Abstract
The title compound, [Cu(C7H3NO5)(C12H8N2)(H2O)]·4.5H2O or [Cu(hypydc)(phen)(H2O)]·4.5H2O (phen is 1,10-phenanthroline and hypydcH2 is 4-hydroxypyridine-2,6-dicarboxylic acid), was obtained by the reaction of copper(II) nitrate hexahydrate with the proton-transfer compound (phenH)2(hypydc) in aqueous solution. Both the cationic and the anionic fragments of the proton-transfer compound are involved in complexation. Each CuII atom has a distorted octahedral geometry. It is hexacoordinated by three O atoms and three N atoms, from one phen fragment (as bidentate ligand), one (hypydc)2− unit (as tridentate ligand) and a water molecule. In the crystal structure, O—H⋯O and C—H⋯O hydrogen bonds, and π–π stacking interactions [centroid-to-centroid distance 3.5642 (11) Å] between the phen ring systems, contribute to the formation of a three-dimensional supramolecular structure.
Related literature
For related literature, see: Aghabozorg, Attar Gharamaleki, Ghadermazi et al. (2007 ▶); Aghabozorg, Attar Gharamaleki, Ghasemikhah et al. (2007 ▶); Aghabozorg, Daneshvar et al. (2007 ▶).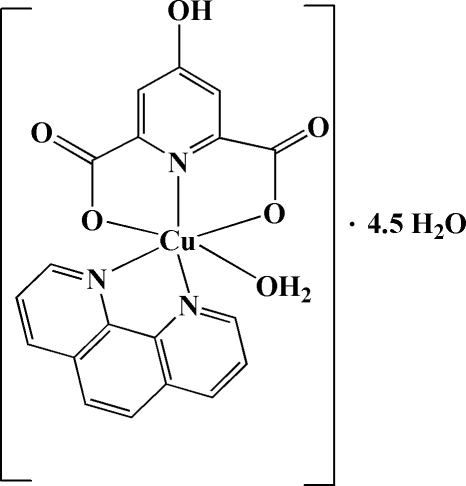
Experimental
Crystal data
[Cu(C7H3NO5)(C12H8N2)(H2O)]·4.5H2O
M r = 523.94
Orthorhombic,
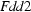
a = 18.686 (4) Å
b = 44.033 (8) Å
c = 10.3812 (18) Å
V = 8542 (3) Å3
Z = 16
Mo Kα radiation
μ = 1.09 mm−1
T = 296 (2) K
0.34 × 0.24 × 0.12 mm
Data collection
Bruker SMART 1000 diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 1998 ▶) T min = 0.709, T max = 0.881
59661 measured reflections
10800 independent reflections
9609 reflections with I > 2σ(I)
R int = 0.032
Refinement
R[F 2 > 2σ(F 2)] = 0.027
wR(F 2) = 0.066
S = 1.01
10800 reflections
300 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.50 e Å−3
Δρmin = −0.33 e Å−3
Absolute structure: Flack (1983 ▶), with 4504 Friedel pairs
Flack parameter: 0.007 (4)
Data collection: SMART (Bruker, 1998 ▶); cell refinement: SAINT (Bruker, 1998 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997 ▶); molecular graphics: SHELXTL (Bruker, 2005 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536807067207/su2032sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807067207/su2032Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O5—H5A⋯O5W | 0.85 | 1.73 | 2.5771 (14) | 174 |
| O6—H6D⋯O1i | 0.85 | 1.88 | 2.7059 (12) | 162 |
| O6—H6C⋯O4ii | 0.85 | 1.76 | 2.6048 (11) | 174 |
| O1W—H1C⋯O1iii | 0.85 | 1.97 | 2.8154 (12) | 171 |
| O2W—H2C⋯O3iv | 0.85 | 2.37 | 3.1062 (16) | 145 |
| O2W—H2D⋯O4W | 0.85 | 1.90 | 2.7470 (16) | 172 |
| O3W—H3D⋯O2v | 0.85 | 1.93 | 2.7757 (13) | 175 |
| O3W—H3C⋯O4 | 0.85 | 1.90 | 2.7496 (13) | 175 |
| O4W—H4D⋯O3W | 0.85 | 1.84 | 2.6834 (15) | 171 |
| O4W—H4C⋯O5i | 0.85 | 2.26 | 3.0835 (16) | 165 |
| O5W—H5C⋯O1W | 0.85 | 2.03 | 2.8604 (15) | 164 |
| O5W—H5D⋯O2Wvi | 0.85 | 1.91 | 2.7204 (15) | 159 |
| C1—H1⋯O5vii | 0.93 | 2.41 | 3.1610 (18) | 137 |
| C3—H3⋯O3viii | 0.93 | 2.25 | 3.130 (2) | 158 |
| C8—H8⋯O3Wix | 0.93 | 2.42 | 3.315 (2) | 161 |
| C10—H10⋯O6 | 0.93 | 2.50 | 2.9995 (17) | 114 |
| C10—H10⋯O5Wx | 0.93 | 2.42 | 3.1809 (19) | 139 |
Symmetry codes: (i) 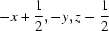 ; (ii)
; (ii) 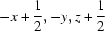 ; (iii)
; (iii) 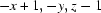 ; (iv)
; (iv) 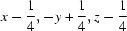 ; (v)
; (v) 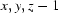 ; (vi)
; (vi) 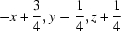 ; (vii)
; (vii) 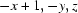 ; (viii)
; (viii) 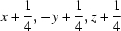 ; (ix)
; (ix) 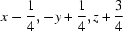 ; (x)
; (x) 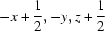 .
.
Acknowledgments
Financial support from Ilam University and the Teacher Training University is gratefully acknowledged.
supplementary crystallographic information
Comment
Non-covalent interactions, including hydrogen bonds, are of great importance in stabilizing structures in the solid state. We have synthesized several proton transfer compounds, some with remaining sites as electron donors that can coordinate to metal ions (Aghabozorg, Attar Gharamaleki, Ghadermazi et al., 2007; Aghabozorg, Attar Gharamaleki, Ghasemikhah et al., 2007; Aghabozorg, Daneshvar et al., 2007 and references therein). A wide range of different hydrogen bonds were observed in these compounds and water molecules of crystallization were also involved in hydrogen bonding. Here, we report on the synthesis and crystal structure of the title compound, (I).
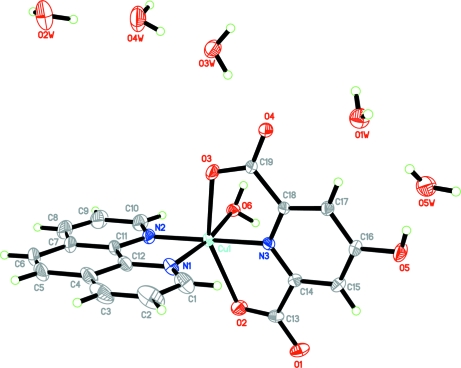
The title complex crystallizes in the orthorhombic space group Fdd2, with sixteen molecules in the unit cell. The molecular structure is shown in Fig. 1. Both the cationic and anionic fragments of the starting proton transfer compound are involved in complexation. Each CuII atom has a distorted octahedral geometry. It is coordinated by one 1,10-phenanthroline ligand, (phen as bidentate ligand), one 4-hydroxypyridine-2,6-dicarboxylate group, [(hypydc)2- as a tridentate ligand] and one coordinated water molecule. The axial bond lengths, Cu1—O2 and Cu1—O3 [2.3679 (9) and 2.3205 (11) Å, respectively] are longer than the equitoral metal-ligand bond lengths [1.9996 (8) - 2.0370 (9) Å], probabaly due to the Jahn-Teller effect. The dihedral angle between the planes passing through the (hypydc)2– and (phen) fragments is 83.41 (4)°, indicating that these to units are almost perpendicular to one another.
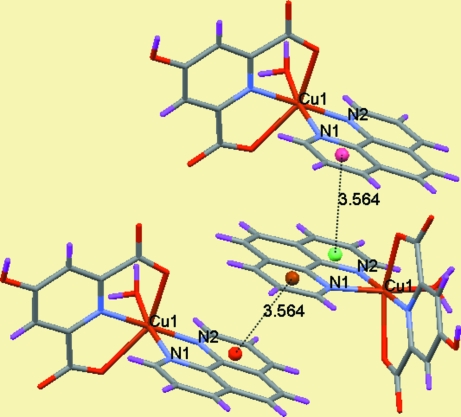
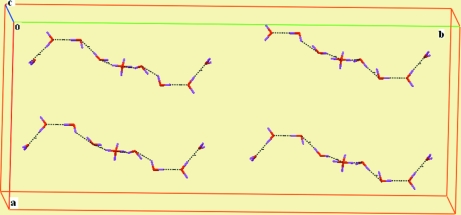
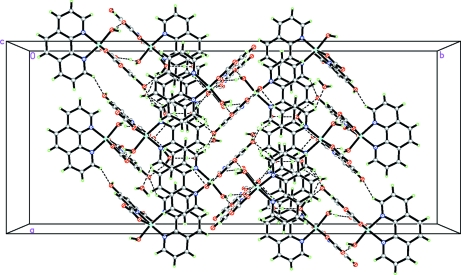
In the crystal weak π-π stacking interactions [3.5642 (11) Å [1/4 + x, 1/4 - y, 1/4 + z] between the aromatic rings of the coordinated (phen) fragments are present (Fig. 2). The water molecules of crystallization are involved in the formation of hydrogen bonds, forming chains (Fig. 3 and Table 1). O—H···O and C—H···O hydrogen bonds, ion pairing and π–π stacking interactions all contribute to the formation of a supramolecular structure (Fig. 4).
Experimental
The proton transfer compound, (phenH)2(hypydc), was prepared by the reaction of 4-hydroxypyridine-2,6-dicarboxylic acid, hypydcH2, with 1,10-phenanthroline, (phen). Cu(NO3)2.6H2O (125 mg, 0.5 mmol) in water (20 ml) and the proton transfer compound, (phenH)2(hypydc) (500 mg, 1.0 mmol) in water (20 ml), in a 1:2 molar ratio, were mixed. Blue crystals of (I) were obtained by the slow evaporation at room temperature.
Refinement
The H-atoms were included in calculated positions and treated as riding atoms: O—H = 0.85 Å and C—H = 0.93 - 0.95 Å with U ĩso~(H) = 1.2U~eq~ (parent O or C-atom).
Figures
Fig. 1.
The molecular structure of compound (I), showing the atom numbering scheme and displacement ellipsoids drawn at the 50% probability level.
Fig. 2.
A view of the π-π stacking interactions between the aromatic rings of the 1,10-phenanthroline (phen) fragments with distances of 3.5639 (11) Å [1/4 + x, 1/4 - y, 1/4 + z].
Fig. 3.
A view of the chain of hydrogen bonded water mlecules in compound (I) with hydrogen bonds shown as dashed lines.
Fig. 4.
The crystal packing of compound (I), with hydrogen bonds shown as dashed lines.
Crystal data
| [Cu(C7H3NO5)(C12H8N2)(H2O)]·4.5H2O | F000 = 4320 |
| Mr = 523.94 | Dx = 1.630 Mg m−3 |
| Orthorhombic, Fdd2 | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: F 2 -2d | Cell parameters from 30613 reflections |
| a = 18.686 (4) Å | θ = 2.3–36.4º |
| b = 44.033 (8) Å | µ = 1.09 mm−1 |
| c = 10.3812 (18) Å | T = 296 (2) K |
| V = 8542 (3) Å3 | Block, pale-blue |
| Z = 16 | 0.34 × 0.24 × 0.12 mm |
Data collection
| Bruker SMART 1000 diffractometer | 10800 independent reflections |
| Radiation source: fine-focus sealed tube | 9609 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.032 |
| Detector resolution: 100 pixels mm-1 | θmax = 38.6º |
| T = 296(2) K | θmin = 1.9º |
| ω scans | h = −30→31 |
| Absorption correction: multi-scan(SADABS; Bruker, 1998) | k = −72→69 |
| Tmin = 0.709, Tmax = 0.881 | l = −17→17 |
| 59661 measured reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.027 | w = 1/[σ2(Fo2) + (0.0367P)2] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.066 | (Δ/σ)max = 0.002 |
| S = 1.01 | Δρmax = 0.50 e Å−3 |
| 10800 reflections | Δρmin = −0.33 e Å−3 |
| 300 parameters | Extinction correction: none |
| 1 restraint | Absolute structure: Flack (1983), 4504 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.007 (4) |
| Secondary atom site location: difference Fourier map |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cu1 | 0.260204 (6) | 0.052661 (3) | 1.031738 (12) | 0.01475 (3) | |
| N1 | 0.33349 (5) | 0.086905 (19) | 1.03093 (10) | 0.02096 (15) | |
| N2 | 0.19432 (5) | 0.08711 (2) | 1.07530 (9) | 0.01972 (16) | |
| N3 | 0.32981 (5) | 0.019941 (19) | 0.98387 (8) | 0.01433 (14) | |
| O1 | 0.37788 (5) | −0.00342 (2) | 1.29870 (8) | 0.02511 (17) | |
| O2 | 0.29904 (5) | 0.03122 (2) | 1.22904 (7) | 0.02473 (16) | |
| O3 | 0.26351 (6) | 0.054290 (19) | 0.80826 (9) | 0.02719 (19) | |
| O4 | 0.30653 (5) | 0.026752 (19) | 0.64661 (7) | 0.02505 (17) | |
| O5 | 0.47567 (5) | −0.04570 (2) | 0.88292 (8) | 0.02523 (17) | |
| H5A | 0.4872 | −0.0449 | 0.8038 | 0.030* | |
| O6 | 0.17682 (4) | 0.024272 (16) | 1.02325 (8) | 0.01793 (12) | |
| H6D | 0.1690 | 0.0174 | 0.9480 | 0.022* | |
| H6C | 0.1854 | 0.0080 | 1.0643 | 0.022* | |
| C1 | 0.40278 (7) | 0.08587 (3) | 1.00820 (13) | 0.0302 (3) | |
| H1 | 0.4249 | 0.0670 | 1.0000 | 0.036* | |
| C2 | 0.44453 (9) | 0.11231 (4) | 0.99596 (15) | 0.0413 (4) | |
| H2 | 0.4934 | 0.1109 | 0.9799 | 0.050* | |
| C3 | 0.41253 (9) | 0.14022 (3) | 1.00797 (14) | 0.0401 (4) | |
| H3 | 0.4394 | 0.1579 | 0.9992 | 0.048* | |
| C4 | 0.33932 (8) | 0.14188 (3) | 1.03349 (13) | 0.0307 (2) | |
| C5 | 0.29961 (11) | 0.16990 (3) | 1.05229 (13) | 0.0414 (4) | |
| H5 | 0.3235 | 0.1884 | 1.0460 | 0.050* | |
| C6 | 0.22947 (10) | 0.16978 (3) | 1.07841 (15) | 0.0397 (4) | |
| H6 | 0.2059 | 0.1882 | 1.0906 | 0.048* | |
| C7 | 0.18977 (8) | 0.14200 (3) | 1.08811 (12) | 0.0312 (3) | |
| C8 | 0.11568 (10) | 0.14002 (4) | 1.11384 (15) | 0.0404 (4) | |
| H8 | 0.0889 | 0.1575 | 1.1272 | 0.048* | |
| C9 | 0.08381 (8) | 0.11233 (4) | 1.11891 (14) | 0.0357 (3) | |
| H9 | 0.0351 | 0.1108 | 1.1360 | 0.043* | |
| C10 | 0.12452 (7) | 0.08614 (3) | 1.09834 (12) | 0.0265 (2) | |
| H10 | 0.1019 | 0.0673 | 1.1009 | 0.032* | |
| C11 | 0.22684 (7) | 0.11447 (2) | 1.07039 (11) | 0.0222 (2) | |
| C12 | 0.30164 (6) | 0.11451 (2) | 1.04432 (10) | 0.02191 (19) | |
| C13 | 0.34558 (6) | 0.01134 (3) | 1.21286 (9) | 0.01798 (18) | |
| C14 | 0.36451 (5) | 0.00388 (2) | 1.07388 (9) | 0.01544 (16) | |
| C15 | 0.41397 (5) | −0.01821 (2) | 1.04245 (10) | 0.01813 (17) | |
| H15 | 0.4377 | −0.0290 | 1.1065 | 0.022* | |
| C16 | 0.42769 (5) | −0.02401 (2) | 0.91269 (10) | 0.01755 (17) | |
| C17 | 0.39083 (6) | −0.00763 (2) | 0.81932 (10) | 0.01653 (16) | |
| H17 | 0.3987 | −0.0112 | 0.7322 | 0.020* | |
| C18 | 0.34222 (6) | 0.01404 (2) | 0.85910 (9) | 0.01394 (16) | |
| C19 | 0.30013 (6) | 0.03315 (2) | 0.76368 (9) | 0.01689 (17) | |
| O1W | 0.5000 | 0.0000 | 0.45689 (13) | 0.0277 (2)* | |
| H1C | 0.5384 | 0.0026 | 0.4143 | 0.033* | |
| O2W | 0.13216 (6) | 0.15930 (2) | 0.41663 (14) | 0.0457 (3) | |
| H2C | 0.0889 | 0.1635 | 0.4349 | 0.055* | |
| H2D | 0.1336 | 0.1403 | 0.4299 | 0.055* | |
| O3W | 0.24433 (5) | 0.05872 (2) | 0.44855 (9) | 0.02822 (19) | |
| H3D | 0.2586 | 0.0500 | 0.3801 | 0.034* | |
| H3C | 0.2628 | 0.0496 | 0.5126 | 0.034* | |
| O4W | 0.13298 (7) | 0.09704 (2) | 0.43343 (14) | 0.0474 (3) | |
| H4D | 0.1683 | 0.0853 | 0.4474 | 0.057* | |
| H4C | 0.0974 | 0.0857 | 0.4167 | 0.057* | |
| O5W | 0.51596 (5) | −0.04686 (2) | 0.64555 (9) | 0.03011 (19) | |
| H5C | 0.5200 | −0.0331 | 0.5886 | 0.036* | |
| H5D | 0.5500 | −0.0593 | 0.6321 | 0.036* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.01572 (5) | 0.01249 (5) | 0.01605 (5) | 0.00093 (4) | 0.00172 (5) | −0.00189 (4) |
| N1 | 0.0232 (4) | 0.0187 (4) | 0.0210 (4) | −0.0040 (3) | 0.0044 (4) | −0.0048 (3) |
| N2 | 0.0230 (4) | 0.0173 (4) | 0.0189 (4) | 0.0048 (3) | 0.0015 (3) | −0.0016 (3) |
| N3 | 0.0163 (4) | 0.0136 (3) | 0.0131 (3) | 0.0001 (3) | 0.0000 (3) | −0.0003 (3) |
| O1 | 0.0224 (4) | 0.0370 (5) | 0.0159 (4) | 0.0013 (3) | −0.0015 (3) | 0.0065 (3) |
| O2 | 0.0276 (4) | 0.0306 (4) | 0.0161 (3) | 0.0069 (3) | 0.0018 (3) | −0.0014 (3) |
| O3 | 0.0394 (5) | 0.0240 (4) | 0.0181 (4) | 0.0176 (3) | −0.0018 (3) | −0.0007 (3) |
| O4 | 0.0408 (5) | 0.0205 (4) | 0.0138 (3) | 0.0097 (3) | −0.0028 (3) | −0.0010 (3) |
| O5 | 0.0265 (4) | 0.0256 (4) | 0.0236 (4) | 0.0144 (3) | 0.0046 (3) | 0.0027 (3) |
| O6 | 0.0200 (3) | 0.0177 (3) | 0.0161 (3) | −0.0011 (2) | −0.0013 (3) | 0.0014 (3) |
| C1 | 0.0249 (5) | 0.0303 (6) | 0.0355 (7) | −0.0083 (4) | 0.0074 (5) | −0.0087 (5) |
| C2 | 0.0321 (7) | 0.0500 (9) | 0.0418 (8) | −0.0241 (6) | 0.0079 (6) | −0.0082 (6) |
| C3 | 0.0560 (9) | 0.0340 (7) | 0.0305 (7) | −0.0281 (6) | 0.0005 (6) | −0.0032 (5) |
| C4 | 0.0498 (7) | 0.0198 (5) | 0.0224 (5) | −0.0125 (4) | −0.0037 (5) | −0.0018 (4) |
| C5 | 0.0805 (12) | 0.0144 (5) | 0.0292 (7) | −0.0099 (6) | −0.0146 (6) | 0.0009 (4) |
| C6 | 0.0702 (11) | 0.0155 (5) | 0.0333 (6) | 0.0079 (6) | −0.0147 (7) | −0.0050 (4) |
| C7 | 0.0498 (8) | 0.0179 (5) | 0.0258 (5) | 0.0117 (5) | −0.0070 (5) | −0.0043 (4) |
| C8 | 0.0524 (9) | 0.0340 (7) | 0.0347 (7) | 0.0262 (7) | −0.0054 (6) | −0.0075 (5) |
| C9 | 0.0300 (7) | 0.0421 (8) | 0.0352 (7) | 0.0180 (6) | 0.0017 (5) | −0.0053 (6) |
| C10 | 0.0229 (5) | 0.0303 (6) | 0.0264 (5) | 0.0083 (4) | 0.0028 (4) | −0.0017 (4) |
| C11 | 0.0333 (6) | 0.0153 (4) | 0.0181 (4) | 0.0040 (4) | −0.0016 (4) | −0.0024 (3) |
| C12 | 0.0327 (5) | 0.0158 (4) | 0.0173 (4) | −0.0043 (4) | −0.0003 (4) | −0.0026 (3) |
| C13 | 0.0176 (5) | 0.0235 (5) | 0.0129 (4) | −0.0025 (4) | 0.0001 (3) | 0.0006 (3) |
| C14 | 0.0147 (4) | 0.0165 (4) | 0.0151 (4) | −0.0011 (3) | −0.0001 (3) | 0.0017 (3) |
| C15 | 0.0165 (4) | 0.0202 (4) | 0.0178 (4) | 0.0027 (3) | −0.0002 (3) | 0.0043 (3) |
| C16 | 0.0164 (4) | 0.0159 (4) | 0.0204 (4) | 0.0031 (3) | 0.0015 (3) | 0.0024 (3) |
| C17 | 0.0174 (4) | 0.0158 (4) | 0.0163 (4) | 0.0019 (3) | 0.0010 (3) | −0.0001 (3) |
| C18 | 0.0166 (4) | 0.0118 (4) | 0.0135 (4) | 0.0005 (3) | −0.0009 (3) | 0.0002 (3) |
| C19 | 0.0220 (4) | 0.0132 (4) | 0.0155 (4) | 0.0028 (3) | −0.0021 (3) | 0.0006 (3) |
| O2W | 0.0372 (6) | 0.0250 (5) | 0.0747 (8) | 0.0000 (4) | 0.0068 (6) | −0.0087 (5) |
| O3W | 0.0340 (5) | 0.0279 (4) | 0.0227 (4) | 0.0101 (3) | 0.0024 (3) | 0.0032 (3) |
| O4W | 0.0441 (7) | 0.0248 (5) | 0.0735 (9) | 0.0095 (4) | −0.0168 (6) | −0.0081 (5) |
| O5W | 0.0278 (5) | 0.0364 (5) | 0.0262 (4) | 0.0049 (4) | 0.0036 (3) | −0.0030 (3) |
Geometric parameters (Å, °)
| Cu1—O6 | 1.9996 (8) | C5—H5 | 0.9300 |
| Cu1—N3 | 2.0036 (9) | C6—C7 | 1.434 (2) |
| Cu1—N2 | 2.0052 (9) | C6—H6 | 0.9300 |
| Cu1—N1 | 2.0370 (9) | C7—C11 | 1.4082 (16) |
| Cu1—O3 | 2.3219 (10) | C7—C8 | 1.413 (2) |
| Cu1—O2 | 2.3693 (9) | C8—C9 | 1.358 (3) |
| N1—C1 | 1.3168 (16) | C8—H8 | 0.9300 |
| N1—C12 | 1.3604 (14) | C9—C10 | 1.3979 (17) |
| N2—C10 | 1.3267 (16) | C9—H9 | 0.9300 |
| N2—C11 | 1.3505 (15) | C10—H10 | 0.9300 |
| N3—C14 | 1.3392 (13) | C11—C12 | 1.4237 (17) |
| N3—C18 | 1.3412 (12) | C13—C14 | 1.5213 (14) |
| O1—C13 | 1.2572 (13) | C14—C15 | 1.3810 (14) |
| O2—C13 | 1.2453 (14) | C15—C16 | 1.3948 (15) |
| O3—C19 | 1.2445 (12) | C15—H15 | 0.9300 |
| O4—C19 | 1.2533 (12) | C16—C17 | 1.3906 (14) |
| O5—C16 | 1.3460 (13) | C17—C18 | 1.3806 (14) |
| O5—H5A | 0.8500 | C17—H17 | 0.9300 |
| O6—H6D | 0.8500 | C18—C19 | 1.5194 (14) |
| O6—H6C | 0.8500 | O1W—H1C | 0.8500 |
| C1—C2 | 1.4072 (18) | O2W—H2C | 0.8501 |
| C1—H1 | 0.9300 | O2W—H2D | 0.8501 |
| C2—C3 | 1.372 (3) | O3W—H3D | 0.8500 |
| C2—H2 | 0.9300 | O3W—H3C | 0.8500 |
| C3—C4 | 1.395 (2) | O4W—H4D | 0.8501 |
| C3—H3 | 0.9300 | O4W—H4C | 0.8498 |
| C4—C12 | 1.4005 (15) | O5W—H5C | 0.8501 |
| C4—C5 | 1.453 (2) | O5W—H5D | 0.8499 |
| C5—C6 | 1.338 (3) | ||
| O6—Cu1—N3 | 92.60 (4) | C5—C6—H6 | 119.2 |
| O6—Cu1—N2 | 90.26 (4) | C7—C6—H6 | 119.2 |
| N3—Cu1—N2 | 176.79 (4) | C11—C7—C8 | 116.97 (13) |
| O6—Cu1—N1 | 170.57 (3) | C11—C7—C6 | 118.07 (14) |
| N3—Cu1—N1 | 95.44 (4) | C8—C7—C6 | 124.96 (13) |
| N2—Cu1—N1 | 81.59 (4) | C9—C8—C7 | 119.51 (12) |
| O6—Cu1—O3 | 89.77 (4) | C9—C8—H8 | 120.2 |
| N3—Cu1—O3 | 75.95 (3) | C7—C8—H8 | 120.2 |
| N2—Cu1—O3 | 102.61 (3) | C8—C9—C10 | 119.75 (14) |
| N1—Cu1—O3 | 87.43 (4) | C8—C9—H9 | 120.1 |
| O6—Cu1—O2 | 91.59 (3) | C10—C9—H9 | 120.1 |
| N3—Cu1—O2 | 74.28 (3) | N2—C10—C9 | 122.41 (13) |
| N2—Cu1—O2 | 107.13 (4) | N2—C10—H10 | 118.8 |
| N1—Cu1—O2 | 95.31 (4) | C9—C10—H10 | 118.8 |
| O3—Cu1—O2 | 150.22 (3) | N2—C11—C7 | 122.80 (12) |
| C1—N1—C12 | 118.64 (10) | N2—C11—C12 | 116.73 (9) |
| C1—N1—Cu1 | 129.51 (8) | C7—C11—C12 | 120.46 (11) |
| C12—N1—Cu1 | 111.52 (7) | N1—C12—C4 | 122.74 (12) |
| C10—N2—C11 | 118.55 (10) | N1—C12—C11 | 116.62 (9) |
| C10—N2—Cu1 | 128.30 (9) | C4—C12—C11 | 120.64 (11) |
| C11—N2—Cu1 | 112.97 (8) | O2—C13—O1 | 127.08 (10) |
| C14—N3—C18 | 119.19 (9) | O2—C13—C14 | 116.23 (9) |
| C14—N3—Cu1 | 121.39 (7) | O1—C13—C14 | 116.69 (10) |
| C18—N3—Cu1 | 119.41 (7) | N3—C14—C15 | 122.08 (9) |
| C13—O2—Cu1 | 112.17 (7) | N3—C14—C13 | 115.80 (9) |
| C19—O3—Cu1 | 111.29 (7) | C15—C14—C13 | 122.12 (9) |
| C16—O5—H5A | 111.1 | C14—C15—C16 | 118.69 (9) |
| Cu1—O6—H6D | 113.4 | C14—C15—H15 | 120.7 |
| Cu1—O6—H6C | 111.0 | C16—C15—H15 | 120.7 |
| H6D—O6—H6C | 101.1 | O5—C16—C17 | 122.54 (10) |
| N1—C1—C2 | 122.19 (13) | O5—C16—C15 | 118.30 (9) |
| N1—C1—H1 | 118.9 | C17—C16—C15 | 119.16 (9) |
| C2—C1—H1 | 118.9 | C18—C17—C16 | 118.41 (9) |
| C3—C2—C1 | 119.41 (15) | C18—C17—H17 | 120.8 |
| C3—C2—H2 | 120.3 | C16—C17—H17 | 120.8 |
| C1—C2—H2 | 120.3 | N3—C18—C17 | 122.45 (9) |
| C2—C3—C4 | 119.43 (12) | N3—C18—C19 | 115.64 (8) |
| C2—C3—H3 | 120.3 | C17—C18—C19 | 121.90 (9) |
| C4—C3—H3 | 120.3 | O3—C19—O4 | 125.54 (10) |
| C3—C4—C12 | 117.59 (12) | O3—C19—C18 | 117.16 (9) |
| C3—C4—C5 | 124.80 (12) | O4—C19—C18 | 117.27 (9) |
| C12—C4—C5 | 117.60 (13) | H2C—O2W—H2D | 102.1 |
| C6—C5—C4 | 121.61 (12) | H3D—O3W—H3C | 108.3 |
| C6—C5—H5 | 119.2 | H4D—O4W—H4C | 106.6 |
| C4—C5—H5 | 119.2 | H5C—O5W—H5D | 106.2 |
| C5—C6—C7 | 121.61 (13) | ||
| O6—Cu1—N1—C1 | −149.7 (2) | C7—C8—C9—C10 | −0.2 (2) |
| N3—Cu1—N1—C1 | −1.22 (12) | C11—N2—C10—C9 | −0.67 (18) |
| N2—Cu1—N1—C1 | −179.99 (13) | Cu1—N2—C10—C9 | −175.40 (10) |
| O3—Cu1—N1—C1 | −76.84 (12) | C8—C9—C10—N2 | 0.9 (2) |
| O2—Cu1—N1—C1 | 73.44 (12) | C10—N2—C11—C7 | −0.28 (16) |
| O6—Cu1—N1—C12 | 23.5 (3) | Cu1—N2—C11—C7 | 175.23 (9) |
| N3—Cu1—N1—C12 | 171.95 (8) | C10—N2—C11—C12 | −179.47 (11) |
| N2—Cu1—N1—C12 | −6.81 (8) | Cu1—N2—C11—C12 | −3.95 (13) |
| O3—Cu1—N1—C12 | 96.34 (8) | C8—C7—C11—N2 | 0.96 (17) |
| O2—Cu1—N1—C12 | −113.39 (8) | C6—C7—C11—N2 | −178.83 (11) |
| O6—Cu1—N2—C10 | 5.57 (10) | C8—C7—C11—C12 | −179.88 (12) |
| N3—Cu1—N2—C10 | 158.3 (7) | C6—C7—C11—C12 | 0.32 (17) |
| N1—Cu1—N2—C10 | −179.17 (11) | C1—N1—C12—C4 | 0.74 (18) |
| O3—Cu1—N2—C10 | 95.39 (10) | Cu1—N1—C12—C4 | −173.26 (9) |
| O2—Cu1—N2—C10 | −86.17 (10) | C1—N1—C12—C11 | −179.24 (11) |
| O6—Cu1—N2—C11 | −169.41 (8) | Cu1—N1—C12—C11 | 6.76 (13) |
| N3—Cu1—N2—C11 | −16.7 (7) | C3—C4—C12—N1 | −0.05 (19) |
| N1—Cu1—N2—C11 | 5.85 (8) | C5—C4—C12—N1 | −178.80 (11) |
| O3—Cu1—N2—C11 | −79.58 (8) | C3—C4—C12—C11 | 179.93 (12) |
| O2—Cu1—N2—C11 | 98.86 (8) | C5—C4—C12—C11 | 1.18 (18) |
| O6—Cu1—N3—C14 | −93.65 (8) | N2—C11—C12—N1 | −2.01 (16) |
| N2—Cu1—N3—C14 | 113.7 (7) | C7—C11—C12—N1 | 178.79 (10) |
| N1—Cu1—N3—C14 | 91.27 (8) | N2—C11—C12—C4 | 178.01 (10) |
| O3—Cu1—N3—C14 | 177.24 (8) | C7—C11—C12—C4 | −1.19 (17) |
| O2—Cu1—N3—C14 | −2.73 (7) | Cu1—O2—C13—O1 | 177.76 (10) |
| O6—Cu1—N3—C18 | 86.62 (8) | Cu1—O2—C13—C14 | −3.11 (12) |
| N2—Cu1—N3—C18 | −66.1 (7) | C18—N3—C14—C15 | 1.41 (15) |
| N1—Cu1—N3—C18 | −88.46 (8) | Cu1—N3—C14—C15 | −178.33 (7) |
| O3—Cu1—N3—C18 | −2.49 (8) | C18—N3—C14—C13 | −178.14 (9) |
| O2—Cu1—N3—C18 | 177.53 (8) | Cu1—N3—C14—C13 | 2.13 (12) |
| O6—Cu1—O2—C13 | 95.47 (8) | O2—C13—C14—N3 | 1.10 (14) |
| N3—Cu1—O2—C13 | 3.22 (8) | O1—C13—C14—N3 | −179.67 (10) |
| N2—Cu1—O2—C13 | −173.77 (8) | O2—C13—C14—C15 | −178.44 (10) |
| N1—Cu1—O2—C13 | −90.96 (8) | O1—C13—C14—C15 | 0.78 (15) |
| O3—Cu1—O2—C13 | 3.16 (12) | N3—C14—C15—C16 | −0.40 (15) |
| O6—Cu1—O3—C19 | −86.68 (9) | C13—C14—C15—C16 | 179.12 (10) |
| N3—Cu1—O3—C19 | 6.06 (8) | C14—C15—C16—O5 | −179.62 (10) |
| N2—Cu1—O3—C19 | −176.89 (8) | C14—C15—C16—C17 | −0.57 (15) |
| N1—Cu1—O3—C19 | 102.33 (9) | O5—C16—C17—C18 | 179.53 (10) |
| O2—Cu1—O3—C19 | 6.11 (13) | C15—C16—C17—C18 | 0.52 (15) |
| C12—N1—C1—C2 | −0.7 (2) | C14—N3—C18—C17 | −1.47 (15) |
| Cu1—N1—C1—C2 | 172.03 (11) | Cu1—N3—C18—C17 | 178.27 (8) |
| N1—C1—C2—C3 | 0.0 (2) | C14—N3—C18—C19 | 179.53 (9) |
| C1—C2—C3—C4 | 0.7 (2) | Cu1—N3—C18—C19 | −0.73 (12) |
| C2—C3—C4—C12 | −0.7 (2) | C16—C17—C18—N3 | 0.50 (15) |
| C2—C3—C4—C5 | 178.00 (14) | C16—C17—C18—C19 | 179.45 (9) |
| C3—C4—C5—C6 | −178.98 (15) | Cu1—O3—C19—O4 | 173.59 (10) |
| C12—C4—C5—C6 | −0.3 (2) | Cu1—O3—C19—C18 | −8.18 (12) |
| C4—C5—C6—C7 | −0.5 (2) | N3—C18—C19—O3 | 6.64 (14) |
| C5—C6—C7—C11 | 0.5 (2) | C17—C18—C19—O3 | −172.37 (10) |
| C5—C6—C7—C8 | −179.23 (14) | N3—C18—C19—O4 | −174.98 (10) |
| C11—C7—C8—C9 | −0.7 (2) | C17—C18—C19—O4 | 6.01 (15) |
| C6—C7—C8—C9 | 179.07 (14) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O5—H5A···O5W | 0.85 | 1.73 | 2.5771 (14) | 174 |
| O6—H6D···O1i | 0.85 | 1.88 | 2.7059 (12) | 162 |
| O6—H6C···O4ii | 0.85 | 1.76 | 2.6048 (11) | 174 |
| O1W—H1C···O1iii | 0.85 | 1.97 | 2.8154 (12) | 171 |
| O2W—H2C···O3iv | 0.85 | 2.37 | 3.1062 (16) | 145 |
| O2W—H2D···O4W | 0.85 | 1.90 | 2.7470 (16) | 172 |
| O3W—H3D···O2v | 0.85 | 1.93 | 2.7757 (13) | 175 |
| O3W—H3C···O4 | 0.85 | 1.90 | 2.7496 (13) | 175 |
| O4W—H4D···O3W | 0.85 | 1.84 | 2.6834 (15) | 171 |
| O4W—H4C···O5i | 0.85 | 2.26 | 3.0835 (16) | 165 |
| O5W—H5C···O1W | 0.85 | 2.03 | 2.8604 (15) | 164 |
| O5W—H5D···O2Wvi | 0.85 | 1.91 | 2.7204 (15) | 159 |
| C1—H1···O5vii | 0.93 | 2.41 | 3.1610 (18) | 137 |
| C3—H3···O3viii | 0.93 | 2.25 | 3.130 (2) | 158 |
| C8—H8···O3Wix | 0.93 | 2.42 | 3.315 (2) | 161 |
| C10—H10···O6 | 0.93 | 2.50 | 2.9995 (17) | 114 |
| C10—H10···O5Wx | 0.93 | 2.42 | 3.1809 (19) | 139 |
Symmetry codes: (i) −x+1/2, −y, z−1/2; (ii) −x+1/2, −y, z+1/2; (iii) −x+1, −y, z−1; (iv) x−1/4, −y+1/4, z−1/4; (v) x, y, z−1; (vi) −x+3/4, y−1/4, z+1/4; (vii) −x+1, −y, z; (viii) x+1/4, −y+1/4, z+1/4; (ix) x−1/4, −y+1/4, z+3/4; (x) −x+1/2, −y, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2032).
References
- Aghabozorg, H., Attar Gharamaleki, J., Ghadermazi, M., Ghasemikhah, P. & Soleimannejad, J. (2007). Acta Cryst. E63, m1803–m1804.
- Aghabozorg, H., Attar Gharamaleki, J., Ghasemikhah, P., Ghadermazi, M. & Soleimannejad, J. (2007). Acta Cryst. E63, m1710–m1711.
- Aghabozorg, H., Daneshvar, S., Motyeian, E., Ghadermazi, M. & Attar Gharamaleki, J. (2007). Acta Cryst. E63, m2468–m2469. [DOI] [PMC free article] [PubMed]
- Bruker (1998). SADABS (Version 2004/1), SAINT (Version 6.01) and SMART (Version 5.059). Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2005). SHELXTL Version 6.10. Bruker AXS Inc., Madison, Wisconsin, USA.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Sheldrick, G. M. (1997). SHELXS97 and SHELXL97 University of Göttingen, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536807067207/su2032sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807067207/su2032Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report