | 54,59,196 |
| NSCLC | 40−60% indeterminate |
40−60% indeterminate |
10−20% indeterminate |
Poor outcome |
31,196–203 |
| Bladder | 20−40%poor outcome |
20–80% favourable outcome |
30−70% poor outcome |
Not assessed | 199,204−208 |
| Prostate | 10−40% indeterminate |
0−35% indeterminate |
30−60% indeterminate |
Poor outcome |
31,78,109, 111,144−148, 150, 209−213 |
| Melanoma | 40−60% poor outcome |
30−60% no influence |
10−40% no influence |
Not assessed | 214−219 |
| Pancreas | 40−70% poor outcome |
50−70% indeterminate |
<5% ND | Not assessed | 220−225 |
| Colorectal | 30−60% no influence |
40−60% no influence |
0−20% no influence |
Not assessed | 203,226−228 |
| Liver | 30−70% indeterminate |
20−40% indeterminate |
20−40% indeterminate |
Poor outcome |
174,229−237 |
| Breast | 20−40% indeterminate |
30−70% favourable outcome (ER-positive) |
10−30% indeterminate (ER-negative) |
Poor outcome |
31,104,118, 127,168,174, 229−237 |
The percent of lesions in each pathway constituent is reflective of reports in the published literature and where possible represents a consensus range. The effect of the lesion in regard to survival is noted. In certain contexts a consensus based on tumours analysed has not been reached (indeterminate). Unless otherwise noted outcome is measured as survival.
Functional RB status was determined by indirect measures in these analyses. ER, oestrogen receptor; ND, not determined; NSCLC, non-small-cell lung cancer; SCLC, small-cell lung cancer.
Diverse impact of RB dysfunction
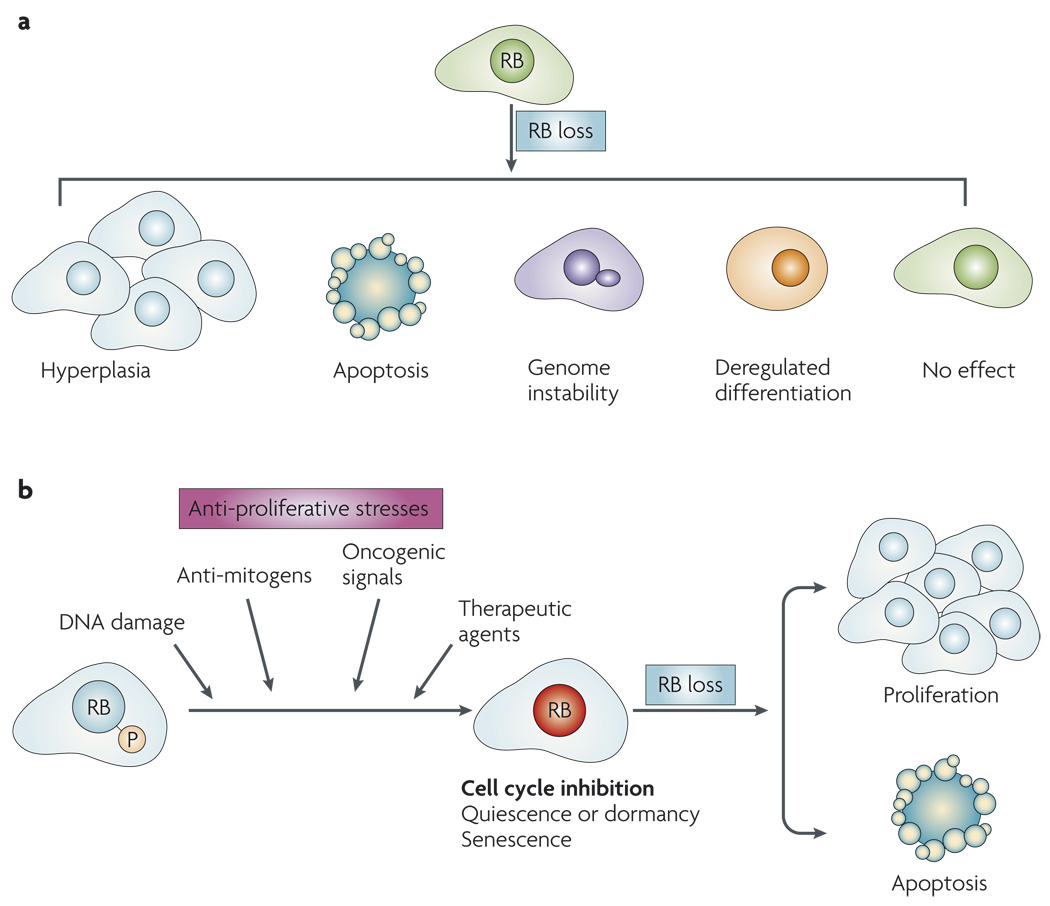
It is vital to appreciate that the effect of disrupting the RB pathway is contextdependent and can lead to diverse outcomes (FIG. 2a). For example, deletion of Rb1 in murine fibroblasts has only a minor effect on cell-cycle kinetics, but can compromise differentiation programmes or lead to genome instability9,61–65. Similar observations have been made in other cell culture systems and specific mouse tissues, such as liver, melanocytes and ovary66–68. These findings contrast with the crucial role for RB in controlling proliferation in the pituitary and several other murine organs (for example, epidermis), with similar results also shown in specific cell culture models69–71. Importantly, there are also significant differences in the consequence of RB loss between murine and human models. For example, Rb1 heterozygosity is associated with pituitary and thyroid cancer in mice, whereas humans with a defective RB1 gene develop paediatric retinoblastoma followed by increased risk for multiple secondary tumours in adulthood72–74. Such heterogeneous results become more complex if one takes into account the disparate effects of p16INK4A loss or cyclin D1 overexpression in culture models and mouse tissues. Thus, it is crucial to consider the overall significance of RB disruption in a manner that is highlight modified by genetic, tissue and organismal context. Crucially, this specificity significantly changes when conditions that inhibit proliferation and mediate the dephosphorylation of RB are considered.
Figure 2. Influence of RB loss: context dependence.
a | RB loss leads to diverse outcomes, such as hyperplasia or genome instability, in different cellular and tissue contexts. b | A diverse class of signals leads to the dephosphorylation and activation of RB. Activated RB can mediate cell-cycle inhibition leading to quiescence and/or dormancy or senescence. With the loss of RB, it is possible to escape cell-cycle inhibition and progress to either proliferation or apoptotic cell death.
Pathway specificity
Many signals and pathways lead to G1 arrest and the dephosphorylation and activation of RB1,15; of these, only a minor subset can be bypassed by cyclin D1 overexpression or p16INK4A loss. There are several proposed mechanisms that underlie these observations. First, relatively few anti-proliferative pathways function through p16INK4A (REF. 75). Second, cyclin D1 levels are often reduced by protein degradation or other mechanisms that are dominant to overexpression and/or amplification of CCND1 (REF 19,REF 76). In addition, cyclin D1 has been reported to be mislocalized to the cytoplasm in a subset of tumours, thus potentially limiting its effect on the nuclear RB protein77,78. Third, there is evidence that RB dephosphorylation induced by cell stress can emerge through mechanisms other than attenuation of CDK4 and CDK6 activity12,36,37. Thus, for many stress responses RB loss is distinct from upstream alterations in the pathway.
From studies using murine fibroblasts it has been shown that specific deletion of Rb1 alters the cell cycle-inhibitory effect of DNA-damaging agents, transforming growth factor β (TGFβ1) signalling, oncogene signalling and inhibition of the Ras pathway35,79–81 (FIG. 2b). This effect of Rb1 deficiency can lead to disparate outcomes that are highly relevant to cancer treatment. For example, Rb1 deficiency leads to enhanced cell death following genotoxic DNA damage versus ongoing proliferation in the case of Ras inhibition or TGFβ1 exposure35,82,83. Thus, RB status can serve as a crucial determinant in bypassing cell cycle-inhibitory pathways and/or promoting cell death.
Cell death
A multitude of studies in cell culture and mouse models support the concept that RB loss is associated with enhanced susceptibility to cell death. Although numerous cell types and mouse tissues can tolerate RB deficiency84,85, there is also a general predilection toward cell death that is apparent in specific contexts22,71,86,87. Two complementary mechanisms are invoked to explain this facet of RB function. First, inappropriate cell cycle progression can sensitize to cell death. A key example of this involves DNA-damaging agents or microtubule poisons wherein deregulated cell-cycle progression can lead to enhanced damage burden by inducing secondary replication-associated lesions or mitotic catastrophe88. Second, unrestrained E2F activity can lead to the aberrant expression of pro-apoptotic genes that will predispose RB-deficient cells to death22,82,89,90. Consistent with the latter concept, RB loss sensitizes cells to a variety of pro-apoptotic stimuli that do not intrinsically act on the cell cycle86,87,91,92, and the effects of RB on cell cycle and apoptosis can be dissociated91. Additionally, loss of RB can contribute to p53-mediated cell death through a mechanism that is at least partially dependent on the ARF tumour suppressor93. ARF expression can be induced by inappropriate E2F activity, as occurs with the disruption of RB, and this triggers activation of p53 and apoptosis94,95. As such there can be clear cross-talk between the RB and p53 pathways. However, loss of RB also contributes to both ARF- and p53-independent cell death96–98. Therefore, an appealing goal is to target signals and pathways that will specifically induce death in ectopically dividing RB-deficient cells.
Senescence
Akin to apoptosis, senescence is viewed as a terminal fate for cells and involves ‘irreversible’ cell-cycle arrest. Senescence can be induced by a panoply of signals, including activation of oncogenic signals, withdrawal of oncogenic signals, restoration of p53 activity, DNA damage and specific therapeutic regimens, thus serving as an intrinsic barrier to tumorigenesis47,48,99–101. p16INK4A is a marker for senescence and is required for the establishment of cell-cycle arrest, whereas RB loss is sufficient to mediate escape from senescence102,103. Thus, aberrations in the RB pathway may be particularly relevant to escape the engagement of this pathway.
Although many of these functional findings have been generated using murine fibroblasts in vitro or analyses of mouse development, it seems logical to consider whether the status of the RB pathway could influence the response of a tumour to specific therapeutic modalities.
A case for hormone independence
RB pathway aberrations are found with significant frequency in hormone-dependent cancers, including those of the breast104–108 and prostate109–111. As such, several studies assessed the impact of RB status on endocrine-based therapeutic regimens (TABLE 2).
Table 2.
Preclinical assessment of RB loss and therapeutic response
| Agent | Model | RB modulation | Outcome | Refs |
|---|---|---|---|---|
| Hormonal antagonism | ||||
| Oestrogen withdrawal |
ER+ breast cancer | Knockdown | Bypass | 118 |
| Tamoxifen | ER+ breast cancer | Knockdown, viral oncoproteins |
Bypass | 118,119 |
| ICI182780 | ER+ breast cancer | Knockdown, viral oncoproteins |
Bypass | 118 |
| Androgen withdrawal | Prostate cancer | Knockdown, viral oncoproteins |
Bypass |
136,137, 143 |
| Casodex | Prostate cancer | Knockdown | Bypass | 143 |
| Cytotoxic chemlotherapy | ||||
| Cisplatin | Breast cancer, lung cancer, murine fibroblasts, prostate cancer* |
Knockout, knockdown | Sensitivity/ Resistance* |
35,81, 118,151 |
| 5-Fluorouracil |
Lung cancer, murine fibroblasts |
Knockout, knockdown, viral oncoproteins |
Sensitivity |
16,151, 238,239 |
| Ionizing radiation |
Breast cancer, murine fibroblasts |
Knockout, knockdown, viral oncoproteins |
Sensitivity |
81,118, 240 |
| Doxorubicin | Lung cancer, murine fibroblasts |
Knockout, knockdown, oncoproteins |
Sensitivity |
16,81, 82,151 |
| Kinase inhibitors | ||||
| U0126 | Murine fibroblasts, transformed murine fibroblasts |
Knockout, spontaneous loss |
Bypass | 155 |
| Staurosporine and UCN01 |
Murine fibroblasts, breast cancer, prostate cancer |
Knockout, knockdown | Bypass | 156−158 |
| PD 332991 | Breast cancer, lung cancer |
Spontaneous loss | Bypass | 159,160 |
Tumour lines or the use of murine fibroblasts are indicated. The model through which RB deficiency was produced is indicated (knockout reflects use of gene-targeted murine cells, knockdown reflects use of RNA-interference-based approach and viral oncoproteins reflect E1A or SV40 large T-antigen reagents that target pocket proteins). Bypass indicates a proliferative advantage with RB deficiency, whereas sensitivity indicates that RB deficiency decreased cellular viability.
Resistance to cisplatin treatment in RB-deficient cells was observed in prostate cancer models. ER, oestrogen receptor.
Breast cancer
Breast cancers that are oestrogen receptor (ER)-positive are frequently treated using hormone therapies that deplete oestrogen (such as GnRH agonists) or directly antagonize the transactivation function of ER (such as tamoxifen or ICI182780). The effects of oestrogen antagonists on the cell-cycle response in ER-positive models of breast cancer have been analysed extensively112–114. These cell culture studies demonstrated that ER antagonism leads to cell-cycle cessation in G1 and the dephosphorylation and activation of RB. p16INK4A activity seems not to be required, as commonly used ER-positive tamoxifen-sensitive cell lines lack p16INK4A expression115. Furthermore, cyclin D1 levels are effectively attenuated in ER-positive cell lines, including those that express high levels of endogenous cyclin D1 (for example, MCF7 cells)116. Indeed, overexpression of cyclin D1 is not sufficient to maintain RB phosphorylation or promote proliferation in the face of prolonged exposure to oestrogen antagonists117. Thus, irrespective of upstream lesions in the RB pathway, the RB protein can be activated and cell-cycle progression impeded. Presumably, this is because ER antagonists affect multiple facets of cell-cycle machinery that coalesce in the regulation of RB114,117. In such cell culture models, knockdown of RB1 by RNA-interference approaches or functional inactivation through the use of viral oncoproteins leads to an effective bypass of the effects of tamoxifen and related therapeutics118,119. These studies were extended to xenograft models, where RB deficiency leads to therapeutic failure with tamoxifen119. These findings suggest that disruption of RB function, but not loss of p16INK4A or cyclin D1 overexpression, has a deleterious effect on the treatment of ER-positive breast cancer.
The relevance of the RB pathway to tamoxifen response in cancer biopsies has been widely analysed. Although cyclin D1 is overexpressed in a large fraction of ER-positive breast cancers, the relevance of this event to the response to tamoxifen remains uncertain. In several large studies, high-level expression of cyclin D1 was not observed to influence the response to tamoxifen therapy120–122; however, other similarly devised studies suggest that overexpression of cyclin D1 is associated with tumour recurrence for tamoxifen therapy123,124. This controversy remains unresolved, but could be related to the observation that overexpression of cyclin D1 is associated with a form of breast cancer that has intrinsically improved prognosis123,125–127. Surprisingly, overexpression of p16INK4A is generally associated with a poor prognosis in breast cancer128,129; moreover, loss of p16INK4A has been only been marginally studied in the context of tamoxifen therapy, and appears to hold little prognostic significance130,131.
Consistent with the lack of clear relationship with upstream aberrations in the RB pathway, hyperphosphorylation of RB, as detected using phospho-specific antibodies, is not associated with response to endocrine therapy132. As discussed in more detail below, loss of RB is difficult to evaluate histologically. Furthermore, the percentage of ER-positive breast cancers that lack RB expression is rather limited (10–20%). However, histological loss of RB has been associated with a poor response to endocrine therapy132,133. Furthermore, indirect analyses of RB loss, using gene-profiling approaches, further support this concept31,118. Thus, combined with the data from preclinical models, there is evidence to suggest that RB loss is associated with therapeutic failure and that heterogeneity within the RB pathway could be of particular significance in specifying response to oestrogen antagonists.
Prostate cancer
Prostate cancers are exquisitely dependent on androgen receptor (AR) activity for growth and progression134. Given the poor response of this tumour type to cytotoxic therapeutic agents, strategies to ablate AR function (achieved through the use of androgen depletion strategies or direct AR antagonists) are the first line of treatment for disseminated prostate tumours135.
In cell culture models of androgen-dependent prostate cancer, such therapeutics lead to cell-cycle arrest in G1 that is accompanied by reduced expression of D-cyclins and efficient RB dephosphorylation136. Given these observations, there has been a concerted interest in delineating the mechanisms by which the cyclin D1–p16INK4A–RB axis may be perturbed in the transition to androgen independence and thus promote therapeutic resistance. With regard to p16INK4A, although overexpression can potently arrest prostate cancer cell lines137, there is little evidence that p16INK4A induction participates in cell-cycle exit following AR antagonism. Similarly, although D-cyclins are induced by androgen through post-translational mechanisms138, cyclin D1 expression is not sufficient to restore cellular proliferation in cultured prostate cancer cells challenged with androgen ablation or androgen antagonists137.In this cell type, accumulated cyclin D1 markedly antagonizes AR function and can impede subsequent rounds of cellular proliferation through kinase-independent mechanisms that have been well defined139–142. Thus, cyclin D1 has a general anti-proliferative role in such models. By contrast, emerging evidence suggests that disruption of RB itself may have a significant consequence on prostate cancer therapies. Initially, it was observed that viral oncoproteins with the capacity to inhibit RB function were sufficient to promote cell-cycle progression in the absence of androgen or presence of androgen antagonists136. Subsequent analyses of isogenic, AR-positive cancer cells revealed that RB depletion alone rendered no discernable proliferative advantage to prostate cancer cells in the presence of androgen143. However, RB depletion in prostate cancer cells was sufficient to sustain cell-cycle progression after challenge with androgen ablation and/or AR antagonist strategies that mimic therapeutic intervention136,143. Based on these observations, it was suspected that aberrations in RB itself (rather than loss of p16INK4A or overexpression of cyclin D1) may contribute to hormone resistance.
Despite these findings, clinical studies examining the impact of cyclin D1, p16INK4A or RB as determinants of therapeutic outcome have been only preliminarily considered. p16INK4A loss is infrequently observed; conversely, increased p16INK4A levels are associated with a poor prognosis, similar to that observed in breast cancer144–146. Thus, p16INK4A function appears to be maintained in the majority of prostate cancers, and p16INK4A loss does not appear to have a role in the transition to androgen independence. A role for cyclin D1 in this process is similarly obscure, and the function(s) of cyclin D1 in this tissue type incompletely defined. Several reports have demonstrated that cyclin D1 expression is rare or infrequent in primary disease, supporting the idea that excessive cyclin D1 expression is probably not a major factor in disease development or progression147–150. Furthermore, recent analyses showed that cyclin D1 is low or mislocalized to the cytoplasm in a significant fraction of prostate cancers obtained from radical prostatectomy78. However, a subset of tumours, either associated with high p21CIP1 levels or associated with bone metastases, do show enhanced nuclear cyclin D1, suggesting that cyclin D1 function is complex and dependent on molecular milieu78,149. Surprisingly, there have been few studies of RB loss in prostate cancer specimens; however, RB loss is overrepresented in recurrent prostate cancers, which are resistant to hormone therapy109. Furthermore, the gene expression profile generated by viral oncoproteins that disrupt RB function is associated with poor disease outcome in prostate cancer31. These findings are consistent with functional analyses of RB loss in cultured prostate cancer cells, and indicate that RB loss may have a specific role in the acquisition of androgen independence.
Based on these collective findings, it is predicted that the efficacy of endocrine-based therapies for breast and prostate cancers is particularly reliant on RB activation, and that loss of RB serves as a mechanism to bypass therapeutic intervention.
Priming to kill: cytotoxics
Contrary to results observed with hormone-based regimens, preclinical models in a multitude of systems suggest that RB loss can actually sensitize cells to specific cytotoxic or genotoxic agents, but this response is dependent on tissue type (TABLE 2). DNA damage will effectively and rapidly lead to the degradation of cyclin D1 (REF. 76), and RB-dependent DNA damage checkpoints are operable in the absence of p16INK4A (REF 118,REF 151). Thus, upstream lesions in the RB pathway are not a priori crucial determinants of the response to chemotherapeutic agents. Whereas breast cancer cells or xenografts depleted of RB can successfully evade ER antagonists, these cells are more susceptible to elimination induced by cisplatin or ionizing radiation than isogenic RB-proficient counterparts118. Similar effects were observed in lung cancer xenografts, where RB depletion sensitized cells to the cytotoxic effects of cisplatin, doxorubicin and 5-fluorouracil151. In both cases, failure to arrest after chemotherapeutic intervention was noted118,151. Interestingly, these effects were observed both in the presence and in the absence of p53, suggesting that RB can function as a determinant of cytotoxic therapies independently of p53. However, these outcomes were not uniformly observed with all tumour types or cytotoxic agents. For example, in the context of prostate cancer, RB-depleted cells were sensitized to cell death induced by microtubule poisons (docetaxel and paclitaxel) and etoposide but, in contrast to breast and lung cancer cells, were notably more resistant to cisplatin143. The basis of this divergent sensitivity is a focus of current investigation, and will provide a platform through which to delineate the underpinning mechanisms of divergent chemotherapeutic responses.
The effect of RB deficiency on the response to cytotoxic therapies has also been observed in tumour specimens. Specific loss of RB is associated with improved response to cytotoxic therapeutics in bladder cancer152,153, and recently published studies have demonstrated that although hyperphosphorylation of RB does not affect the response to cytotoxic agents in breast cancer, RB loss is directly associated with improved outcome with chemotherapy132. In this study the effect of RB status was independent of p53 and other pathophysiological markers in multivariate analyses. Thus, loss of RB in tumours could be a specific determinant of sensitivity to cytotoxic therapies.
Although these findings are promising, it has yet to be conclusively determined whether RB deficiency and resultant sensitivity to cytotoxic agents broadly translates into improved long-term survival. Analyses in RB-deficient lung cancer xenograft models showed that the initial dramatic response to cisplatin and doxorubicin did not lead to a durable effect on tumour burden. Rather, RB-deficient tumours recurred readily with the completion of the treatment cycle151. Such a troubling trend is observed in specific tumour types, such as small-cell lung cancer (which often loses RB), where the initial response to therapy can be quite effective but recurrence of resistant disease occurs at a high frequency154. Therefore, additional studies will be required to define how RB loss translates into overall survival in additional tumour types following specific treatment regimens.
RB and kinase inhibitors
In addition to hormonal and cytotoxic agents, a number of kinase inhibitors have been shown to depend on RB protein function for activity (TABLE 2). Although not yet in routine clinical use, these agents are viewed as foundations for ongoing therapeutic development or have been used in the context of clinical trials. The kinase inhibitors that have been interrogated fall into three principle categories as discussed below.
Erk/MEK inhibitors
Based on the finding that RB-negative murine fibroblasts can proliferate in the presence of Ras antagonists79, it is perhaps not surprising that they are less sensitive to the effects of MEK inhibition by U0126. This has been demonstrated in both primary and transformed rodent lines and has been associated with RB status in human tumour lines155. Although U0126 is not used clinically, the compounds PD 0325901 and AZD6244, which are in clinical trials, would be expected to function in a similar fashion. Thus, loss of RB may have a significant impact on the response to such agents.
UCNO1 and staurosporine
These agents are broad-functioning kinase inhibitors with targets that include protein kinase C (PKC) enzymes and checkpoint kinases. Irrespective of function, both agents lead to RB-dependent suppression of cell-cycle progression 156–158. As such, RB loss enables tumour cells to proliferate in the presence of these agents.
CDK inhibitors
Given the central role of RB in cell cycle control, there is an obvious need to determine the impact of RB status on the cellular response to CDK inhibitors. Agents such as roscovitine that target CDK2 activity also target CDK1 and can inhibit cell cycle progression independent of RB. However, consistent with RB being a downstream target of CDK4, RB is required for the effect of the compound PD 0332991, as determined in cell culture and xenograft experiments159,160. Thus, different classes of CDK inhibitor elicit distinct dependence on the RB pathway for efficacy.
Combined, the analyses of these agents suggest that RB deficiency could represent a bypass mechanism for a relatively large range of ‘molecularly-targeted therapies’ that function through a cytostatic mechanism.
RB status in clinical application
Given the preclinical indications that RB status might be considered for development of tailored therapy, it is imperative to identify hurdles associated with the development of RB as a prognostic marker. Although genetic consideration of RB status in the context of retinoblastoma diagnoses is routine and deletions of the locus at 13q14 are used to define severity of multiple myeloma161,162, few treatment regimens interrogate RB status when considering efficacy. At present there has been only one National Cancer Institute-sponsored trial that considered the presence of RB for inclusion, which used the CDK4-specific inhibitor PD 332991 (NCT00141297). This agent is critically dependent on RB for tumour-static activity in xenograft models159,160; thus the eligibility criteria are important for the logical direction of therapy. The importance of targeting RB has also been appreciated retrospectively in several studies, where reduced levels of RB phosphorylation (suggesting effective engagement of RB) have been used as a means to assess drug efficacy163,164.
To most effectively challenge the contribution of RB for therapeutic response in prospective studies, it must first be determined how RB status might be effectively and economically determined (FIG. 3). While genetic loss of RB1 would be considered the gold standard, this approach fails to detect the myriad of additional mechanisms through which RB is functionally inactivated in cancer. Traditional immunohistochemical analyses also have limitations, in that RB levels fluctuate as a function of cellular proliferation165, and the protein can be expressed yet be inactive in tumour cells. These facets of RB protein expression may in part explain the somewhat confused picture of RB status that emerges from histological studies. Although phospho-specific antibodies have been used to assess RB phosphorylation in biopsy specimens163,164,166,167, as discussed above, the basal state of RB phosphorylation is not necessarily indicative of altered response to therapeutic agents.
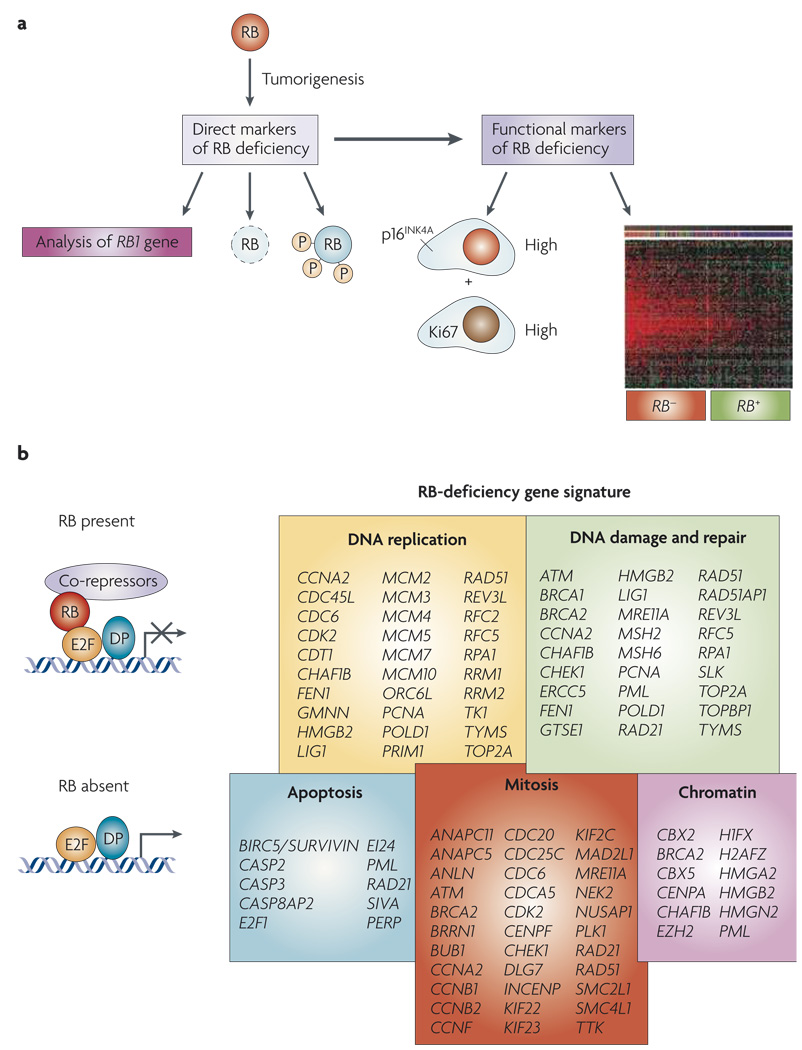
Figure 3. Mutiple markers for RB dysfunction.
a | Direct analyses of the status of RB1 are used in genetic tests to establish the presence of heritable retinoblastoma. RB levels can be directly interrogated by immunohistochemical analyses and the use of phospho-specific RB antibodies provides additional information as to the status of RB. However, as RB can be inactivated through multiple mechanisms, indirect markers for RB function can also be particularly informative. In this context, gene expression signatures reveal the downstream consequence of RB loss. p16INK4A levels are increased in specific RB-negative tumours, and these cells can be discerned from pockets of senescent pre-neo-plastic cells by the inclusion of a proliferative marker such as Ki67. b | Gene-signature analyses have shown the functional groups of genes that are deregulated by the loss of RB1 through either deletion or expression of viral oncoproteins. This gene expression signature overlaps with the gene expression grade index and the Oncotype Dx signatures by 65% and 80% respectively.
Owing to such limitations, it is appealing to use indirect methods for monitoring ‘RB activity’ in tumours. An unexpected marker for loss of RB activity may be high-level expression of p16INK4A (REF. 168) (FIG. 3). The p16INK4A protein is induced by oncogenic stresses to suppress tumorigenesis; however, tumours lacking RB are able to proliferate despite high levels of p16INK4A expression169,170. Thus, scoring for p16INK4A and proliferative markers (for example, Ki67) would be expected to define RB-deficient tumours. This combination proved effective in defining ductal carcinomas in situ that are likely to progress168, and could be useful in deciphering the ability of established tumours to respond to therapy. Consistent with this supposition, high levels of p16INK4A in breast and prostate tumours are an indicator of poor prognosis128,129,171. Additionally, high levels of p16INK4A are used to define cancerous cervical lesions that arise as a consequence of HPV-mediated RB inactivation172,173.
An alternative strategy makes use of the known programme of genes that are controlled by RB–E2F8–11,31. Although there is some heterogeneity between tissues, the RB gene expression signature has been readily observed in numerous tumour types, including breast, lung and hepatocellular carcinoma31,118,174. The RB gene expression signature shares significant features with other gene expression signatures that have been associated with disease outcomes including, the ‘proliferation signature’, the ‘chromosome instability signature’, ‘genomic grade index’ (GGI), and ‘recurrence score’ (Oncotype Dx)175–179 (FIG. 4). Of these signatures, the GGI and recurrence score have been shown to have significant utility in differentiating tumours that will respond to tamoxifen therapy or will require more aggressive therapy176,177,180–186. Such prognostics are currently being analysed in the context of prospective breast cancer clinical treatment, and the signature of RB–E2F–regulated proliferative genes contributes to the predictive nature of virtually all such gene expression prognostics176,185. Therefore, it could be argued that RB activity is already being considered in the context of breast cancer prognosis, and that the recent preclinical investigations may be useful in refining and directing the use of such predictive signatures.
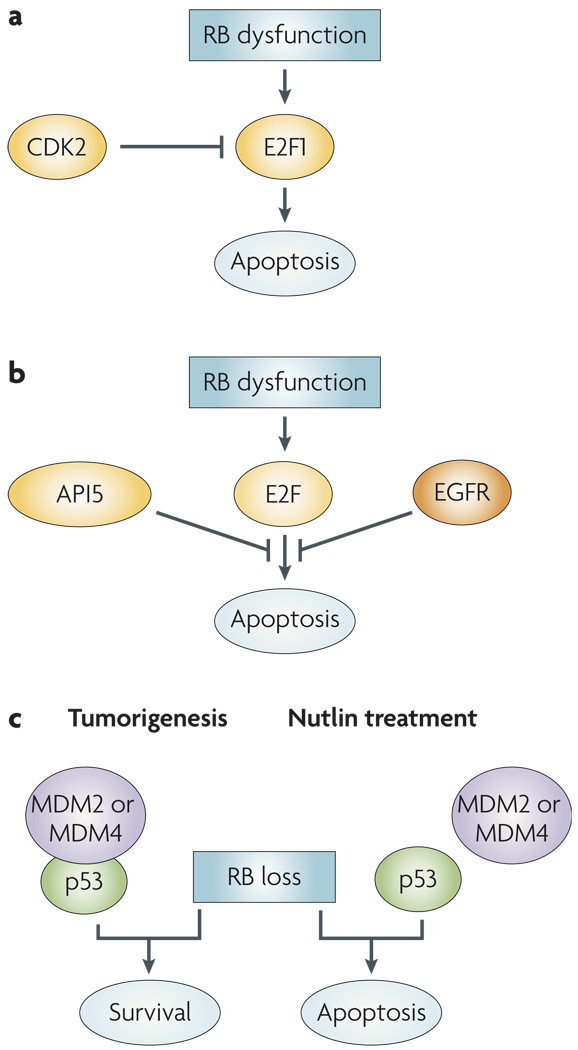
Figure 4. Exploiting RB deficiency therapeutically.
a | RB limits the pro-apoptotic activity of E2F1, so RB-deficient cells are more prone to apoptosis. Cyclin-dependent kinase 2 (CDK2) activity further limits E2F1 activity. Thus, CDK2 inhibitors can further increase E2F1 activity and drive RB-deficient cells to apoptosis. b | In Drosophila melanogaster models, E2F activity can induce apoptosis; however, this is limited by the action of epidermal growth factor receptor (EGFR) signalling and the apoptosis inhibitor 5 (API5, also known as AAC11). c | In mouse models of retinoblastoma, p53 inactivation is required for tumour development and the survival of RB-deficient cells. In tumorigenesis, this inactivation occurs owing to an upregulation of MDM2 or MDM4. The activity of these oncogenes can be targeted by the nutlins, a group of drugs that disrupt the interaction between MDM2 and p53.
Exploiting RB loss therapeutically
Directly exploiting the intrinsic properties of RB deficiency to therapeutic end could have utility in a large range of human cancers. Whereas other targets have been the subject of intense screening efforts, only a single unbiased chemical screen that was directed at specific elimination of RB-defective tumour cells has been reported. In this system, RB activity was compromised by viral oncoproteins in an isogenic human transformed fibroblast system187. Although viral oncoproteins do efficiently target RB, a caveat of these studies is the impact of viral oncoproteins on additional factors of importance in cancer. These potentially confounding factors aside, in a total of >25,000 compounds screened, the only compounds that selected for RB-deficient cell death were established topoisomerase II poisons (for example, doxorubicin)187. Strikingly, doxorubicin has significantly enhanced effect in treating RB-deficient lung tumour xenografts151. Thus, relatively simple chemical screens could be effective in defining compounds that exploit the impact of RB loss, and should be considered for tissue-specific analyses of cytotoxicity as a function of RB status.
An alternative therapeutic approach to target RB-deficient cells may be to harness the latent pro-apoptotic activity of E2F (FIG. 4). One such approach takes advantage of the fact that E2F1 activity, which is particularly pro-apoptotic, is restricted by CDK2-mediated phosphorylation in latter phases of the cell cycle188,189. For example, in the absence of functional RB, CDK2 inhibition leads to increased E2F1 function and resultant cell death189. Conversely, a number of synthetic lethal screens have been performed in Drosophila melanogaster models to identify factors that restrain E2F–mediated apoptosis190,191. In these studies, the epidermal growth factor receptor (EGFR) and apoptosis inhibitor 5 (API5, also known as AAC11) were identified as required to suppress apoptosis arising from deregulated E2F activity. These studies suggest that EGFR antagonists may have significant utility in those tumours that harbour loss of RB function191. Furthermore, API5 is upregulated in a number of tumour cell types, and depletion of API5 specifically mediated cell death of tumour cells190. Thus, drugs targeting these ‘survival’ factors could be highly effective in RB-deficient tumours.
Although these screens clearly have the power to define genetic and functional interactions in model systems, it is equally important to decipher tumour-specific relationships that can be targeted therapeutically. Retinoblastoma is a tumour type where such relationships can be investigated against a high background of RB loss. Analyses of a combination of mouse models and human tumours suggested that inhibition of p53 function is a crucial determinant in the genesis of retinoblastoma192 (FIG. 4). Furthermore, in human tumours this event was associated with the amplification of MDM2 or MDM4; therefore, targeting MDM2 or MDM4 function may be of benefit in the treatment of retinoblastoma. Consistent with this concept, therapeutic agents that antagonize MDM2 and MDM4 function (for example, nutlins) show promise in murine models of retinoblastoma192–194. Additional studies in other tumours support the general concept that loss of RB may be particularly important in facilitating the cytotoxic effects of nutlin-3a195. Based on these findings, it is evident that tissue- and context-specific functions of RB provide a rich foundation for the development of future targeted therapeutic strategies.
Summary
Although RB was identified as the first bona fide tumour suppressor over 20 years ago, the implications of RB loss for cancer progression and tumour management are only now being uncovered. It is clear that RB serves as a gatekeeper of proliferative control, and that perturbations of RB function can occur through multiple tumour-specific alterations. These lesions arise in a non-overlapping fashion suggesting that they similarly perturb RB function; however, heterogeneity in the RB pathway is clearly relevant to both the aetiology of specific tumour types and disease behaviour. The specific import of RB deficiency is unmasked in the presence of cellular stress, including those introduced during cancer therapy. In hormone-dependent cancers, evidence suggests that RB depletion is sufficient to bypass endocrine-based therapeutic regimens. Similar results are observed with kinase inhibitors that antagonize distinct signalling pathways. Given the importance of these strategies in current and future tumour management, these findings may have significant clinical ramification. By contrast, RB deficiency can sensitize tumour cells to a subset of cytotoxic agents. Together, these findings identify RB as a crucial node that could be developed as a component of tailored anticancer strategies. However, formidable questions remain. First, what is the nature of tissue-specific and lesion-specific RB perturbation, and how do the differential mechanisms used to compromise RB function affect its predictive value? Second, how can RB deficiency be expediently identified in human tumour specimens? Third, can the predictive value of RB status be validated in prospective studies? At present, few clinical studies have considered RB status as a predictive factor for therapeutic response. Last, how can knowledge of RB function and cooperative factors be optimized so as to elicit maximal, tumour-specific cytotoxic response in RB-deficient contexts? Once addressed, it is hoped that these and similar queries will define the denouement of investigation of RB perturbations in human tumours, and will provide the foundation for translating decades of RB-related research to the clinic.
Acknowledgements
The authors would like to thank all of their colleagues for thought-provoking discussions that provide the basis for the current Perspective. In particular the authors appreciate the contributions of W. Cavenee, R. Bremner, and T Tlsty to the preparation of the manuscript. A concerted effort was made to provide a highly inclusive discussion of the field: any omission was accidental and the authors apologize for not being able to cite all of the outstanding studies in the literature. The authors are supported by grants from the NIH: CA10617 and CA104213 to E.S.K. and CA099996 and CA116777 to K.E.K.
Footnotes
DATABASES
ClinicalTrials.gov: http://clinicaltrials.gov/ NCT00141297
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene API5 | CDK2 | CDK4 | CDK6 | CDKN2A | cyclin D1 | cyclin D2 | cyclin D3 | EGFR | MDM2 | MDM4 | RB1 | p53 | TGFβ1
National Cancer Institute: http://www.cancer.gov/ bladder cancer | breast cancer | cervical cancer | lung cancer | mantle cell lymphoma | oesophageal cancer | parathyroid cancer | prostate cancer | retinoblastoma
National Cancer Institute Drug Dictionary: http://www.cancer.gov/drugdictionary/ 5-fluorouracil | Casodex | cisplatin | docetaxel | doxorubicin | etoposide | ICI182780 | paclitaxel | PD 0325901 | PD 0332991 | tamoxifen | UCN01
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Erik S. Knudsen, Department of Cancer Biology, Kimmel Cancer Center, Bluemle Life Science Building-Room 1002, 233, South 10th Street, Thomas Jefferson University, Philadelphia, Pennsylvania 19107, USA. eknudsen@kimmelcancercenter.org
Karen E. Knudsen, Department of Cancer Biology, Kimmel Cancer Center, Bluemle Life Science Building-Room 1002, 233, South 10th Street, Thomas Jefferson University, Philadelphia, Pennsylvania 19107, USA. Department of Urology, Kimmel Cancer Center, Bluemle Life Science Building-Room 1002, 233, South 10th Street, Thomas Jefferson University, Philadelphia, Pennsylvania 19107, USA. kknudsen@kimmelcancercenter.org
References
- 1.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 2.Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nature Rev Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 3.Liu H, Dibling B, Spike B, Dirlam A, Macleod K. New roles for the RB tumor suppressor protein. Curr. Opin . Genet. Dev. 2004;14:55–64. doi: 10.1016/j.gde.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 5.Nevins JR. The Rb/E2F pathway and cancer. Hum. Mol . Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 6.Blais A, Dynlacht BD. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr. Opin. Genet. Dev. 2004;14:527–532. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Blais A, Dynlacht BD. E2F–associated chromatin modifiers and cell cycle control. Curr. Opin. Cell Biol. 2007;19:658–662. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markey MP, et al. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 2002;62:6587–6597. [PubMed] [Google Scholar]
- 9.Markey MP, et al. Loss of the retinoblastoma tumor suppressor: differential action on transcriptional programs related to cell cycle control and immune function. Oncogene. 2007;26:6307–6318. doi: 10.1038/sj.onc.1210450. [DOI] [PubMed] [Google Scholar]
- 10.Black EP, et al. Distinct gene expression phenotypes of cells lacking Rb and Rb family members. Cancer Res. 2003;63:3716–3723. [PubMed] [Google Scholar]
- 11.Vernell R, Helin K, Muller H. Identification of target genes of the p16INK4A–pRB-E2F pathway. J. Biol . Chem. 2003;278:46124–46137. doi: 10.1074/jbc.M304930200. [DOI] [PubMed] [Google Scholar]
- 12.Mittnacht S. Control of pRB phosphorylation. Curr. Opin . Genet. Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 13.Vietri M, Bianchi M, Ludlow JW, Mittnacht S, Villa-Moruzzi E. Direct interaction between the catalytic subunit of Protein Phosphatase 1 and pRb. Cancer Cell Int. 2006;6:3. doi: 10.1186/1475-2867-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broceno C, Wilkie S, Mittnacht S. RB activation defect in tumor cell lines. Proc. Natl Acad. Sci. USA. 2002;99:14200–14205. doi: 10.1073/pnas.212519499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JY, Knudsen ES, Welch PJ. The retinoblastoma tumor suppressor protein. Adv. Cancer Res. 1994;64:25–85. doi: 10.1016/s0065-230x(08)60834-9. [DOI] [PubMed] [Google Scholar]
- 16.Mayhew CN, et al. Discrete signaling pathways participate in RB-dependent responses to chemotherapeutic agents. Oncogene. 2004;23:4107–4120. doi: 10.1038/sj.onc.1207503. [DOI] [PubMed] [Google Scholar]
- 17.Cavenee WK, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 18.Cavenee WK, et al. Genetic origin of mutations predisposing to retinoblastoma. Science. 1985;228:501–503. doi: 10.1126/science.3983638. [DOI] [PubMed] [Google Scholar]
- 19.Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol. Ther. 2002;1:226–231. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- 20.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 21.Sherr CJ. Cell cycle control and cancer. Harvey Lect. 2000;96:73–92. [PubMed] [Google Scholar]
- 22.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr. Opin. Cell Biol. 2007;19:649–657. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 24.Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv. Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- 25.Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 2002;14:684–691. doi: 10.1016/s0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- 26.Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 27.Knudsen ES, Wang JY. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol. Cell. Biol. 1997;17:5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinds PW, et al. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 29.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 30.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclincdk complexes. Mol. Cell. Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deeb KK, et al. Identification of an integrated SV40 T/t-antigen cancer signature in aggressive human breast, prostate, and lung carcinomas with poor prognosis. Cancer Res. 2007;67:8065–8080. doi: 10.1158/0008-5472.CAN-07-1515. [DOI] [PubMed] [Google Scholar]
- 32.Ishida S, et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren B, et al. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamrakar S, Rubin E, Ludlow JW. Role of pRB dephosphorylation in cell cycle regulation. Front. Biosci. 2000;5:D121–D137. doi: 10.2741/tamrakar. [DOI] [PubMed] [Google Scholar]
- 35.Knudsen KE, et al. RB-dependent S-phase response to DNA damage. Mol. Cell. Biol. 2000;20:7751–7763. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avni D, et al. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol. Cell. 2003;12:735–746. doi: 10.1016/s1097-2765(03)00355-1. [DOI] [PubMed] [Google Scholar]
- 37.Dou QP, An B, Will PL. Induction of a retinoblastoma phosphatase activity by anticancer drugs accompanies p53-independent G1 arrest and apoptosis. Proc. Natl Acad. Sci. USA. 1995;92:9019–9023. doi: 10.1073/pnas.92.20.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold A, Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J. Clin. Oncol. 2005;23:4215–4224. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 39.Benzeno S, et al. Identification of mutations that disrupt phosphorylation-dependent nuclear export of cyclin D1. Oncogene. 2006;25:6291–6303. doi: 10.1038/sj.onc.1209644. [DOI] [PubMed] [Google Scholar]
- 40.Palmero I, Peters G. Perturbation of cell cycle regulators in human cancer. Cancer Surv. 1996;27:351–367. [PubMed] [Google Scholar]
- 41.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nature Rev. Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 42.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 43.Kamb A, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 44.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 45.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, et al. Somatic cell type specific gene transfer reveals a tumor-promoting function for p21(Waf1/ Cip1) EMBO J. 2007;26:4683–4693. doi: 10.1038/sj.emboj.7601886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 48.Collado M, et al. Tumour biology: senescence in premalignant tumours. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 49.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 50.Wolfel T, et al. A p16INK4a–insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 51.Malumbres M, et al. Driving the cell cycle to cancer. Adv. Exp. Med. Biol. 2003;532:1–11. doi: 10.1007/978-1-4615-0081-0_1. [DOI] [PubMed] [Google Scholar]
- 52.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim. Biophys. Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 53.Horowitz JM, et al. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc. Natl Acad. Sci. USA. 1990;87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaye FJ. RB and cyclin dependent kinase pathways: defining a distinction between RB and p16 loss in lung cancer. Oncogene. 2002;21:6908–6914. doi: 10.1038/sj.onc.1205834. [DOI] [PubMed] [Google Scholar]
- 55.Munger K. The role of human papillomaviruses in human cancers. Front. Biosci. 2002;7:d641–d649. doi: 10.2741/a800. [DOI] [PubMed] [Google Scholar]
- 56.Munger K, et al. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene. 2001;20:7888–7898. doi: 10.1038/sj.onc.1204860. [DOI] [PubMed] [Google Scholar]
- 57.Lukas J, et al. DNA tumor virus oncoproteins and retinoblastoma gene mutations share the ability to relieve the cell’s requirement for cyclin D1 function in G1. J. Cell Biol. 1994;125:625–638. doi: 10.1083/jcb.125.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Cell. Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan J, et al. Expression of p16 and lack of pRB in primary small cell lung cancer. J. Pathol. 1999;189:358–362. doi: 10.1002/(SICI)1096-9896(199911)189:3<358::AID-PATH452>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 60.Otterson GA, Kratzke RA, Coxon A, Kim YW, Kaye FJ. Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB. Oncogene. 1994;9:3375–3378. [PubMed] [Google Scholar]
- 61.Herrera RE. Altered cell cycle kinetics, gene expression,and G1 restriction point regulation in Rb-deficient fibroblasts. Mol. Cell. Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mayhew CN, Bosco EE, Solomon DA, Knudsen ES, Angus SP. Analysis of RB action in DNA damage checkpoint response. Methods Mol. Biol. 2004;281:3–16. doi: 10.1385/1-59259-811-0:003. [DOI] [PubMed] [Google Scholar]
- 63.Novitch BG, Spicer DB, Kim PS, Cheung WL, Lassar AB. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr. Biol. 1999;9:449–459. doi: 10.1016/s0960-9822(99)80210-3. [DOI] [PubMed] [Google Scholar]
- 64.Knudsen ES, Sexton CR, Mayhew CN. Role of the retinoblastoma tumor suppressor in the maintenance of genome integrity. Curr. Mol. Med. 2006;6:749–757. doi: 10.2174/1566524010606070749. [DOI] [PubMed] [Google Scholar]
- 65.Hansen JB, et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc. Natl Acad. Sci. USA. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayhew CN, et al. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- 67.Tonks ID, et al. Melanocytes in conditional Rb−/− mice are normal in vivo but exhibit proliferation and pigmentation defects in vitro. Pigment Cell Res. 2005;18:252–264. doi: 10.1111/j.1600-0749.2005.00245.x. [DOI] [PubMed] [Google Scholar]
- 68.Flesken-Nikitin A, Choi KC, Eng JP, Shmidt EN, Nikitin AY. Induction of carcinogenesis by concurrent inactivation of p53 and Rb1 in the mouse ovarian surface epithelium. Cancer Res. 2003;63:3459–3463. [PubMed] [Google Scholar]
- 69.Balsitis S, et al. Examiniation of the pRB-dependent and pRb-independent functions of E7 in vivo. J. Virol. 2005;79:11392–11402. doi: 10.1128/JVI.79.17.11392-11402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wikenheiser-Brokamp KA. Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development. 2004;131:4299–4310. doi: 10.1242/dev.01232. [DOI] [PubMed] [Google Scholar]
- 71.Wikenheiser-Brokamp KA. Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell. Mol. Life Sci. 2006;63:767–780. doi: 10.1007/s00018-005-5487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaye FJ, Harbour JW. For whom the bell tolls: susceptibility to common adult cancers in retinoblastoma survivors. J. Natl Cancer Inst. 2004;96:342–343. doi: 10.1093/jnci/djh080. [DOI] [PubMed] [Google Scholar]
- 73.Kleinerman RA, et al. Risk of soft tissue sarcomas by individual subtype in survivors of hereditary retinoblastoma. J. Natl Cancer Inst. 2007;99:24–31. doi: 10.1093/jnci/djk002. [DOI] [PubMed] [Google Scholar]
- 74.Yamasaki L, et al. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1+/− mice. Nature Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 75.Gil J, Peters G. Regulation of the INK4b–ARF- INK4a tumour suppressor locus: all for one or one for all. Nature Rev. Mol. Cell. Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 76.Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- 77.Gladden AB, Diehl JA. Location, location, location: the role of cyclin D1 nuclear localization in cancer. J. Cell Biochem. 2005;96:906–913. doi: 10.1002/jcb.20613. [DOI] [PubMed] [Google Scholar]
- 78.Comstock CE, Revelo MP, Buncher CR, Knudsen KE. Impact of differential cyclin D1 expression and localisation in prostate cancer. Br. J. Cancer. 2007;96:970–979. doi: 10.1038/sj.bjc.6603615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peeper DS, et al. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997;386:177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- 80.Herrera RE, Makela TP, Weinberg RA. TGF beta-induced growth inhibition in primary fibroblasts requires the retinoblastoma protein. Mol. Biol. Cell. 1996;7:1335–1342. doi: 10.1091/mbc.7.9.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harrington EA, Bruce JL, Harlow E, Dyson N. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc. Natl Acad. Sci. USA. 1998;95:11945–11950. doi: 10.1073/pnas.95.20.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nahle Z, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nature Cell Biol. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 83.Almasan A, et al. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F–responsive genes, and apoptosis. Proc. Natl Acad. Sci. USA. 1995;92:5436–5440. doi: 10.1073/pnas.92.12.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen D, et al. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004;5:539–551. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 85.Maandag EC, et al. Developmental rescue of an embryonic-lethal mutation in the retinoblastoma gene in chimeric mice. EMBO J. 1994;13:4260–4268. doi: 10.1002/j.1460-2075.1994.tb06746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 87.Chau BN, Wang JY. Coordinated regulation of life and death by RB. Nature Rev. Cancer. 2003;3:130–138. doi: 10.1038/nrc993. [DOI] [PubMed] [Google Scholar]
- 88.Bosco EE, et al. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res. 2004;32:25–34. doi: 10.1093/nar/gkg919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qin XQ, Livingston DM, Kaelin WG, Jr, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl Acad. Sci. USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson DG, Degregori J. Putting the oncogenic and tumor suppressive activities of E2F into context. Curr. Mol. Med. 2006;6:731–738. doi: 10.2174/1566524010606070731. [DOI] [PubMed] [Google Scholar]
- 91.Chau BN, Pan CW, Wang JY. Separation of anti-proliferation and anti-apoptotic functions of retinoblastoma protein through targeted mutations of its A/B domain. PLoS ONE. 2006;1:e82. doi: 10.1371/journal.pone.0000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rogoff HA, Kowalik TF. Life, death and E2F: linking proliferation control and DNA damage signaling via E2F1. Cell Cycle. 2004;3:845–846. doi: 10.4161/cc.3.7.976. [DOI] [PubMed] [Google Scholar]
- 93.Sherr CJ, Weber JD. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 94.Ruiz S, Santos M, Paramio JM. Is the loss of pRb essential for the mouse skin carcinogenesis? Cell Cycle. 2006;5:625–629. doi: 10.4161/cc.5.6.2580. [DOI] [PubMed] [Google Scholar]
- 95.Tsai KY, et al. ARF mutation accelerates pituitary tumor development in Rb+/− mice. Proc. Natl Acad. Sci. USA. 2002;99:16865–16870. doi: 10.1073/pnas.262499599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsai KY, MacPherson D, Rubinson DA, Crowley D, Jacks T. ARF is not required for apoptosis in Rb mutant mouse embryos. Curr. Biol. 2002;12:159–163. doi: 10.1016/s0960-9822(01)00659-5. [DOI] [PubMed] [Google Scholar]
- 97.MacPherson D, et al. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18:1681–1694. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marino S, Hoogervoorst D, Brandner S, Berns A. Rb and p107 are required for normal cerebellar development and granule cell survival but not for Purkinje cell persistence. Development. 2003;130:3359–3368. doi: 10.1242/dev.00553. [DOI] [PubMed] [Google Scholar]
- 99.Braig M, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 100.Serrano M, Blasco MA. Putting the stress on senescence. Curr. Opin. Cell Biol. 2001;13:748–753. doi: 10.1016/s0955-0674(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 101.Wu CH, et al. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc. Natl Acad. Sci. USA. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 103.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 104.Bieche I, Lidereau R. Loss of heterozygosity at 13q14 correlates with RB1 gene underexpression in human breast cancer. Mol. Carcinog. 2000;29:151–158. [PubMed] [Google Scholar]
- 105.Oliveira AM, Ross JS, Fletcher JA. Tumor suppressor genes in breast cancer: the gatekeepers and the caretakers. Am. J. Clin. Pathol. 2005;124 Suppl:S16–S28. doi: 10.1309/5XW3L8LU445QWGQR. [DOI] [PubMed] [Google Scholar]
- 106.Bosco EE, Knudsen ES. RB in breast cancer: at the crossroads of tumorigenesis and treatment. Cell Cycle. 2007;6:667–671. doi: 10.4161/cc.6.6.3988. [DOI] [PubMed] [Google Scholar]
- 107.Ceccarelli C, et al. Retinoblastoma (RB1) gene product expression in breast carcinoma. Correlation with Ki-67 growth fraction and biopathological profile. J. Clin. Pathol. 1998;51:818–824. doi: 10.1136/jcp.51.11.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pietilainen T, et al. Expression of retinoblastoma gene protein (Rb) in breast cancer as related to established prognostic factors and survival. Eur. J. Cancer. 1995;31A:329–333. doi: 10.1016/0959-8049(94)00463-f. [DOI] [PubMed] [Google Scholar]
- 109.Mack PC, et al. Increased RB1 abnormalities in human primary prostate cancer following combined androgen blockade. Prostate. 1998;34:145–151. doi: 10.1002/(sici)1097-0045(19980201)34:2<145::aid-pros10>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 110.Phillips SM, et al. Loss of the retinoblastoma susceptibility gene (RB1) is a frequent and early event in prostatic tumorigenesis. Br. J. Cancer. 1994;70:1252–1257. doi: 10.1038/bjc.1994.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ittmann MM, Wieczorek R. Alterations of the retinoblastoma gene in clinically localized, stage B prostate adenocarcinomas. Hum. Pathol. 1996;27:28–34. doi: 10.1016/s0046-8177(96)90134-3. [DOI] [PubMed] [Google Scholar]
- 112.Sutherland RL, Musgrove EA. Cyclins and breast cancer. Mammary Gland Biol. Neoplasia. 2004;9:95–104. doi: 10.1023/B:JOMG.0000023591.45568.77. [DOI] [PubMed] [Google Scholar]
- 113.Watts CK, et al. Antiestrogen inhibition of cell cycle progression in breast cancer cells in associated with inhibition of cyclin-dependent kinase activity and decreased retinoblastoma protein phosphorylation. Mol. Endocrinol. 1995;9:1804–1813. doi: 10.1210/mend.9.12.8614416. [DOI] [PubMed] [Google Scholar]
- 114.Foster JS, Henley D, Bukovsky A, Seth P, Wimalasena J. Multifaceted regulation of cell cycle progression by estrogen: regulation of Cdk inhibitors and Cdc25A independent of cyclin D1-Cdk4 function. Mol. Cell. Biol. 2001;21:794–810. doi: 10.1128/MCB.21.3.794-810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Craig C, et al. Effects of adenovirus-mediated p16INK4A expression on cell cycle arrest are determined by endogenous p16 and Rb status in human cancer cells. Oncogene. 1998;16:265–272. doi: 10.1038/sj.onc.1201493. [DOI] [PubMed] [Google Scholar]
- 116.Watts CK, Sweeney KJ, Warlters A, Musgrove EA, Sutherland RL. Antiestrogen regulation of cell cycle progression and cyclin D1 gene expression in MCF-7 human breast cancer cells. Breast Cancer Res. Treat. 1994;31:95–105. doi: 10.1007/BF00689680. [DOI] [PubMed] [Google Scholar]
- 117.Hui R, et al. Constitutive overexpression of cyclin D1 but not cyclin E confers acute resistance to antiestrogens in T-47D breast cancer cells. Cancer Res. 2002;62:6916–6923. [PubMed] [Google Scholar]
- 118.Bosco EE, et al. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J. Clin. Invest. 2007;117:218–228. doi: 10.1172/JCI28803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Varma H, Conrad SE. Reversal of an antiestrogen-mediated cell cycle arrest of MCF-7 cells by viral tumor antigens requires the retinoblastoma protein-binding domain. Oncogene. 2000;19:4746–4753. doi: 10.1038/sj.onc.1203827. [DOI] [PubMed] [Google Scholar]
- 120.Linke SP, Bremer TM, Herold CD, Sauter G, Diamond C. A multimarker model to predict outcome in tamoxifen-treated breast cancer patients. Clin. Cancer Res. 2006;12:1175–1183. doi: 10.1158/1078-0432.CCR-05-1562. [DOI] [PubMed] [Google Scholar]
- 121.Michalides R, et al. Cyclin A is a prognostic indicator in early stage breast cancer with and without tamoxifen treatment. Br. J. Cancer. 2002;86:402–408. doi: 10.1038/sj.bjc.6600072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Han S, et al. Cyclin D1 expression and patient outcome after tamoxifen therapy in estrogen receptor positive metastatic breast cancer. Oncol. Rep. 2003;10:141–144. [PubMed] [Google Scholar]
- 123.Stendahl M, et al. Cyclin D1 overexpression is a negative predictive factor for tamoxifen response in postmenopausal breast cancer patients. Br. J. Cancer. 2004;90:1942–1948. doi: 10.1038/sj.bjc.6601831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rudas M, et al. Cyclin D1 expression in breast cancer patients receiving adjuvant tamoxifen-based therapy. Clin. Cancer Res. 2008;14:1767–1774. doi: 10.1158/1078-0432.CCR-07-4122. [DOI] [PubMed] [Google Scholar]
- 125.Barnes DM, Gillett CE. Cyclin D1 in breast cancer. Breast Cancer Res. Treat. 1998;52:1–15. doi: 10.1023/a:1006103831990. [DOI] [PubMed] [Google Scholar]
- 126.Gillett C, et al. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 1994;54:1812–1817. [PubMed] [Google Scholar]
- 127.Gillett C, et al. Cyclin D1 and prognosis in human breast cancer. Int. J. Cancer. 1996;69:92–99. doi: 10.1002/(SICI)1097-0215(19960422)69:2<92::AID-IJC4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 128.Hui R, et al. INK4a gene expression and methylation in primary breast cancer: overexpression of p16INK4a messenger RNA is a marker of poor prognosis. Clin. Cancer Res. 2000;6:2777–2787. [PubMed] [Google Scholar]
- 129.Dublin EA, et al. Retinoblastoma and p16 proteins in mammary carcinoma: their relationship to cyclin D1 and histopathological parameters. Int. J. Cancer. 1998;79:71–75. doi: 10.1002/(sici)1097-0215(19980220)79:1<71::aid-ijc14>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 130.Nielsen NH, Loden M, Cajander J, Emdin SO, Landberg G. G1-S transition defects occur in most breast cancers and predict outcome. Breast Cancer Res. Treat. 1999;56:105–112. doi: 10.1023/a:1006208419350. [DOI] [PubMed] [Google Scholar]
- 131.Kroger N, et al. Prognostic and predictive effects of immunohistochemical factors in high-risk primary breast cancer patients. Clin. Cancer Res. 2006;12:159–168. doi: 10.1158/1078-0432.CCR-05-1340. [DOI] [PubMed] [Google Scholar]
- 132.Derenzini M, et al. Loss of retinoblastoma tumor suppressor protein makes human breast cancer cells more sensitive to antimetabolite exposure. Clin. Cancer Res. 2008;14:2199–2209. doi: 10.1158/1078-0432.CCR-07-2065. [DOI] [PubMed] [Google Scholar]
- 133.Anderson JJ, et al. Retinoblastoma protein in human breast carcinoma: immunohistochemical study using a new monoclonal antibody effective on routinely processed tissues. J. Pathol. 1996;180:65–70. doi: 10.1002/(SICI)1096-9896(199609)180:1<65::AID-PATH607>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 134.Balk SP, Knudsen KE. AR, the cell cycle, and prostate cancer. Nucl. Recept. Signal. 2008;6:e001. doi: 10.1621/nrs.06001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sowery RD, So AI, Gleave ME. Therapeutic options in advanced prostate cancer: present and future. Curr. Urol. Rep. 2007;8:53–59. doi: 10.1007/s11934-007-0021-9. [DOI] [PubMed] [Google Scholar]
- 136.Knudsen KE, Arden KC, Cavenee WK. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. J. Biol. Chem. 1998;273:20213–20222. doi: 10.1074/jbc.273.32.20213. [DOI] [PubMed] [Google Scholar]
- 137.Fribourg AF, Knudsen KE, Strobeck MW, Lindhorst CM, Knudsen ES. Differential requirements for ras and the retinoblastoma tumor suppressor protein in the androgen dependence of prostatic adenocarcinoma cells. Cell Growth Differ. 2000;11:361–372. [PubMed] [Google Scholar]
- 138.Xu Y, Chen SY, Ross KN, Balk SP. Androgens induce prostate cancer cell proliferation through mammalian target of rapamycin activation and post-transcriptional increases in cyclin D proteins. Cancer Res. 2006;66:7783–7792. doi: 10.1158/0008-5472.CAN-05-4472. [DOI] [PubMed] [Google Scholar]
- 139.Knudsen KE, Cavenee WK, Arden KC. D-type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res. 1999;59:2297–2301. [PubMed] [Google Scholar]
- 140.Petre CE, Wetherill YB, Danielsen M, Knudsen KE. Cyclin D1: mechanism and consequence of androgen receptor co-repressor activity. J. Biol. Chem. 2002;277:2207–2215. doi: 10.1074/jbc.M106399200. [DOI] [PubMed] [Google Scholar]
- 141.Knudsen KE. The cyclin D1b splice variant: an old oncogene learns new tricks. Cell Div. 2006;1:15. doi: 10.1186/1747-1028-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reutens AT, et al. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol. Endocrinol. 2001;15:797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 143.Sharma A, et al. Retinoblastoma tumor suppressor status is a critical determinant of therapeutic response in prostate cancer cells. Cancer Res. 2007;67:6192–6203. doi: 10.1158/0008-5472.CAN-06-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jarrard DF, et al. Alterations in the p16/pRb cell cycle checkpoint occur commonly in primary and metastatic human prostate cancer. Cancer Lett. 2002;185:191–199. doi: 10.1016/s0304-3835(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 145.Gaddipati JP, et al. Mutations of the p16 gene product are rare in prostate cancer. Prostate. 1997;30:188–194. doi: 10.1002/(sici)1097-0045(19970215)30:3<188::aid-pros7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 146.Chakravarti A, et al. Prognostic value of p16 in locally advanced prostate cancer: a study based on Radiation Therapy Oncology Group Protocol 9202. J. Clin. Oncol. 2007;25:3082–3089. doi: 10.1200/JCO.2006.08.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kallakury BV, et al. The prognostic significance of p34cdc2 and cyclin D1 protein expression in prostate adenocarcinoma. Cancer. 1997;80:753–763. doi: 10.1002/(sici)1097-0142(19970815)80:4<753::aid-cncr15>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 148.Aaltomaa S, et al. Expression of Ki-67, cyclin D1 and apoptosis markers correlated with survival in prostate cancer patients treated by radical prostatectomy. Anticancer Res. 2006;26:4873–4878. [PubMed] [Google Scholar]
- 149.Drobnjak M, Osman I, Scher HI, Fazzari M, Cordon-Cardo C. Overexpression of cyclin D1 is associated with metastatic prostate cancer to bone. Clin. Cancer Res. 2000;6:1891–1895. [PubMed] [Google Scholar]
- 150.Gumbiner LM, et al. Overexpression of cyclin D1 is rare in human prostate carcinoma. Prostate. 1999;38:40–45. doi: 10.1002/(sici)1097-0045(19990101)38:1<40::aid-pros5>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 151.Zagorski WA, Knudsen ES, Reed MF. Retinoblastoma deficiency increases chemosensitivity in lung cancer. Cancer Res. 2007;67:8264–8273. doi: 10.1158/0008-5472.CAN-06-4753. [DOI] [PubMed] [Google Scholar]
- 152.Pollack A, et al. Retinoblastoma protein expression and radiation response in muscle-invasive bladder cancer. Int. J. Radiat. Oncol. Biol. Phys. 1997;39:687–695. doi: 10.1016/s0360-3016(97)00147-8. [DOI] [PubMed] [Google Scholar]
- 153.Agerbaek M, Alsner J, Marcussen N, Lundbeck F, von der Maase H. Retinoblastoma protein expression is an independent predictor of both radiation response and survival in muscle-invasive bladder cancer. Br. J. Cancer. 2003;89:298–304. doi: 10.1038/sj.bjc.6601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Simon GR, Turrisi A. Management of small cell lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 2007;132:324S–339S. doi: 10.1378/chest.07-1385. [DOI] [PubMed] [Google Scholar]
- 155.D’Abaco GM, Hooper S, Paterson H, Marshall CJ. Loss of Rb overrides the requirement for ERK activity for cell proliferation. J. Cell Sci. 2002;115:4607–4616. doi: 10.1242/jcs.00161. [DOI] [PubMed] [Google Scholar]
- 156.Mack PC, et al. RB status as a determinant of response to UCN-01 in non-small cell lung carcinoma. Clin. Cancer Res. 1999;5:2596–2604. [PubMed] [Google Scholar]
- 157.McGahren-Murray M, Terry NH, Keyomarsi K. The differential staurosporine-mediated G1 arrest in normal versus tumor cells is dependent on the retinoblastoma protein. Cancer Res. 2006;66:9744–9753. doi: 10.1158/0008-5472.CAN-06-1809. [DOI] [PubMed] [Google Scholar]
- 158.Chen X, Lowe M, Keyomarsi K. UCN-01-mediated G1 arrest in normal but not tumor breast cells is pRb-dependent and p53-independent. Oncogene. 1999;18:5691–5702. [PubMed] [Google Scholar]
- 159.Fry DW, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 160.Toogood PL, et al. Discovery of a potent and selective inhibitor of cyclin-dependent kinase 4/6. J. Med. Chem. 2005;48:2388–2406. doi: 10.1021/jm049354h. [DOI] [PubMed] [Google Scholar]
- 161.Gallie BL. Predictive testing for retinoblastoma comes of age. Am. J. Hum. Genet. 1997;61:279–281. doi: 10.1086/514861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liebisch P, Dohner H. Cytogenetics and molecular cytogenetics in multiple myeloma. Eur. J. Cancer. 2006;42:1520–1529. doi: 10.1016/j.ejca.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 163.Haddad RI, et al. A phase II clinical and pharmacodynamic study of E7070 in patients with metastatic, recurrent, or refractory squamous cell carcinoma of the head and neck: modulation of retinoblastoma protein phosphorylation by a novel chloroindolyl sulfonamide cell cycle inhibitor. Clin. Cancer Res. 2004;10:4680–4687. doi: 10.1158/1078-0432.CCR-04-0229. [DOI] [PubMed] [Google Scholar]
- 164.Tan AR, et al. Phase I trial of the cyclin-dependent kinase inhibitor flavopiridol in combination with docetaxel in patients with metastatic breast cancer. Clin. Cancer Res. 2004;10:5038–5047. doi: 10.1158/1078-0432.CCR-04-0025. [DOI] [PubMed] [Google Scholar]
- 165.Shan B, Chang CY, Jones D, Lee WH. The transcription factor E2F-1 mediates the autoregulation of RB gene expression. Mol. Cell. Biol. 1994;14:299–309. doi: 10.1128/mcb.14.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Montanaro L, et al. Controversial relationship between the expression of the RB pathway components and RB protein phosphorylation in human breast cancer. Histol. Histopathol. 2007;22:769–775. doi: 10.14670/HH-22.769. [DOI] [PubMed] [Google Scholar]
- 167.Derenzini M, et al. Relationship between the RB1 mRNA level and the expression of phosphorylated RB protein in human breast cancers: their relevance in cell proliferation activity and patient clinical outcome. Histol. Histopathol. 2007;22:505–513. doi: 10.14670/HH-22.505. [DOI] [PubMed] [Google Scholar]
- 168.Gauthier ML, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lukas J, et al. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 170.Medema RH, Herrera RE, Lam F, Weinberg RA. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc. Natl Acad. Sci. USA. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang Z, Rosen DG, Yao JL, Huang J, Liu J. Expression of p14ARF, p15INK4b, p16INK4a, and DCR2 increases during prostate cancer progression. Mod. Pathol. 2006;19:1339–1343. doi: 10.1038/modpathol.3800655. [DOI] [PubMed] [Google Scholar]
- 172.Lambert AP, Anschau F, Schmitt VM. p16INK4A expression in cervical premalignant and malignant lesions. Exp. Mol. Pathol. 2006;80:192–196. doi: 10.1016/j.yexmp.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 173.Andersson S, et al. Expression of E6/E7 mRNA from ‘high risk’ human papillomavirus in relation to CIN grade, viral load and p16INK4a. Int. J. Oncol. 2006;29:705–711. [PubMed] [Google Scholar]
- 174.Mayhew CN, et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976–984. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 175.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nature Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 176.Desmedt C, Sotiriou C. Proliferation: the most prominent predictor of clinical outcome in breast cancer. Cell Cycle. 2006;5:2198–2202. doi: 10.4161/cc.5.19.3254. [DOI] [PubMed] [Google Scholar]
- 177.Sotiriou C, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 178.Perreard L, et al. Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay. Breast Cancer Res. 2006;8:R23. doi: 10.1186/bcr1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Naderi A, et al. A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene. 2007;26:1507–1516. doi: 10.1038/sj.onc.1209920. [DOI] [PubMed] [Google Scholar]
- 180.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J. Clin. Oncol. 2008;26:721–728. doi: 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 181.Paik S. Development and clinical utility of a 21-gene recurrence score prognostic assay in patients with early breast cancer treated with tamoxifen. Oncologist. 2007;12:631–635. doi: 10.1634/theoncologist.12-6-631. [DOI] [PubMed] [Google Scholar]
- 182.Paik S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 183.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 184.Loi S, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J. Clin. Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 185.Ignatiadis M, Desmedt C. Predicting risk of breast cancer recurrence using gene-expression profiling. Pharmacogenomics. 2007;8:101–111. doi: 10.2217/14622416.8.1.101. [DOI] [PubMed] [Google Scholar]
- 186.Sotiriou C, Desmedt C. Gene expression profiling in breast cancer. Ann. Oncol. 2006;17 Suppl. 10:x259–x262. doi: 10.1093/annonc/mdl270. [DOI] [PubMed] [Google Scholar]
- 187.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 188.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nature Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 189.Chen YN, et al. Selective killing of transformed cells by cyclin/cyclin-dependent kinase 2 antagonists. Proc. Natl Acad. Sci. USA. 1999;96:4325–4329. doi: 10.1073/pnas.96.8.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Morris EJ, et al. Functional identification of Api5 as a suppressor of E2F–dependent apoptosis in vivo. PLoS Genet. 2006;2:e196. doi: 10.1371/journal.pgen.0020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Moon NS, Di Stefano L, Dyson NA. A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F–dependent apoptosis. Mol. Cell. Biol. 2006;26:7601–7615. doi: 10.1128/MCB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Laurie NA, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 193.Laurie NA, Schin-Shih C, Dyer MA. Targeting MDM2 and MDMX in retinoblastoma. Curr. Cancer Drug Targets. 2007;7:689–695. doi: 10.2174/156800907782418266. [DOI] [PubMed] [Google Scholar]
- 194.Elison JR, Cobrinik D, Claros N, Abramson DH, Lee TC. Small molecule inhibition of HDM2 leads to p53-mediated cell death in retinoblastoma cells. Arch. Ophthalmol. 2006;124:1269–1275. doi: 10.1001/archopht.124.9.1269. [DOI] [PubMed] [Google Scholar]
- 195.Kitagawa M, Aonuma M, Lee SH, Fukutake S, McCormick F. E2F-1 transcriptional activity is a critical determinant of Mdm2 antagonist-induced apoptosis in human tumor cell lines. Oncogene. 2008 Jun 2; doi: 10.1038/onc.2008.164. (doi:10.1038/onc.2008.164) [DOI] [PubMed] [Google Scholar]
- 196.Salgia R, Skarin AT. Molecular abnormalities in lung cancer. J. Clin. Oncol. 1998;16:1207–1217. doi: 10.1200/JCO.1998.16.3.1207. [DOI] [PubMed] [Google Scholar]
- 197.Kelley MJ, et al. Differential inactivation of CDKN2 and Rb protein in non-small-cell and small-cell lung cancer cell lines. J. Natl Cancer Inst. 1995;87:756–761. doi: 10.1093/jnci/87.10.756. [DOI] [PubMed] [Google Scholar]
- 198.Kratzke RA, et al. Rb and p16INK4a expression in resected non-small cell lung tumors. Cancer Res. 1996;56:3415–3420. [PubMed] [Google Scholar]
- 199.Wang J, et al. Value of p16INK4a and RASSF1A promoter hypermethylation in prognosis of patients with resectable non-small cell lung cancer. Clin. Cancer Res. 2004;10:6119–6125. doi: 10.1158/1078-0432.CCR-04-0652. [DOI] [PubMed] [Google Scholar]
- 200.Niklinski J, Niklinska W, Laudanski J, Chyczewska E, Chyczewski L. Prognostic molecular markers in non-small cell lung cancer. Lung Cancer. 2001;34 Suppl. 2:S53–S58. doi: 10.1016/s0169-5002(01)00345-2. [DOI] [PubMed] [Google Scholar]
- 201.Au NH, et al. Evaluation of immunohistochemical markers in non-small cell lung cancer by unsupervised hierarchical clustering analysis: a tissue microarray study of 284 cases and 18 markers. J. Pathol. 2004;204:101–109. doi: 10.1002/path.1612. [DOI] [PubMed] [Google Scholar]
- 202.Nguyen VN, Mirejovsky N, Mirejovsky T, Melinova L, Mandys V. Expression of cyclin D1, Ki-67 and PCNA in non-small cell lung cancer: prognostic significance and comparison with p53 and bcl-2. Acta Histochem. 2000;102:323–338. doi: 10.1078/s0065-1281(04)70039-2. [DOI] [PubMed] [Google Scholar]
- 203.McKay JA, et al. Cyclin D1 protein expression and gene polymorphism in colorectal cancer. Aberdeen Colorectal Initiative. Int. J. Cancer. 2000;88:77–81. doi: 10.1002/1097-0215(20001001)88:1<77::aid-ijc12>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 204.Sgambato A, et al. Cyclin D1 expression in papillary superficial bladder cancer: its association with other cell cycle-associated proteins, cell proliferation and clinical outcome. Int. J. Cancer. 2002;97:671–678. doi: 10.1002/ijc.10055. [DOI] [PubMed] [Google Scholar]
- 205.Tut VM, et al. Cyclin D1 expression in transitional cell carcinoma of the bladder: correlation with p53, waf1, pRb and Ki67. Br. J. Cancer. 2001;84:270–275. doi: 10.1054/bjoc.2000.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mitra AP, Datar RH, Cote RJ. Molecular pathways in invasive bladder cancer: new insights into mechanisms, progression, and target identification. J. Clin. Oncol. 2006;24:5552–5564. doi: 10.1200/JCO.2006.08.2073. [DOI] [PubMed] [Google Scholar]
- 207.Cote RJ, et al. Elevated and absent pRb expression is associated with bladder cancer progression and has cooperative effects with p53. Cancer Res. 1998;58:1090–1094. [PubMed] [Google Scholar]
- 208.Chatterjee SJ, et al. Combined effects of p53, p21, and pRb expression in the progression of bladder transitional cell carcinoma. J. Clin. Oncol. 2004;22:1007–1013. doi: 10.1200/JCO.2004.05.174. [DOI] [PubMed] [Google Scholar]
- 209.Brooks JD, Bova GS, Isaacs WB. Allelic loss of the retinoblastoma gene in primary human prostatic adenocarcinomas. Prostate. 1995;26:35–39. doi: 10.1002/pros.2990260108. [DOI] [PubMed] [Google Scholar]
- 210.Cooney KA, et al. Distinct regions of allelic loss on 13q in prostate cancer. Cancer Res. 1996;56:1142–1145. [PubMed] [Google Scholar]
- 211.Kaltz-Wittmer C, et al. FISH analysis of gene aberrations (MYC, CCND1, ERBB2, RB, and AR) in advanced prostatic carcinomas before and after androgen deprivation therapy. Lab. Invest. 2000;80:1455–1464. doi: 10.1038/labinvest.3780152. [DOI] [PubMed] [Google Scholar]
- 212.Li C, et al. Identification of two distinct deleted regions on chromosome 13 in prostate cancer. Oncogene. 1998;16:481–487. doi: 10.1038/sj.onc.1201554. [DOI] [PubMed] [Google Scholar]
- 213.Melamed J, Einhorn JM, Ittmann MM. Allelic loss on chromosome 13q in human prostate carcinoma. Clin. Cancer Res. 1997;3:1867–1872. [PubMed] [Google Scholar]
- 214.Vuhahula E, Straume O, Akslen LA. Frequent loss of p16 protein expression and high proliferative activity (Ki-67) in malignant melanoma from black Africans. Anticancer Res. 2000;20:4857–4862. [PubMed] [Google Scholar]
- 215.Straume O, Sviland L, Akslen LA. Loss of nuclear p16 protein expression correlates with increased tumor cell proliferation (Ki-67) and poor prognosis in patients with vertical growth phase melanoma. Clin. Cancer Res. 2000;6:1845–1853. [PubMed] [Google Scholar]
- 216.Straume O, Akslen LA. Alterations and prognostic significance of p16 and p53 protein expression in subgroups of cutaneous melanoma. Int. J. Cancer. 1997;74:535–539. doi: 10.1002/(sici)1097-0215(19971021)74:5<535::aid-ijc10>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 217.Alonso SR, et al. Progression in cutaneous malignant melanoma is associated with distinct expression profiles: a tissue microarray-based study. Am. J. Pathol. 2004;164:193–203. doi: 10.1016/s0002-9440(10)63110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bachmann IM, Straume O, Akslen LA. Altered expression of cell cycle regulators Cyclin D1, p14, p16, CDK4 and Rb in nodular melanomas. Int. J. Oncol. 2004;25:1559–1565. [PubMed] [Google Scholar]
- 219.Korabiowska M, et al. Downregulation of the retinoblastoma gene expression in the progression of malignant melanoma. Pathobiology. 2001;69:274–280. doi: 10.1159/000064338. [DOI] [PubMed] [Google Scholar]
- 220.Garcea G, Neal CP, Pattenden CJ, Steward WP, Berry DP. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur. J. Cancer. 2005;41:2213–2236. doi: 10.1016/j.ejca.2005.04.044. [DOI] [PubMed] [Google Scholar]
- 221.Gerdes B, et al. p16INK4a is a prognostic marker in resected ductal pancreatic cancer: an analysis of p16INK4a, p53, MDM2, an Rb. Ann. Surg. 2002;235:51–59. doi: 10.1097/00000658-200201000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hu YX, et al. Frequent loss of p16 expression and its correlation with clinicopathological parameters in pancreatic carcinoma. Clin. Cancer Res. 1997;3:1473–1477. [PubMed] [Google Scholar]
- 223.Kawesha A, et al. K-ras oncogene subtype mutations are associated with survival but not expression of p53, p16INK4A, p21WAF-1, cyclin D1, erbB-2 and erbB-3 in resected pancreatic ductal adenocarcinoma. Int. J. Cancer. 2000;89:469–474. doi: 10.1002/1097-0215(20001120)89:6<469::aid-ijc1>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 224.Gansauge F, Gansauge S, Schmidt E, Muller J, Beger HG. Prognostic significance of molecular alterations in human pancreatic carcinoma—an immunohistological study. Langenbecks Arch. Surg. 1998;383:152–155. doi: 10.1007/pl00008076. [DOI] [PubMed] [Google Scholar]
- 225.Gansauge S, et al. Overexpression of cyclin D1 in human pancreatic carcinoma is associated with poor prognosis. Cancer Res. 1997;57:1634–1637. [PubMed] [Google Scholar]
- 226.McKay JA, et al. Analysis of key cell-cycle checkpoint proteins in colorectal tumours. J. Pathol. 2002;196:386–393. doi: 10.1002/path.1053. [DOI] [PubMed] [Google Scholar]
- 227.Poller DN, Baxter KJ, Shepherd NA. p53 and Rb1 protein expression: are they prognostically useful in colorectal cancer? Br. J. Cancer. 1997;75:87–93. doi: 10.1038/bjc.1997.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Esteller M, et al. K-ras and p16 aberrations confer poor prognosis in human colorectal cancer. J. Clin. Oncol. 2001;19:299–304. doi: 10.1200/JCO.2001.19.2.299. [DOI] [PubMed] [Google Scholar]
- 229.Azechi H, et al. Disruption of the p16/cyclin D1/ retinoblastoma protein pathway in the majority of human hepatocellular carcinomas. Oncology. 2001;60:346–354. doi: 10.1159/000058531. [DOI] [PubMed] [Google Scholar]
- 230.Lee TJ, et al. Abrogation of the p16-Rb pathway in Korean hepatocellular carcinomas. Hepatogastroenterology. 2000;47:1663–1668. [PubMed] [Google Scholar]
- 231.Zucman-Rossi J, Laurent-Puig P. Genetic diversity of hepatocellular carcinomas and its potential impact on targeted therapies. Pharmacogenomics. 2007;8:997–1003. doi: 10.2217/14622416.8.8.997. [DOI] [PubMed] [Google Scholar]
- 232.Laurent-Puig P, Zucman-Rossi J. Genetics of hepatocellular tumors. Oncogene. 2006;25:3778–3786. doi: 10.1038/sj.onc.1209547. [DOI] [PubMed] [Google Scholar]
- 233.Anzola M, Cuevas N, Lopez-Martinez M, Martinez de, Pancorbo M, Burgos JJ. p16INK4A gene alterations are not a prognostic indicator for survival in patients with hepatocellular carcinoma undergoing curative hepatectomy. J. Gastroenterol. Hepatol. 2004;19:397–405. doi: 10.1111/j.1440-1746.2003.03305.x. [DOI] [PubMed] [Google Scholar]
- 234.Fujimoto Y, et al. Alterations of tumor suppressor genes and allelic losses in human hepatocellular carcinomas in China. Cancer Res. 1994;54:281–285. [PubMed] [Google Scholar]
- 235.Lee JS, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nature Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 236.Thorgeirsson SS, Lee JS, Grisham JW. Molecular prognostication of liver cancer: end of the beginning. J. Hepatol. 2006;44:798–805. doi: 10.1016/j.jhep.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 237.Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004;127:S51–S55. doi: 10.1053/j.gastro.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 238.Angus SP, et al. Retinoblastoma tumor suppressor targets dNTP metabolism to regulate DNA replication. J. Biol. Chem. 2002;277:44376–44384. doi: 10.1074/jbc.M205911200. [DOI] [PubMed] [Google Scholar]
- 239.Cole SL, Tevethia MJ. Simian virus 40 large T antigen and two independent T-antigen segments sensitize cells to apoptosis following genotoxic damage. J. Virol. 2002;76:8420–8432. doi: 10.1128/JVI.76.16.8420-8432.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Balsitis SJ, et al. Recapitulation of the effects of the human papillomavirus type 16 E7 oncogene on mouse epithelium by somatic Rb deletion and detection of pRb-independent effects of E7 in vivo. Mol. Cell. Biol. 2003;23:9094–9103. doi: 10.1128/MCB.23.24.9094-9103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]