Abstract
In the title compound, C17H15N5O3S, the plane of the triazolo–thiadiazole system forms dihedral angles of 15.68 and 4.46° with the planes of the pyridine and furan rings, respectively. In the molecule, there is an intramolecular C—H⋯N interaction. The crystal structure also contains other intermolecular interactions, such as C—H⋯O hydrogen bonds, π–π stacking (centroid–centroid distances = 3.746 and 3.444 Å), non-bonded S⋯N [3.026 (2) Å] and C—H⋯π interactions.
Related literature
For related literature, see: Borbulevych et al. (1998 ▶); Bruno et al. (2003 ▶); Collin et al. (2003 ▶); Cooper & Steele (1990 ▶); Dinçer et al. (2005 ▶); Golgolab et al. (1973 ▶); Holla et al. (1994 ▶, 1998 ▶, 2002 ▶); Lu et al. (2007 ▶); Shen et al. (2006 ▶); Tsukuda et al. (1998 ▶); Wagner et al. (2005 ▶); Witkoaski et al. (1972 ▶); Zhang & Wen (1998 ▶); Özbey et al. (2000 ▶).
Experimental
Crystal data
C17H15N5O3S
M r = 369.40
Triclinic,

a = 8.094 (4) Å
b = 10.535 (5) Å
c = 10.996 (5) Å
α = 66.699 (5)°
β = 73.683 (5)°
γ = 80.998 (5)°
V = 825.4 (7) Å3
Z = 2
Mo Kα radiation
μ = 0.23 mm−1
T = 296 (2) K
0.30 × 0.30 × 0.25 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.935, T max = 0.946
4297 measured reflections
2860 independent reflections
2531 reflections with I > 2σ(I)
R int = 0.017
Refinement
R[F 2 > 2σ(F 2)] = 0.038
wR(F 2) = 0.100
S = 1.06
2860 reflections
235 parameters
H-atom parameters constrained
Δρmax = 0.22 e Å−3
Δρmin = −0.25 e Å−3
Data collection: SMART (Bruker, 2000 ▶); cell refinement: SAINT (Bruker, 2000 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997 ▶); molecular graphics: SHELXTL (Bruker, 2000 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536807061144/av3123sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807061144/av3123Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg is the centroid of the furan ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3—H3⋯N5 | 0.93 | 2.31 | 2.982 (3) | 128 |
| C13—H13⋯O2i | 0.93 | 2.51 | 3.285 (3) | 141 |
| C17—H17A⋯O1ii | 0.96 | 2.56 | 3.424 (3) | 149 |
| C17—H17A⋯Cgiii | 0.96 | 3.29 | 4.008 (3) | 134 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii) 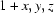 .
.
Acknowledgments
The authors thank the Instrument Analysis and Research Center of Shanghai University for the crystal structure analysis.
supplementary crystallographic information
Comment
Antimicrobials reduce or completely block the growth and multiplication of bacteria. This has made them unique for the control of deadly infectious diseases caused by a variety of pathogens. They have transformed our ability to treat infectious diseases such as pneumonia, meningitis, tuberculosis, malaria, and AIDS. Derivatives of 1,2,4-triazole and 1,3,4-thiadiazole condensed nucleus systems are found to have diverse pharmacological activities (Collin et al., 2003) such as fungicidal, insecticidal, bactericidal, herbicidal, anti-tumor (Holla et al., 2002), and anti-inflammatory (Cooper & Steele, 1990), and CNS stimulant properties (Holla et al., 1994). They also find applications as dyes, lubricants and analytical reagents (Zhang & Wen, 1998), antiviral agents (Witkoaski et al., 1972), heterocyclic ligands (Shen et al., 2006). Examples of such compounds bearing the 1,2,4-triazole moieties are fluconazole, a powerful azole antifungal agent (Tsukuda et al., 1998) as well as the potent antiviral N-nucleoside ribavirin (Witkoaski et al., 1972). Also, a number of 1,3,4-thiadiazoles show antibacterial properties similar to those of well known sulfonamide drugs (Golgolab et al., 1973). The thiadiazole nucleus with N—C—S linkage exhibits a large number of biological activities (Holla et al., 1998). Prompted by these findings and in continuation of our efforts in synthesizing some condensed bridge bioactive molecules bearing multifunctional and pharmaceutically active groups, herein, we report a new 1,2,4-triazolo[3,4-b][1,3,4]thiadiazole derivative which has been firstly prepared. In view of these important properties, the present single-crystal X-ray diffraction study of the title compound, (I), was carried out in order to investigate this bicyclic system and to confirm the assigned structure.
Compound (I) (Fig. 1) is composed of a fused triazolo-thiadiazole system, one pyridine ring attached to the triazole ring, the other furan ring is bonded to the thiadiazole ring, respectively. The fused triazolo-thiadiazole ring and furan ring nearly adopt coplanar form due to formation of a large π-π conjugation system. The plane of the triazolo-thiadiazole system forms dihedral angles of 15.68 and 4.46 ° with the planes of the pyridine and furan rings, respectively. Remarkably, the dihedral angle of pyridine and triazole-thiadiazole planes is significantly larger than this in the literature [1.53 ° (Dinçer et al., 2005)]. This should be attributed to the steric hindrance caused by the methyl group at α-position of pyridine ring, resulting in the enlargement of dihedral angle.
Moreover, the bond distance N2—N3 (1.398 (2) Å) is in accordance with those found for structures containing the 1,2,4-triazole ring (Lu et al., 2007; Bruno et al., 2003), indicating that the bond distance N2—N3 is relevant to the resonance effect of fused triazole-thiadizole ring, other than the dihedral angle between the pyridine ring and a fused ring, although two molecules display different dihedral angles. Apparently, it is longer than that of 5-ammo-3-trifluoromethyl-1-H-1,2,4-triazole [1.371 Å (Borbulevych et al., 1998)], because the latter contains an electron-withdrawing CF3 group attached to the 3-position of the triazole ring. In the thiadiazole moiety, the S1—C10 (1.730 (8) Å) and S1—C11 (1.761 (8) Å) bond distances show the similar pattern as those in the literature (Dinçer et al., 2005). The difference between S1—C10 and S1—C11 indicates that the bond distances mainly depend on the nature of resonance effort caused by fused triazolo-thiadiazole, rather than the aromatic ring attached to the 5-position of thiadiazole ring, because in our case, the aromatic ring is not the phenyl group, but the furan-2-yl group. The bond distances and angles of the furan ring are comparable with those in the literature (Wagner et al., 2005).
In the molecular structure, intramolecular C3—H3···N5 contacts lead to the formation of a five-membered ring fused with the pyridine ring. In addition, the C3—N5 distance is 2.982 (3) Å, a little shorter than that in the literature [3.140 (2)Å Özbey et al., 2000)].
The structure contains two weak π-π stacking interactions. The first of them is between the triazole ring and the thiadiazol ring, its symmetry related partner at (1 - x, -y, 1 - z), with a distance of 3.746 Å between the ring centroids and a perpendicular distance between the rings of 3.444 Å. The second one is between the triazole ring and the pyridine ring at (1 - x, 1 - y, -z), with a distance of 3.701Å between the ring centroids and a perpendicular distance between the rings of 3.541 Å. In addition to these interactions, there are two types of intermolecular hydrogen bonds C13—H13···O2 (1 - x, 1 - y, 1 - z) and C17—H17A···O1(1 - x, -y, 1 - z) which make the crystal structure to be more stable. The hydrogen-bonding geometry is listed in Table 2. Some short-contact distances are not listed in the table. Yet noteworthy, is S1···N3 (1 - x, -y, -z) of 3.026 (2) Å, and a π-ring interaction at (1 + x, y, z) with the distance 3.285 (9) Å between H17A and furan ring, both of them may cause steric hindrance.
Experimental
(1 mmol, 0.293 g)ethyl- 5-(4-amino-5-mercapto-4H-1,2,4-triazol-3-yl) -2,6-dimethylnicotinate, and (1 mmol, 0.112 g) furan-2- carboxylic acid were dissloved in 3.5 ml phosphorous oxychloride. The mixture was refluxed for 4 h, and allowed to cool to room temperature. The reaction mixture was poured into crushed ice, then, neutralized with dilute sodium bicarbonate solution. The solid separated was filtered off, washed with water, dried and recrystallized from chloroform afford pure product in a yield of 86% (m.p. 501–502 K). Single crystals suitable for X-ray analysis were obtained from chloroform by slow evaporation at room temperature. IR(ν, cm-1): 3101–3001 (Furan CH), 1725 (C=O), 1586–1444 (C=C, C=N), 1270 (C—S—C); 1H-NMR (500 MHz, CD3Cl): δ 8.82 (s, 1H, Pyridine CH), 6.67–7.70 (m, 3H, Furan CH), 4.41 (q, J=7.0 Hz, 2H, CH), 2.92 (s, 3H, Pyridine CH), 2.87 (s, 3H, Pyridine CH), 1.42 (t, J=7.0 Hz, 3H, CH);Element analysis, required for C17H15N5O3S: C 55.27, H 4.09, N 18.96%; Found: C 55.29, H 4.08, N 18.68%.
Refinement
All H atoms were placed in calculated positions, with C—H=0.93–0.97 Å, and included in the final cycles of refinement using a riding model, Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of the title compound with the atom-numbering scheme. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
The crystal packing of (I) showing the intermolecular S···N interactions.
Crystal data
| C17H15N5O3S | Z = 2 |
| Mr = 369.40 | F000 = 384 |
| Triclinic, P1 | Dx = 1.486 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71073 Å |
| a = 8.094 (4) Å | Cell parameters from 2601 reflections |
| b = 10.535 (5) Å | θ = 2.4–27.3º |
| c = 10.996 (5) Å | µ = 0.23 mm−1 |
| α = 66.699 (5)º | T = 296 (2) K |
| β = 73.683 (5)º | Block, brown |
| γ = 80.998 (5)º | 0.30 × 0.30 × 0.25 mm |
| V = 825.4 (7) Å3 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2860 independent reflections |
| Radiation source: fine-focus sealed tube | 2531 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.017 |
| T = 296(2) K | θmax = 25.0º |
| φ and ω scans | θmin = 2.4º |
| Absorption correction: multi-scan(SADABS; Sheldrick, 1996) | h = −9→9 |
| Tmin = 0.935, Tmax = 0.946 | k = −12→12 |
| 4297 measured reflections | l = −13→11 |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.038 | w = 1/[σ2(Fo2) + (0.0502P)2 + 0.2861P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.100 | (Δ/σ)max < 0.001 |
| S = 1.06 | Δρmax = 0.22 e Å−3 |
| 2860 reflections | Δρmin = −0.25 e Å−3 |
| 235 parameters | Extinction correction: SHELXL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.013 (3) |
| Secondary atom site location: difference Fourier map |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.82163 (19) | 0.64436 (15) | −0.06646 (15) | 0.0380 (4) | |
| N2 | 0.6722 (2) | 0.25738 (17) | −0.06479 (16) | 0.0448 (4) | |
| N3 | 0.5890 (2) | 0.13509 (17) | −0.02295 (17) | 0.0473 (4) | |
| N4 | 0.54173 (18) | 0.20294 (14) | 0.15000 (14) | 0.0320 (3) | |
| N5 | 0.46583 (19) | 0.19007 (14) | 0.28238 (15) | 0.0349 (3) | |
| O1 | 0.19535 (18) | −0.07982 (13) | 0.53282 (14) | 0.0469 (3) | |
| O2 | 0.7628 (2) | 0.63716 (16) | 0.33760 (15) | 0.0646 (5) | |
| O3 | 0.7023 (2) | 0.41744 (14) | 0.41061 (14) | 0.0572 (4) | |
| S1 | 0.37936 (6) | −0.01525 (5) | 0.23778 (5) | 0.04173 (16) | |
| C1 | 0.8076 (2) | 0.65217 (18) | 0.05519 (19) | 0.0361 (4) | |
| C2 | 0.7476 (2) | 0.54134 (18) | 0.17662 (18) | 0.0337 (4) | |
| C3 | 0.6956 (2) | 0.42634 (18) | 0.16808 (18) | 0.0342 (4) | |
| H3 | 0.6540 | 0.3523 | 0.2477 | 0.041* | |
| C4 | 0.7047 (2) | 0.42024 (17) | 0.04290 (18) | 0.0322 (4) | |
| C5 | 0.7739 (2) | 0.53224 (18) | −0.07487 (18) | 0.0340 (4) | |
| C6 | 0.8008 (3) | 0.5362 (2) | −0.21624 (19) | 0.0442 (5) | |
| H6A | 0.8492 | 0.6220 | −0.2808 | 0.066* | |
| H6B | 0.6922 | 0.5286 | −0.2315 | 0.066* | |
| H6C | 0.8782 | 0.4604 | −0.2274 | 0.066* | |
| C7 | 0.8615 (3) | 0.7848 (2) | 0.0491 (2) | 0.0515 (5) | |
| H7A | 0.8978 | 0.8454 | −0.0444 | 0.077* | |
| H7B | 0.9552 | 0.7655 | 0.0925 | 0.077* | |
| H7C | 0.7660 | 0.8284 | 0.0955 | 0.077* | |
| C8 | 0.7393 (2) | 0.54110 (19) | 0.31403 (19) | 0.0385 (4) | |
| C9 | 0.6430 (2) | 0.29802 (18) | 0.03893 (18) | 0.0340 (4) | |
| C10 | 0.5122 (2) | 0.10682 (18) | 0.10579 (19) | 0.0366 (4) | |
| C11 | 0.3772 (2) | 0.07975 (17) | 0.33896 (18) | 0.0352 (4) | |
| C12 | 0.2798 (2) | 0.04004 (17) | 0.47908 (18) | 0.0363 (4) | |
| C13 | 0.2505 (3) | 0.0984 (2) | 0.5722 (2) | 0.0456 (5) | |
| H13 | 0.2936 | 0.1803 | 0.5607 | 0.055* | |
| C14 | 0.1413 (3) | 0.0111 (2) | 0.6913 (2) | 0.0501 (5) | |
| H14 | 0.0985 | 0.0245 | 0.7734 | 0.060* | |
| C15 | 0.1114 (3) | −0.0941 (2) | 0.6634 (2) | 0.0492 (5) | |
| H15 | 0.0428 | −0.1670 | 0.7245 | 0.059* | |
| C16 | 0.7021 (3) | 0.3943 (2) | 0.5502 (2) | 0.0541 (5) | |
| H16A | 0.6072 | 0.3378 | 0.6130 | 0.065* | |
| H16B | 0.6864 | 0.4822 | 0.5622 | 0.065* | |
| C17 | 0.8677 (3) | 0.3237 (2) | 0.5807 (2) | 0.0533 (5) | |
| H17A | 0.8826 | 0.2366 | 0.5692 | 0.080* | |
| H17B | 0.8664 | 0.3083 | 0.6730 | 0.080* | |
| H17C | 0.9611 | 0.3807 | 0.5195 | 0.080* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0420 (9) | 0.0342 (8) | 0.0382 (9) | −0.0062 (6) | −0.0090 (7) | −0.0126 (7) |
| N2 | 0.0553 (10) | 0.0447 (9) | 0.0382 (9) | −0.0173 (7) | −0.0012 (7) | −0.0207 (7) |
| N3 | 0.0605 (11) | 0.0479 (9) | 0.0407 (9) | −0.0211 (8) | −0.0009 (8) | −0.0249 (8) |
| N4 | 0.0370 (8) | 0.0317 (7) | 0.0311 (8) | −0.0054 (6) | −0.0067 (6) | −0.0152 (6) |
| N5 | 0.0403 (8) | 0.0340 (8) | 0.0323 (8) | −0.0061 (6) | −0.0056 (6) | −0.0148 (6) |
| O1 | 0.0598 (9) | 0.0393 (7) | 0.0412 (7) | −0.0186 (6) | −0.0027 (6) | −0.0150 (6) |
| O2 | 0.1001 (13) | 0.0548 (9) | 0.0505 (9) | −0.0282 (9) | −0.0098 (8) | −0.0283 (8) |
| O3 | 0.0959 (12) | 0.0451 (8) | 0.0385 (8) | −0.0120 (8) | −0.0231 (8) | −0.0158 (6) |
| S1 | 0.0537 (3) | 0.0365 (3) | 0.0392 (3) | −0.0147 (2) | −0.0047 (2) | −0.0183 (2) |
| C1 | 0.0346 (9) | 0.0340 (9) | 0.0425 (10) | −0.0045 (7) | −0.0107 (8) | −0.0151 (8) |
| C2 | 0.0320 (9) | 0.0357 (9) | 0.0372 (10) | −0.0038 (7) | −0.0091 (7) | −0.0164 (8) |
| C3 | 0.0358 (9) | 0.0333 (9) | 0.0348 (9) | −0.0067 (7) | −0.0067 (7) | −0.0135 (7) |
| C4 | 0.0326 (9) | 0.0321 (9) | 0.0350 (9) | −0.0032 (7) | −0.0079 (7) | −0.0151 (7) |
| C5 | 0.0324 (9) | 0.0349 (9) | 0.0356 (9) | −0.0013 (7) | −0.0076 (7) | −0.0147 (8) |
| C6 | 0.0554 (12) | 0.0417 (10) | 0.0341 (10) | −0.0073 (9) | −0.0054 (9) | −0.0144 (8) |
| C7 | 0.0695 (14) | 0.0388 (11) | 0.0503 (12) | −0.0178 (10) | −0.0155 (10) | −0.0146 (9) |
| C8 | 0.0373 (10) | 0.0418 (10) | 0.0422 (10) | −0.0069 (8) | −0.0076 (8) | −0.0214 (9) |
| C9 | 0.0373 (9) | 0.0348 (9) | 0.0322 (9) | −0.0064 (7) | −0.0063 (7) | −0.0145 (8) |
| C10 | 0.0438 (10) | 0.0349 (9) | 0.0382 (10) | −0.0068 (8) | −0.0093 (8) | −0.0196 (8) |
| C11 | 0.0395 (9) | 0.0318 (9) | 0.0371 (10) | −0.0041 (7) | −0.0093 (8) | −0.0151 (8) |
| C12 | 0.0402 (10) | 0.0311 (9) | 0.0382 (10) | −0.0058 (7) | −0.0082 (8) | −0.0129 (8) |
| C13 | 0.0534 (12) | 0.0414 (11) | 0.0457 (11) | −0.0087 (9) | −0.0059 (9) | −0.0218 (9) |
| C14 | 0.0568 (13) | 0.0534 (12) | 0.0380 (11) | −0.0045 (10) | −0.0011 (9) | −0.0211 (9) |
| C15 | 0.0525 (12) | 0.0483 (11) | 0.0378 (11) | −0.0122 (9) | −0.0014 (9) | −0.0095 (9) |
| C16 | 0.0740 (15) | 0.0556 (13) | 0.0356 (11) | −0.0058 (11) | −0.0124 (10) | −0.0195 (10) |
| C17 | 0.0652 (14) | 0.0550 (12) | 0.0472 (12) | −0.0138 (10) | −0.0126 (10) | −0.0236 (10) |
Geometric parameters (Å, °)
| N1—C5 | 1.343 (2) | C4—C5 | 1.405 (2) |
| N1—C1 | 1.345 (2) | C4—C9 | 1.472 (2) |
| N2—C9 | 1.320 (2) | C5—C6 | 1.493 (3) |
| N2—N3 | 1.398 (2) | C6—H6A | 0.9600 |
| N3—C10 | 1.307 (2) | C6—H6B | 0.9600 |
| N4—C10 | 1.360 (2) | C6—H6C | 0.9600 |
| N4—N5 | 1.372 (2) | C7—H7A | 0.9600 |
| N4—C9 | 1.376 (2) | C7—H7B | 0.9600 |
| N5—C11 | 1.300 (2) | C7—H7C | 0.9600 |
| O1—C15 | 1.364 (2) | C11—C12 | 1.443 (3) |
| O1—C12 | 1.365 (2) | C12—C13 | 1.342 (3) |
| O2—C8 | 1.193 (2) | C13—C14 | 1.411 (3) |
| O3—C8 | 1.328 (2) | C13—H13 | 0.9300 |
| O3—C16 | 1.455 (2) | C14—C15 | 1.332 (3) |
| S1—C10 | 1.7308 (19) | C14—H14 | 0.9300 |
| S1—C11 | 1.7618 (18) | C15—H15 | 0.9300 |
| C1—C2 | 1.401 (3) | C16—C17 | 1.485 (3) |
| C1—C7 | 1.499 (3) | C16—H16A | 0.9700 |
| C2—C3 | 1.388 (2) | C16—H16B | 0.9700 |
| C2—C8 | 1.492 (3) | C17—H17A | 0.9600 |
| C3—C4 | 1.384 (3) | C17—H17B | 0.9600 |
| C3—H3 | 0.9300 | C17—H17C | 0.9600 |
| C5—N1—C1 | 121.07 (15) | O2—C8—O3 | 122.87 (18) |
| C9—N2—N3 | 109.45 (15) | O2—C8—C2 | 125.99 (18) |
| C10—N3—N2 | 105.44 (14) | O3—C8—C2 | 111.14 (15) |
| C10—N4—N5 | 118.56 (14) | N2—C9—N4 | 107.67 (15) |
| C10—N4—C9 | 106.04 (15) | N2—C9—C4 | 128.65 (16) |
| N5—N4—C9 | 135.33 (14) | N4—C9—C4 | 123.68 (15) |
| C11—N5—N4 | 107.20 (14) | N3—C10—N4 | 111.39 (16) |
| C15—O1—C12 | 105.97 (15) | N3—C10—S1 | 139.04 (14) |
| C8—O3—C16 | 118.99 (15) | N4—C10—S1 | 109.55 (13) |
| C10—S1—C11 | 87.13 (9) | N5—C11—C12 | 120.11 (16) |
| N1—C1—C2 | 120.65 (16) | N5—C11—S1 | 117.56 (14) |
| N1—C1—C7 | 115.27 (16) | C12—C11—S1 | 122.33 (13) |
| C2—C1—C7 | 124.07 (17) | C13—C12—O1 | 110.19 (17) |
| C3—C2—C1 | 118.28 (17) | C13—C12—C11 | 133.14 (17) |
| C3—C2—C8 | 118.83 (16) | O1—C12—C11 | 116.66 (15) |
| C1—C2—C8 | 122.88 (16) | C12—C13—C14 | 106.50 (18) |
| C4—C3—C2 | 121.00 (16) | C12—C13—H13 | 126.7 |
| C4—C3—H3 | 119.5 | C14—C13—H13 | 126.7 |
| C2—C3—H3 | 119.5 | C15—C14—C13 | 106.87 (18) |
| C3—C4—C5 | 117.68 (16) | C15—C14—H14 | 126.6 |
| C3—C4—C9 | 119.08 (16) | C13—C14—H14 | 126.6 |
| C5—C4—C9 | 123.23 (16) | C14—C15—O1 | 110.47 (17) |
| N1—C5—C4 | 121.21 (16) | C14—C15—H15 | 124.8 |
| N1—C5—C6 | 115.29 (16) | O1—C15—H15 | 124.8 |
| C4—C5—C6 | 123.50 (16) | O3—C16—C17 | 109.93 (18) |
| C5—C6—H6A | 109.5 | O3—C16—H16A | 109.7 |
| C5—C6—H6B | 109.5 | C17—C16—H16A | 109.7 |
| H6A—C6—H6B | 109.5 | O3—C16—H16B | 109.7 |
| C5—C6—H6C | 109.5 | C17—C16—H16B | 109.7 |
| H6A—C6—H6C | 109.5 | H16A—C16—H16B | 108.2 |
| H6B—C6—H6C | 109.5 | C16—C17—H17A | 109.5 |
| C1—C7—H7A | 109.5 | C16—C17—H17B | 109.5 |
| C1—C7—H7B | 109.5 | H17A—C17—H17B | 109.5 |
| H7A—C7—H7B | 109.5 | C16—C17—H17C | 109.5 |
| C1—C7—H7C | 109.5 | H17A—C17—H17C | 109.5 |
| H7A—C7—H7C | 109.5 | H17B—C17—H17C | 109.5 |
| H7B—C7—H7C | 109.5 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3—H3···N5 | 0.93 | 2.31 | 2.982 (3) | 128 |
| C13—H13···O2i | 0.93 | 2.51 | 3.285 (3) | 141 |
| C17—H17A···O1ii | 0.96 | 2.56 | 3.424 (3) | 149 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1, −y, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: AV3123).
References
- Borbulevych, O. Y., Shishkin, O. V., Desenko, S. M., Chernenko, V. N. & Orlov, V. D. (1998). Acta Cryst. C54, 442–444.
- Buker (2000). SMART, SAINT and SHELXTL Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruno, G., Nicoló, F., Puntoriero, F., Giuffrida, G., Ricevuto, V. & Rosace, G. (2003). Acta Cryst. C59, o390–o391. [DOI] [PubMed]
- Collin, X., Sauleau, A. & Coulon, J. (2003). Bioorg. Med. Chem. Lett.13, 2601–2605. [DOI] [PubMed]
- Cooper, K. & Steele, J. (1990). Chem. Abstr.112, 76957.
- Dinçer, M., Özdemir, N., Çetin, A., Cansız, A. & Büyükgüngör, O. (2005). Acta Cryst. C61, o665–o667. [DOI] [PubMed]
- Golgolab, H., Lalezari, I., Hosseini, L. & Gohari, L. (1973). J. Heterocycl. Chem.10, 387–390.
- Holla, B. S., Gonsalves, R. & Shenoy, S. (1998). Il Farmaco, 53, 574–578. [DOI] [PubMed]
- Holla, B. S., Kalluraya, B., Sridhar, K. R., Drake, E., Thomas, L. M., Bhandary, K. K. & Levine, M. J. (1994). Eur. J. Med. Chem.29, 301–308.
- Holla, B. S., Poojary, K. N., Rao, B. S. & Shivananda, M. K. (2002). Eur. J. Med. Chem.37, 511–517. [DOI] [PubMed]
- Lu, D.-L., Zhang, M., Song, L.-P., Huang, P.-G. & Zhang, J. (2007). Acta Cryst. E63, o3846.
- Özbey, S., Ulusoy, N. & Kendi, E. (2000). Acta Cryst. C56, 222–224. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (1997). SHELXS97 and SHELXL97 University of Göttingen, Germany.
- Shen, X.-Q., Yang, R., Yao, H.-C., Zhang, H.-Y., Li, G., Zhang, H.-Q., Chen, P.-K. & Hou, H.-W. (2006). J. Coord. Chem., 59, 2031–2038.
- Tsukuda, T., Shiratori, Y., Watanabe, M. H., Ontsuka, K., Hattori, M., Shirai, N. & Shimma, N. (1998). Bioorg. Med. Chem. Lett.8, 1819–1824. [DOI] [PubMed]
- Wagner, A., Hannu-Kuure, M. S., Oilunkaniemi, R. & Laitinen, R. S. (2005). Acta Cryst. E61, m2198–m2200.
- Witkoaski, J. T., Robins, R. K., Sidwell, R. W. & Simon, L. N. (1972). J. Med. Chem.15, 150–154. [DOI] [PubMed]
- Zhang, Z.-Y. & Wen, S.-X. (1998). Heterocycles, 48, 561–584.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536807061144/av3123sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807061144/av3123Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




