Abstract
In the title compound, [Cu(C2O4)(C5H8N2)2(H2O)], the CuII atom is coordinated in a slightly distorted square-pyramidal geometry by two N atoms belonging to the two 3,5-dimethyl-1H-pyrazole ligands, two O atoms of the oxalate anion providing an O,O′-chelating coordination mode, and an O atom of the water molecule occupying the apical position. The crystal packing shows a well defined layer structure. Intra-layer connections are realised through a system of hydrogen bonds while the nature of the inter-layer interactions is completely hydrophobic, including no hydrogen-bonding interactions.
Related literature
For related literature on metal oxalates and 1H-pyrazole complexes, see: Abdeljalil et al. (2006 ▶); Bataille & Louër (1999 ▶); Castillo et al. (2001 ▶); Naumov et al. (1995 ▶); Raptis et al. (1999 ▶); Strotmeyer et al. (2003 ▶); Tomyn et al. (2007 ▶); Warda (1998 ▶).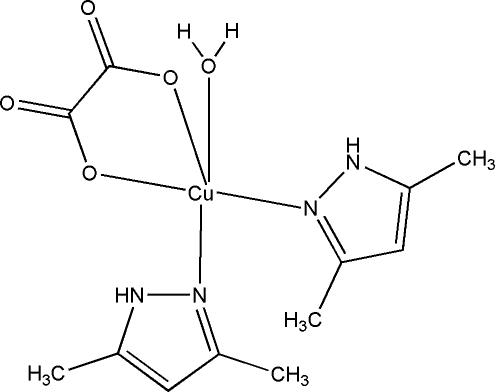
Experimental
Crystal data
[Cu(C2O4)(C5H8N2)2(H2O)]
M r = 361.84
Triclinic,

a = 8.2597 (6) Å
b = 8.4010 (8) Å
c = 12.2288 (11) Å
α = 77.007 (4)°
β = 89.189 (6)°
γ = 62.436 (5)°
V = 729.00 (11) Å3
Z = 2
Mo Kα radiation
μ = 1.53 mm−1
T = 120 (2) K
0.23 × 0.13 × 0.08 mm
Data collection
Nonius KappaCCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2003 ▶) T min = 0.720, T max = 0.888
11529 measured reflections
3765 independent reflections
3186 reflections with I > 2σ(I)
R int = 0.069
Refinement
R[F 2 > 2σ(F 2)] = 0.070
wR(F 2) = 0.175
S = 1.12
3765 reflections
204 parameters
H-atom parameters constrained
Δρmax = 0.80 e Å−3
Δρmin = −1.02 e Å−3
Data collection: COLLECT (Nonius, 1998 ▶); cell refinement: DENZO (Otwinowski & Minor, 1997 ▶); data reduction: DENZO; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536807058928/hy2100sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807058928/hy2100Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected geometric parameters (Å, °).
| Cu1—O2 | 1.946 (2) |
| Cu1—O5 | 1.971 (2) |
| Cu1—N2 | 1.975 (3) |
| Cu1—N3 | 2.002 (2) |
| Cu1—O1 | 2.283 (2) |
| O2—Cu1—O5 | 84.54 (9) |
| O2—Cu1—N2 | 172.35 (10) |
| O5—Cu1—N2 | 92.59 (9) |
| O2—Cu1—N3 | 88.41 (10) |
| O5—Cu1—N3 | 170.11 (10) |
| N2—Cu1—N3 | 93.53 (10) |
| O2—Cu1—O1 | 89.05 (9) |
| O5—Cu1—O1 | 91.85 (9) |
| N2—Cu1—O1 | 98.15 (10) |
| N3—Cu1—O1 | 94.96 (9) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O3i | 0.99 | 1.88 | 2.798 (3) | 153 |
| O1—H2⋯O5ii | 0.97 | 1.97 | 2.923 (3) | 168 |
| N1—H3⋯O3iii | 0.88 | 2.03 | 2.857 (3) | 156 |
| N4—H18⋯O3i | 0.88 | 1.97 | 2.845 (3) | 175 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
The authors thank NATO for financial support (grant CBP. NUKR. CLG 982019). AIB thanks the DAAD for a scholarship under the Leonhard-Euler-Stipendium Programme.
supplementary crystallographic information
Comment
1H-Pyrazole and its 3,5-substituted derivatives have been widely used as bridging ligands in molecular magnetism and supramolecular chemistry for obtaining discrete oligonuclear complexes of high nuclearity and coordination polymers (Abdeljalil et al., 2006; Raptis et al., 1999; Warda, 1998). On the other hand, oxalate is an important polynucleative ligand as it can exhibit various bridging modes, which, together with the varied coordination preferences of metal ions, can result in the formation of oligonuclear species or compounds containing one-, two- and three-dimensional coordination polymers and frameworks (Castillo et al., 2001; Naumov et al., 1995; Bataille & Louër, 1999; Strotmeyer et al., 2003; Tomyn et al., 2007). Simultanetous use of 1H-pyrazole derivatives and oxalates can result in the creation of new molecular topologies or in obtaining mononuclear complexes with vacant donor atoms, which can be used as building blocks for the preparation of oligonuclear assemblies or coordination polymers.
The molecular structure of the title compound (Fig. 1) consists of a CuII ion as the central atom possessing a slightly distorted square-pyramidal geometry. The four equatorial positions are occupied by two N atoms belonging to the two monodentately coordinated 3,5-dimethyl-1H-pyrazole molecules and two O atoms of the oxalate anion coordinated in an O,O'-chelate mode forming a five-membered chelate ring. The axial position is occupied by the O atom of the water molecule (Table 1). A crystal packing diagram (Fig. 2) depicts a well defined layer structure along the c-axis direction. Each layer is formed with the help of O1—H···O and N—H···O hydrogen bonds (Table 2) while the nature of inter-layer interactions is utterly hydrophobic including no hydrogen bonding interactions.
Experimental
Cu(NO3)2.3H2O (0.242 g, 1 mmol) and 3,5-dimethyl-1H-pyrazole (0.961 g, 1 mmol) were dissolved in water (10 ml), and then a powder of K2C2O4.H2O (0.184 g, 1 mmol) was added to the obtained solution. The resulting mixture was stirred at 358 K for 25 min and filtered. Blue needle-like crystals suitable for X-ray analysis were formed from the filtrate in several minutes. They were filtered off and washed with diethyl ester (yield 67%). Analysis calculated for C12H18CuN4O5: C 39.83, H 5.01, N 15.48%; found: C 39.11, H 5.13, N 15.42%.
Refinement
H atoms on the ligand were positioned geometrically and refined as riding atoms, with C—H = 0.95Å (CH), 0.98Å (CH3), N—H = 0.88Å and with Uiso(H) = 1.5Ueq(C) for methyl groups and Uiso(H) = 1.2Ueq(C, N) for the others. H atoms of the water molecule were located from a difference Fourier map and fixed with Uiso(H) = 1.5Ueq(O). The structure was refined as twinned. BASF parameter was refined to 0.307.
Figures
Fig. 1.
The molecular structure of the title compound. Displacement ellipsoids are shown at the 30% probability level.
Fig. 2.
A packing diagram for the title compound, showing the layers along the c-axis direction. Hydrogen bonds are indicated by dashed lines. H atoms not included in hydrogen bonds are omitted for clarity.
Crystal data
| [Cu(C2O4)(C5H8N2)2(H2O)] | Z = 2 |
| Mr = 361.84 | F000 = 374 |
| Triclinic, P1 | Dx = 1.648 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71073 Å |
| a = 8.2597 (6) Å | Cell parameters from 33067 reflections |
| b = 8.4010 (8) Å | θ = 1.0–27.5º |
| c = 12.2288 (11) Å | µ = 1.53 mm−1 |
| α = 77.007 (4)º | T = 120 (2) K |
| β = 89.189 (6)º | Plate, blue |
| γ = 62.436 (5)º | 0.23 × 0.13 × 0.08 mm |
| V = 729.00 (11) Å3 |
Data collection
| Nonius Kappa CCD diffractometer | 3765 independent reflections |
| Radiation source: fine-focus sealed tube | 3186 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.069 |
| Detector resolution: 9 pixels mm-1 | θmax = 28.7º |
| T = 120(2) K | θmin = 2.8º |
| φ and ω scans | h = −11→11 |
| Absorption correction: multi-scan(SADABS; Sheldrick, 2003) | k = −11→11 |
| Tmin = 0.720, Tmax = 0.888 | l = −16→16 |
| 11529 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.070 | H-atom parameters constrained |
| wR(F2) = 0.175 | w = 1/[σ2(Fo2) + 4.1563P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.12 | (Δ/σ)max = 0.001 |
| 3765 reflections | Δρmax = 0.80 e Å−3 |
| 204 parameters | Δρmin = −1.02 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cu1 | 0.14007 (5) | 0.12441 (5) | 0.63070 (3) | 0.01550 (12) | |
| O5 | −0.0055 (3) | 0.3021 (3) | 0.48971 (18) | 0.0169 (5) | |
| O2 | 0.3431 (3) | 0.1554 (3) | 0.56547 (17) | 0.0177 (5) | |
| O4 | 0.0332 (3) | 0.4796 (3) | 0.33510 (18) | 0.0224 (5) | |
| O3 | 0.3977 (3) | 0.3252 (3) | 0.41587 (18) | 0.0185 (5) | |
| O1 | 0.2189 (3) | −0.1213 (3) | 0.55296 (19) | 0.0228 (5) | |
| H1 | 0.3490 | −0.1861 | 0.5383 | 0.034* | |
| H2 | 0.1570 | −0.1934 | 0.5482 | 0.034* | |
| N2 | −0.0797 (3) | 0.1276 (3) | 0.6984 (2) | 0.0158 (5) | |
| N1 | −0.2437 (3) | 0.2848 (4) | 0.6778 (2) | 0.0180 (6) | |
| H3 | −0.2596 | 0.3944 | 0.6406 | 0.022* | |
| N3 | 0.3058 (3) | −0.0261 (4) | 0.7738 (2) | 0.0160 (5) | |
| N4 | 0.4574 (3) | −0.1886 (3) | 0.7756 (2) | 0.0163 (5) | |
| H18 | 0.4959 | −0.2313 | 0.7156 | 0.020* | |
| C2 | −0.3788 (4) | 0.2518 (4) | 0.7215 (3) | 0.0171 (6) | |
| C1 | −0.5724 (4) | 0.4006 (5) | 0.7112 (3) | 0.0237 (7) | |
| H4 | −0.5811 | 0.4835 | 0.7587 | 0.036* | |
| H6 | −0.6521 | 0.3449 | 0.7358 | 0.036* | |
| H5 | −0.6112 | 0.4715 | 0.6324 | 0.036* | |
| C3 | −0.2992 (4) | 0.0638 (4) | 0.7711 (3) | 0.0185 (7) | |
| H7 | −0.3591 | −0.0028 | 0.8086 | 0.022* | |
| C4 | −0.1140 (4) | −0.0081 (4) | 0.7551 (2) | 0.0146 (6) | |
| C5 | 0.0343 (4) | −0.2053 (4) | 0.7949 (3) | 0.0202 (7) | |
| H10 | 0.1196 | −0.2364 | 0.7372 | 0.030* | |
| H8 | −0.0210 | −0.2881 | 0.8083 | 0.030* | |
| H9 | 0.1011 | −0.2201 | 0.8652 | 0.030* | |
| C9 | 0.5419 (4) | −0.2766 (4) | 0.8815 (3) | 0.0183 (7) | |
| C10 | 0.7136 (4) | −0.4587 (4) | 0.9037 (3) | 0.0229 (7) | |
| H15 | 0.6822 | −0.5597 | 0.9263 | 0.034* | |
| H16 | 0.7949 | −0.4659 | 0.9642 | 0.034* | |
| H17 | 0.7761 | −0.4698 | 0.8349 | 0.034* | |
| C8 | 0.4410 (4) | −0.1676 (4) | 0.9506 (3) | 0.0189 (7) | |
| H14 | 0.4654 | −0.1924 | 1.0300 | 0.023* | |
| C7 | 0.2938 (4) | −0.0112 (4) | 0.8806 (3) | 0.0172 (6) | |
| C6 | 0.1419 (4) | 0.1515 (5) | 0.9110 (3) | 0.0243 (7) | |
| H12 | 0.0974 | 0.2589 | 0.8460 | 0.036* | |
| H13 | 0.1869 | 0.1792 | 0.9743 | 0.036* | |
| H11 | 0.0414 | 0.1238 | 0.9329 | 0.036* | |
| C11 | 0.2947 (4) | 0.2729 (4) | 0.4716 (2) | 0.0149 (6) | |
| C12 | 0.0885 (4) | 0.3626 (4) | 0.4252 (3) | 0.0176 (7) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.01345 (19) | 0.0151 (2) | 0.0170 (2) | −0.00671 (15) | 0.00082 (14) | −0.00214 (15) |
| O5 | 0.0149 (10) | 0.0147 (11) | 0.0191 (11) | −0.0066 (9) | 0.0009 (8) | −0.0013 (9) |
| O2 | 0.0150 (11) | 0.0167 (12) | 0.0189 (11) | −0.0065 (9) | 0.0008 (9) | −0.0018 (9) |
| O4 | 0.0206 (11) | 0.0199 (12) | 0.0237 (12) | −0.0091 (10) | −0.0018 (9) | −0.0005 (10) |
| O3 | 0.0181 (11) | 0.0157 (12) | 0.0221 (11) | −0.0089 (9) | 0.0044 (9) | −0.0036 (9) |
| O1 | 0.0174 (11) | 0.0246 (13) | 0.0310 (13) | −0.0107 (10) | 0.0065 (9) | −0.0138 (10) |
| N2 | 0.0137 (12) | 0.0110 (13) | 0.0179 (13) | −0.0032 (10) | −0.0006 (10) | −0.0005 (10) |
| N1 | 0.0147 (13) | 0.0114 (13) | 0.0245 (14) | −0.0039 (11) | −0.0004 (11) | −0.0033 (11) |
| N3 | 0.0174 (13) | 0.0135 (13) | 0.0179 (13) | −0.0075 (11) | 0.0032 (10) | −0.0049 (11) |
| N4 | 0.0144 (12) | 0.0152 (13) | 0.0163 (13) | −0.0040 (11) | 0.0003 (10) | −0.0048 (11) |
| C2 | 0.0146 (15) | 0.0147 (16) | 0.0221 (16) | −0.0056 (12) | 0.0029 (12) | −0.0080 (13) |
| C1 | 0.0154 (15) | 0.0176 (17) | 0.0361 (19) | −0.0064 (13) | 0.0028 (14) | −0.0063 (15) |
| C3 | 0.0213 (16) | 0.0181 (16) | 0.0218 (16) | −0.0137 (14) | 0.0056 (13) | −0.0054 (13) |
| C4 | 0.0185 (15) | 0.0122 (15) | 0.0137 (14) | −0.0072 (13) | 0.0013 (12) | −0.0040 (12) |
| C5 | 0.0207 (16) | 0.0095 (15) | 0.0251 (17) | −0.0030 (13) | 0.0013 (13) | −0.0037 (13) |
| C9 | 0.0188 (15) | 0.0168 (16) | 0.0219 (16) | −0.0110 (13) | 0.0035 (13) | −0.0040 (13) |
| C10 | 0.0208 (16) | 0.0145 (16) | 0.0264 (17) | −0.0030 (13) | −0.0009 (13) | −0.0036 (14) |
| C8 | 0.0175 (16) | 0.0168 (16) | 0.0169 (15) | −0.0047 (13) | −0.0010 (12) | −0.0013 (13) |
| C7 | 0.0188 (15) | 0.0166 (16) | 0.0182 (16) | −0.0097 (13) | 0.0026 (12) | −0.0051 (13) |
| C6 | 0.0222 (17) | 0.0233 (18) | 0.0237 (17) | −0.0061 (14) | 0.0024 (14) | −0.0094 (14) |
| C11 | 0.0139 (14) | 0.0128 (15) | 0.0203 (15) | −0.0062 (12) | 0.0036 (12) | −0.0087 (13) |
| C12 | 0.0171 (15) | 0.0197 (17) | 0.0175 (16) | −0.0092 (13) | 0.0025 (12) | −0.0066 (13) |
Geometric parameters (Å, °)
| Cu1—O2 | 1.946 (2) | C1—H4 | 0.9800 |
| Cu1—O5 | 1.971 (2) | C1—H6 | 0.9800 |
| Cu1—N2 | 1.975 (3) | C1—H5 | 0.9800 |
| Cu1—N3 | 2.002 (2) | C3—C4 | 1.390 (4) |
| Cu1—O1 | 2.283 (2) | C3—H7 | 0.9500 |
| O5—C12 | 1.285 (4) | C4—C5 | 1.503 (4) |
| O2—C11 | 1.261 (4) | C5—H10 | 0.9800 |
| O4—C12 | 1.225 (4) | C5—H8 | 0.9800 |
| O3—C11 | 1.256 (4) | C5—H9 | 0.9800 |
| O1—H1 | 0.9902 | C9—C8 | 1.367 (5) |
| O1—H2 | 0.9664 | C9—C10 | 1.497 (4) |
| N2—C4 | 1.341 (4) | C10—H15 | 0.9800 |
| N2—N1 | 1.359 (3) | C10—H16 | 0.9800 |
| N1—C2 | 1.345 (4) | C10—H17 | 0.9800 |
| N1—H3 | 0.8800 | C8—C7 | 1.408 (4) |
| N3—C7 | 1.337 (4) | C8—H14 | 0.9500 |
| N3—N4 | 1.355 (3) | C7—C6 | 1.487 (4) |
| N4—C9 | 1.352 (4) | C6—H12 | 0.9800 |
| N4—H18 | 0.8800 | C6—H13 | 0.9800 |
| C2—C3 | 1.383 (4) | C6—H11 | 0.9800 |
| C2—C1 | 1.490 (4) | C11—C12 | 1.561 (4) |
| O2—Cu1—O5 | 84.54 (9) | C4—C3—H7 | 126.9 |
| O2—Cu1—N2 | 172.35 (10) | N2—C4—C3 | 110.0 (3) |
| O5—Cu1—N2 | 92.59 (9) | N2—C4—C5 | 122.3 (3) |
| O2—Cu1—N3 | 88.41 (10) | C3—C4—C5 | 127.7 (3) |
| O5—Cu1—N3 | 170.11 (10) | C4—C5—H10 | 109.5 |
| N2—Cu1—N3 | 93.53 (10) | C4—C5—H8 | 109.5 |
| O2—Cu1—O1 | 89.05 (9) | H10—C5—H8 | 109.5 |
| O5—Cu1—O1 | 91.85 (9) | C4—C5—H9 | 109.5 |
| N2—Cu1—O1 | 98.15 (10) | H10—C5—H9 | 109.5 |
| N3—Cu1—O1 | 94.96 (9) | H8—C5—H9 | 109.5 |
| C12—O5—Cu1 | 112.55 (18) | N4—C9—C8 | 106.8 (3) |
| C11—O2—Cu1 | 112.86 (18) | N4—C9—C10 | 120.6 (3) |
| Cu1—O1—H1 | 115.9 | C8—C9—C10 | 132.6 (3) |
| Cu1—O1—H2 | 130.8 | C9—C10—H15 | 109.5 |
| H1—O1—H2 | 111.5 | C9—C10—H16 | 109.5 |
| C4—N2—N1 | 105.8 (2) | H15—C10—H16 | 109.5 |
| C4—N2—Cu1 | 132.3 (2) | C9—C10—H17 | 109.5 |
| N1—N2—Cu1 | 121.3 (2) | H15—C10—H17 | 109.5 |
| C2—N1—N2 | 111.5 (3) | H16—C10—H17 | 109.5 |
| C2—N1—H3 | 124.2 | C9—C8—C7 | 106.3 (3) |
| N2—N1—H3 | 124.2 | C9—C8—H14 | 126.9 |
| C7—N3—N4 | 106.3 (2) | C7—C8—H14 | 126.9 |
| C7—N3—Cu1 | 133.1 (2) | N3—C7—C8 | 109.3 (3) |
| N4—N3—Cu1 | 120.31 (19) | N3—C7—C6 | 121.3 (3) |
| C9—N4—N3 | 111.3 (3) | C8—C7—C6 | 129.3 (3) |
| C9—N4—H18 | 124.4 | C7—C6—H12 | 109.5 |
| N3—N4—H18 | 124.4 | C7—C6—H13 | 109.5 |
| N1—C2—C3 | 106.5 (3) | H12—C6—H13 | 109.5 |
| N1—C2—C1 | 122.5 (3) | C7—C6—H11 | 109.5 |
| C3—C2—C1 | 131.0 (3) | H12—C6—H11 | 109.5 |
| C2—C1—H4 | 109.5 | H13—C6—H11 | 109.5 |
| C2—C1—H6 | 109.5 | O3—C11—O2 | 125.0 (3) |
| H4—C1—H6 | 109.5 | O3—C11—C12 | 118.7 (3) |
| C2—C1—H5 | 109.5 | O2—C11—C12 | 116.2 (3) |
| H4—C1—H5 | 109.5 | O4—C12—O5 | 127.2 (3) |
| H6—C1—H5 | 109.5 | O4—C12—C11 | 119.0 (3) |
| C2—C3—C4 | 106.2 (3) | O5—C12—C11 | 113.7 (3) |
| C2—C3—H7 | 126.9 | ||
| O2—Cu1—O5—C12 | −3.4 (2) | C1—C2—C3—C4 | 179.7 (3) |
| N2—Cu1—O5—C12 | 169.5 (2) | N1—N2—C4—C3 | −0.5 (3) |
| O1—Cu1—O5—C12 | −92.3 (2) | Cu1—N2—C4—C3 | −171.4 (2) |
| O5—Cu1—O2—C11 | 3.0 (2) | N1—N2—C4—C5 | −179.4 (3) |
| N3—Cu1—O2—C11 | −170.0 (2) | Cu1—N2—C4—C5 | 9.7 (5) |
| O1—Cu1—O2—C11 | 95.0 (2) | C2—C3—C4—N2 | −0.1 (4) |
| O5—Cu1—N2—C4 | 134.5 (3) | C2—C3—C4—C5 | 178.7 (3) |
| N3—Cu1—N2—C4 | −53.3 (3) | N3—N4—C9—C8 | 0.2 (4) |
| O1—Cu1—N2—C4 | 42.2 (3) | N3—N4—C9—C10 | −179.6 (3) |
| O5—Cu1—N2—N1 | −35.3 (2) | N4—C9—C8—C7 | −0.2 (4) |
| N3—Cu1—N2—N1 | 137.0 (2) | C10—C9—C8—C7 | 179.5 (3) |
| O1—Cu1—N2—N1 | −127.5 (2) | N4—N3—C7—C8 | 0.0 (3) |
| C4—N2—N1—C2 | 0.9 (3) | Cu1—N3—C7—C8 | 173.5 (2) |
| Cu1—N2—N1—C2 | 173.1 (2) | N4—N3—C7—C6 | −179.8 (3) |
| O2—Cu1—N3—C7 | 123.6 (3) | Cu1—N3—C7—C6 | −6.2 (5) |
| N2—Cu1—N3—C7 | −49.0 (3) | C9—C8—C7—N3 | 0.1 (4) |
| O1—Cu1—N3—C7 | −147.5 (3) | C9—C8—C7—C6 | 179.9 (3) |
| O2—Cu1—N3—N4 | −63.5 (2) | Cu1—O2—C11—O3 | 175.7 (2) |
| N2—Cu1—N3—N4 | 123.9 (2) | Cu1—O2—C11—C12 | −2.2 (3) |
| O1—Cu1—N3—N4 | 25.4 (2) | Cu1—O5—C12—O4 | −176.5 (3) |
| C7—N3—N4—C9 | −0.1 (3) | Cu1—O5—C12—C11 | 3.1 (3) |
| Cu1—N3—N4—C9 | −174.7 (2) | O3—C11—C12—O4 | 1.0 (4) |
| N2—N1—C2—C3 | −1.0 (4) | O2—C11—C12—O4 | 179.0 (3) |
| N2—N1—C2—C1 | 179.8 (3) | O3—C11—C12—O5 | −178.6 (3) |
| N1—C2—C3—C4 | 0.7 (3) | O2—C11—C12—O5 | −0.6 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O3i | 0.99 | 1.88 | 2.798 (3) | 153 |
| O1—H2···O5ii | 0.97 | 1.97 | 2.923 (3) | 168 |
| N1—H3···O3iii | 0.88 | 2.03 | 2.857 (3) | 156 |
| N4—H18···O3i | 0.88 | 1.97 | 2.845 (3) | 175 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) −x, −y, −z+1; (iii) −x, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HY2100).
References
- Abdeljalil, E. F., Najib, B. L., Abdelali, K., El Bali, B. & Bolte, M. (2006). Acta Cryst. E62, m551–m552.
- Bataille, T. & Louër, D. (1999). Acta Cryst. C55, 1760–1762.
- Castillo, O., Luque, A., Lloret, F. & Romàn, P. (2001). Inorg. Chem. Commun.4, 350–353.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Naumov, D. Y., Virovets, A. V., Podberezskaya, N. V. & Boldyreva, E. V. (1995). Acta Cryst. C51, 60–62.
- Nonius (1998). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr and R. M. Sweet, pp. 307–326. New York: Academic Press.
- Raptis, R., Georgakaki, I. & Hockless, D. (1999). Angew. Chem. Int. Ed.38, 1632–1634. [DOI] [PubMed]
- Sheldrick, G. M. (1997). SHELXS97 and SHELXL97 University of Göttingen, Germany.
- Sheldrick, G. M. (2003). SADABS University of Göttingen, Germany.
- Strotmeyer, K. P., Fritsky, I. O., Ott, R., Pritzkow, H. & Krämer, R. (2003). Supramol. Chem.15, 529–547.
- Tomyn, S. V., Gumienna-Kontecka, E., Fritsky, I. O., Iskenderov, T. S. & Światek-Kozłowska, J. (2007). Acta Cryst. E63, m438–m440.
- Warda, S. A. (1998). Acta Cryst. C54, 916–918.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536807058928/hy2100sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807058928/hy2100Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




