Abstract
The geometric parameters of the title compound, C4H6N3
+·C6H2N3O7
−, are in the usual ranges. While two nitro groups are almost coplanar with the aromatic picrate ring [dihedral angles 3.0 (2) and 4.4 (3)°], the third is significantly twisted out of this plane [dihedral angle 46.47 (8)°]. Anions and cations are connected via N—H⋯O hydrogen bonds. The molecules crystallize in planes parallel to (1 1).
1).
Related literature
For related literature, see: Barraclough & Smith (1995 ▶); Etter et al. (1990 ▶); Fischer et al. (2007 ▶); Goswami et al. (2000 ▶); Gueiffier et al. (1996 ▶); Katritzky et al. (2003 ▶); Rival et al. (1991 ▶); Sanfilippo et al. (1988 ▶); Scheinbeim & Schempp (1976 ▶); Schlueter et al. (2006 ▶); Tully et al. (1991 ▶); Yathirajan, Bindya et al. (2007a
▶,b
▶); Yathirajan, Mayekar et al. (2007 ▶); Yathirajan, Narayana et al. (2007 ▶).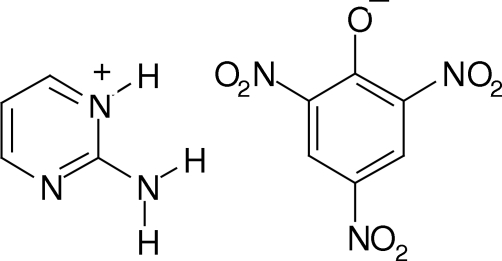
Experimental
Crystal data
C4H6N3 +·C6H2N3O7 −
M r = 324.22
Triclinic,
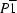
a = 5.8803 (7) Å
b = 8.0025 (10) Å
c = 13.8108 (17) Å
α = 88.021 (10)°
β = 82.322 (9)°
γ = 88.739 (10)°
V = 643.59 (14) Å3
Z = 2
Mo Kα radiation
μ = 0.14 mm−1
T = 173 (2) K
0.26 × 0.22 × 0.09 mm
Data collection
Stoe IPDSII two-circle diffractometer
Absorption correction: none
8757 measured reflections
2402 independent reflections
1927 reflections with I > 2σ(I)
R int = 0.042
Refinement
R[F 2 > 2σ(F 2)] = 0.036
wR(F 2) = 0.099
S = 1.01
2402 reflections
220 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.20 e Å−3
Δρmin = −0.27 e Å−3
Data collection: X-AREA (Stoe & Cie, 2001 ▶); cell refinement: X-AREA; data reduction: X-AREA; program(s) used to solve structure: SHELXS97 (Sheldrick, 1990 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997 ▶); molecular graphics: PLATON (Spek, 2003 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536807062599/at2509sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807062599/at2509Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯N6i | 0.87 (2) | 2.09 (2) | 2.958 (2) | 177.0 (18) |
| N1—H1B⋯O11 | 0.91 (2) | 1.97 (2) | 2.7577 (19) | 143.7 (18) |
| N1—H1B⋯O17 | 0.91 (2) | 2.50 (2) | 3.2488 (18) | 140.0 (17) |
| N2—H2⋯O11 | 0.90 (2) | 1.84 (2) | 2.6501 (16) | 148.6 (19) |
| N2—H2⋯O12 | 0.90 (2) | 2.31 (2) | 2.9792 (18) | 131.6 (17) |
Symmetry code: (i) 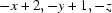 .
.
Acknowledgments
BN thanks Mangalore University for the use of research facilities.
supplementary crystallographic information
Comment
Pyrimidine is a heterocyclic aromatic organic compound similar to benzene and pyridine, containing two nitrogen atoms at positions 1 and 3 of the six-member ring. A pyrimidine has many properties in common with pyridine, as the number of nitrogen atoms in the ring increases the ring π-electrons become less energetic and electrophilic aromatic substitution gets more difficult while nucleophilic aromatic substitution gets easier. Pyrimidines are important compounds in pharmaceutical chemistry as antiviral agents (Gueiffier et al., 1996), inotropic and β-blocking agents (Barraclough & Smith, 1995), antifungal agents (Rival et al. 1991), benzodiazepine receptor agonists (Tully et al.1991), and calcium channel blockers (Sanfilippo et al., 1988). The synthesis of imidazo[1,2-a]pyrimidines has been widely investigated and one of the most common strategies uses 2-aminopyrimidine as the starting material (Katritzky et al., 2003). The crystal structures of the following compounds have been previously reported, viz; 2-aminopyrimidine (Scheinbeim & Schempp, 1976), 1:1 hetero-assembly of 2-aminopyrimidine and (+)-camphoric acid (Goswami, et al., 2000), 2-aminopyrimidine-succinic acid (1:1) cocrystal (Etter et al., 1990), 5-aminopyrimidine (Schlueter et al., 2006), 5-bromopyrimidin-2(1H)-one (Yathirajan, Narayana, Ashalatha et al., 2007), ethyl 7-methyl-2-[4-(methylsulfanyl)benzylidene]-5-[4-(methylsulfanyl)phenyl]-3-oxo-2, 3-dihydro-5H-thiazolo[3,2-a]pyrimidine-6-carboxylate (Fischer et al., 2007), 2-(4-methylbenzoyloxymethyl)-5-(5-methyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1 -yl)tetrahydrofuran-3-yl 4-methylbenzoate (Yathirajan, Mayekar, Sarojini et al., 2007), methyl (4-oxo-1-phenyl-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-5-yl)acetate (Yathirajan, Bindya, Sarojini et al., 2007a), ethyl (4-oxo-1-phenyl-1,4-dihydro-5H-pyrazolo[3,4-d]pyrimidin-5-yl)acetate (Yathirajan, Bindya, Sarojini et al., 2007b). In continuation to our work on picrates of biologically important molecules, we have prepared a new picrate of 2-aminopyrimidine, and its crystal structure is reported.
Geometric parameters of the title compound are in the usual ranges. Whereas two nitrogroups are almost coplanar with the aromatic picrate ring [dihedral angles 3.0 (2)° and 4.4 (3)°] the third one is significantly twisted [dihedral angle 46.47 (8)°] out of this plane Anions and cations are connected via N—H···O hydrogen bonds. The molecules crystallize in planes parallel to (1 - 2 1).
Experimental
2-Aminopyrimidine (0.95 g, 0.01 mol) was dissolved in 20 ml of ethanol. Picric acid (2.29 g, 0.01 mol) was dissolved in 10 ml of water. Both the solutions were mixed and to this, 5 ml of 5 M HCl was added and stirred for few minutes. The formed complex was filtered, dried and recrystallized from ethanol (m.p.: 413–415 K). Composition: Found (calculated): C 37.01(37.05), H 2.46(2.49), N 25.87% (25.92%).
Refinement
H atoms were found in a difference map, but those bonded to C were geometrically positioned and refined with fixed individual displacement parameters [Uiso(H) = 1.2 Ueq(C)] using a riding model with C—H = 0.95 Å. The amino H atoms were freely refined.
Figures
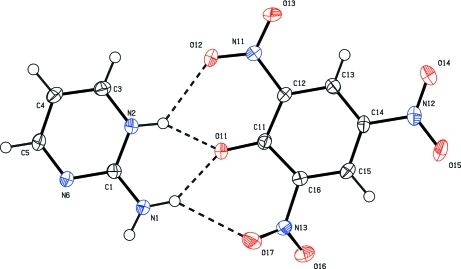
Fig. 1.
Perspective view of the title compound with the atom numbering; displacement ellipsoids are at the 50% probability level. The hydrogen bonds are shown as dashed lines.
Crystal data
| C4H6N3+·C6H2N3O7– | Z = 2 |
| Mr = 324.22 | F000 = 332 |
| Triclinic, P1 | Dx = 1.673 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation λ = 0.71073 Å |
| a = 5.8803 (7) Å | Cell parameters from 8213 reflections |
| b = 8.0025 (10) Å | θ = 3.5–25.8º |
| c = 13.8108 (17) Å | µ = 0.15 mm−1 |
| α = 88.021 (10)º | T = 173 (2) K |
| β = 82.322 (9)º | Plate, yellow |
| γ = 88.739 (10)º | 0.26 × 0.22 × 0.09 mm |
| V = 643.59 (14) Å3 |
Data collection
| Stoe IPDSII two-circle diffractometer | 1927 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.042 |
| Monochromator: graphite | θmax = 25.6º |
| T = 173(2) K | θmin = 3.5º |
| ω scans | h = −7→7 |
| Absorption correction: none | k = −9→9 |
| 8757 measured reflections | l = −16→16 |
| 2402 independent reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.036 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.099 | w = 1/[σ2(Fo2) + (0.0661P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max < 0.001 |
| 2402 reflections | Δρmax = 0.20 e Å−3 |
| 220 parameters | Δρmin = −0.27 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.7277 (3) | 0.40774 (18) | 0.07873 (11) | 0.0279 (3) | |
| H1A | 0.799 (3) | 0.422 (2) | 0.0195 (16) | 0.033 (5)* | |
| H1B | 0.600 (4) | 0.346 (3) | 0.0984 (15) | 0.039 (5)* | |
| C1 | 0.8282 (3) | 0.47463 (18) | 0.14768 (11) | 0.0199 (3) | |
| N2 | 0.7333 (2) | 0.45943 (15) | 0.24319 (9) | 0.0206 (3) | |
| H2 | 0.604 (4) | 0.400 (3) | 0.2556 (15) | 0.039 (5)* | |
| C3 | 0.8321 (3) | 0.52828 (18) | 0.31540 (11) | 0.0229 (3) | |
| H3 | 0.7618 | 0.5183 | 0.3814 | 0.027* | |
| C4 | 1.0334 (3) | 0.61214 (19) | 0.29261 (11) | 0.0241 (3) | |
| H4 | 1.1076 | 0.6609 | 0.3415 | 0.029* | |
| C5 | 1.1247 (3) | 0.62249 (18) | 0.19338 (12) | 0.0229 (3) | |
| H5 | 1.2654 | 0.6791 | 0.1762 | 0.027* | |
| N6 | 1.0274 (2) | 0.55855 (15) | 0.12224 (9) | 0.0228 (3) | |
| C11 | 0.2209 (2) | 0.17922 (17) | 0.24085 (11) | 0.0194 (3) | |
| C12 | 0.0995 (2) | 0.17467 (18) | 0.33896 (11) | 0.0196 (3) | |
| C13 | −0.1064 (2) | 0.09207 (18) | 0.36532 (11) | 0.0201 (3) | |
| H13 | −0.1809 | 0.0931 | 0.4307 | 0.024* | |
| C14 | −0.2010 (2) | 0.00844 (17) | 0.29461 (11) | 0.0197 (3) | |
| C15 | −0.0957 (3) | 0.00409 (18) | 0.19783 (11) | 0.0209 (3) | |
| H15 | −0.1612 | −0.0560 | 0.1506 | 0.025* | |
| C16 | 0.1043 (2) | 0.08881 (18) | 0.17311 (11) | 0.0197 (3) | |
| N11 | 0.1908 (2) | 0.26127 (16) | 0.41664 (10) | 0.0238 (3) | |
| N12 | −0.4142 (2) | −0.08122 (16) | 0.32163 (10) | 0.0250 (3) | |
| N13 | 0.2118 (2) | 0.08312 (16) | 0.07100 (9) | 0.0226 (3) | |
| O11 | 0.41007 (18) | 0.24842 (14) | 0.21384 (8) | 0.0276 (3) | |
| O12 | 0.3751 (2) | 0.33240 (17) | 0.39985 (9) | 0.0371 (3) | |
| O13 | 0.0778 (2) | 0.2607 (2) | 0.49744 (9) | 0.0534 (4) | |
| O14 | −0.5137 (2) | −0.07121 (17) | 0.40512 (9) | 0.0396 (3) | |
| O15 | −0.4859 (2) | −0.16322 (15) | 0.25825 (10) | 0.0366 (3) | |
| O16 | 0.2234 (2) | −0.05385 (14) | 0.03198 (9) | 0.0323 (3) | |
| O17 | 0.2799 (2) | 0.21390 (15) | 0.02874 (9) | 0.0334 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0296 (8) | 0.0360 (8) | 0.0185 (7) | −0.0172 (6) | −0.0015 (6) | −0.0027 (6) |
| C1 | 0.0220 (7) | 0.0191 (7) | 0.0183 (7) | −0.0042 (5) | −0.0013 (6) | −0.0009 (6) |
| N2 | 0.0207 (6) | 0.0211 (6) | 0.0201 (7) | −0.0059 (5) | −0.0014 (5) | −0.0018 (5) |
| C3 | 0.0273 (8) | 0.0228 (7) | 0.0186 (8) | −0.0021 (6) | −0.0026 (6) | −0.0037 (6) |
| C4 | 0.0275 (8) | 0.0234 (7) | 0.0227 (8) | −0.0042 (6) | −0.0063 (6) | −0.0054 (6) |
| C5 | 0.0226 (7) | 0.0211 (7) | 0.0254 (8) | −0.0073 (6) | −0.0034 (6) | −0.0035 (6) |
| N6 | 0.0244 (7) | 0.0235 (6) | 0.0206 (7) | −0.0087 (5) | −0.0015 (5) | −0.0031 (5) |
| C11 | 0.0190 (7) | 0.0181 (7) | 0.0215 (8) | −0.0028 (5) | −0.0030 (6) | −0.0020 (6) |
| C12 | 0.0205 (7) | 0.0205 (7) | 0.0187 (8) | −0.0034 (6) | −0.0049 (6) | −0.0030 (6) |
| C13 | 0.0200 (7) | 0.0199 (7) | 0.0199 (8) | −0.0011 (5) | −0.0007 (6) | 0.0001 (6) |
| C14 | 0.0158 (7) | 0.0179 (7) | 0.0255 (8) | −0.0042 (5) | −0.0022 (6) | −0.0012 (6) |
| C15 | 0.0221 (7) | 0.0193 (7) | 0.0226 (8) | −0.0019 (5) | −0.0063 (6) | −0.0048 (6) |
| C16 | 0.0208 (7) | 0.0208 (7) | 0.0176 (8) | −0.0014 (6) | −0.0025 (6) | −0.0018 (6) |
| N11 | 0.0244 (7) | 0.0276 (7) | 0.0196 (7) | −0.0063 (5) | −0.0023 (5) | −0.0039 (5) |
| N12 | 0.0193 (6) | 0.0241 (6) | 0.0316 (8) | −0.0048 (5) | −0.0031 (5) | −0.0008 (6) |
| N13 | 0.0212 (6) | 0.0270 (7) | 0.0201 (7) | −0.0026 (5) | −0.0035 (5) | −0.0052 (5) |
| O11 | 0.0226 (6) | 0.0368 (6) | 0.0233 (6) | −0.0141 (5) | 0.0006 (4) | −0.0050 (5) |
| O12 | 0.0342 (7) | 0.0518 (8) | 0.0267 (6) | −0.0260 (6) | −0.0030 (5) | −0.0070 (5) |
| O13 | 0.0449 (8) | 0.0925 (12) | 0.0221 (7) | −0.0335 (7) | 0.0095 (6) | −0.0245 (7) |
| O14 | 0.0298 (7) | 0.0545 (8) | 0.0322 (7) | −0.0175 (6) | 0.0075 (5) | −0.0038 (6) |
| O15 | 0.0306 (6) | 0.0386 (7) | 0.0425 (8) | −0.0170 (5) | −0.0067 (5) | −0.0107 (6) |
| O16 | 0.0375 (7) | 0.0321 (6) | 0.0272 (6) | −0.0035 (5) | −0.0006 (5) | −0.0137 (5) |
| O17 | 0.0422 (7) | 0.0339 (6) | 0.0227 (6) | −0.0089 (5) | 0.0017 (5) | 0.0014 (5) |
Geometric parameters (Å, °)
| N1—C1 | 1.320 (2) | C12—C13 | 1.392 (2) |
| N1—H1A | 0.87 (2) | C12—N11 | 1.4631 (19) |
| N1—H1B | 0.91 (2) | C13—C14 | 1.385 (2) |
| C1—N6 | 1.3622 (19) | C13—H13 | 0.9500 |
| C1—N2 | 1.3643 (19) | C14—C15 | 1.397 (2) |
| N2—C3 | 1.3572 (19) | C14—N12 | 1.4568 (18) |
| N2—H2 | 0.90 (2) | C15—C16 | 1.368 (2) |
| C3—C4 | 1.368 (2) | C15—H15 | 0.9500 |
| C3—H3 | 0.9500 | C16—N13 | 1.4682 (19) |
| C4—C5 | 1.404 (2) | N11—O13 | 1.2202 (18) |
| C4—H4 | 0.9500 | N11—O12 | 1.2262 (17) |
| C5—N6 | 1.324 (2) | N12—O14 | 1.2258 (18) |
| C5—H5 | 0.9500 | N12—O15 | 1.2349 (17) |
| C11—O11 | 1.2588 (18) | N13—O17 | 1.2289 (17) |
| C11—C12 | 1.444 (2) | N13—O16 | 1.2345 (17) |
| C11—C16 | 1.452 (2) | ||
| C1—N1—H1A | 115.1 (13) | C13—C12—N11 | 116.45 (13) |
| C1—N1—H1B | 117.1 (13) | C11—C12—N11 | 120.10 (12) |
| H1A—N1—H1B | 127.5 (19) | C14—C13—C12 | 118.89 (14) |
| N1—C1—N6 | 119.19 (14) | C14—C13—H13 | 120.6 |
| N1—C1—N2 | 120.24 (13) | C12—C13—H13 | 120.6 |
| N6—C1—N2 | 120.57 (13) | C13—C14—C15 | 122.04 (13) |
| C3—N2—C1 | 121.42 (13) | C13—C14—N12 | 119.44 (14) |
| C3—N2—H2 | 122.1 (13) | C15—C14—N12 | 118.50 (13) |
| C1—N2—H2 | 116.5 (13) | C16—C15—C14 | 118.09 (13) |
| N2—C3—C4 | 119.58 (14) | C16—C15—H15 | 121.0 |
| N2—C3—H3 | 120.2 | C14—C15—H15 | 121.0 |
| C4—C3—H3 | 120.2 | C15—C16—C11 | 124.88 (14) |
| C3—C4—C5 | 116.62 (14) | C15—C16—N13 | 117.24 (13) |
| C3—C4—H4 | 121.7 | C11—C16—N13 | 117.85 (12) |
| C5—C4—H4 | 121.7 | O13—N11—O12 | 121.68 (13) |
| N6—C5—C4 | 124.25 (14) | O13—N11—C12 | 118.02 (12) |
| N6—C5—H5 | 117.9 | O12—N11—C12 | 120.30 (13) |
| C4—C5—H5 | 117.9 | O14—N12—O15 | 123.36 (13) |
| C5—N6—C1 | 117.53 (13) | O14—N12—C14 | 118.93 (13) |
| O11—C11—C12 | 125.89 (13) | O15—N12—C14 | 117.70 (13) |
| O11—C11—C16 | 121.46 (13) | O17—N13—O16 | 123.85 (13) |
| C12—C11—C16 | 112.63 (13) | O17—N13—C16 | 118.75 (12) |
| C13—C12—C11 | 123.45 (13) | O16—N13—C16 | 117.38 (12) |
| N1—C1—N2—C3 | −179.40 (14) | C14—C15—C16—C11 | 2.1 (2) |
| N6—C1—N2—C3 | 0.7 (2) | C14—C15—C16—N13 | −179.82 (13) |
| C1—N2—C3—C4 | −1.3 (2) | O11—C11—C16—C15 | 176.85 (14) |
| N2—C3—C4—C5 | 0.6 (2) | C12—C11—C16—C15 | −1.5 (2) |
| C3—C4—C5—N6 | 0.7 (2) | O11—C11—C16—N13 | −1.2 (2) |
| C4—C5—N6—C1 | −1.3 (2) | C12—C11—C16—N13 | −179.53 (12) |
| N1—C1—N6—C5 | −179.32 (15) | C13—C12—N11—O13 | −2.7 (2) |
| N2—C1—N6—C5 | 0.6 (2) | C11—C12—N11—O13 | 176.73 (15) |
| O11—C11—C12—C13 | −178.11 (14) | C13—C12—N11—O12 | 177.68 (14) |
| C16—C11—C12—C13 | 0.2 (2) | C11—C12—N11—O12 | −2.9 (2) |
| O11—C11—C12—N11 | 2.5 (2) | C13—C14—N12—O14 | 4.9 (2) |
| C16—C11—C12—N11 | −179.19 (12) | C15—C14—N12—O14 | −176.38 (14) |
| C11—C12—C13—C14 | 0.4 (2) | C13—C14—N12—O15 | −175.46 (14) |
| N11—C12—C13—C14 | 179.81 (13) | C15—C14—N12—O15 | 3.2 (2) |
| C12—C13—C14—C15 | 0.2 (2) | C15—C16—N13—O17 | 133.79 (15) |
| C12—C13—C14—N12 | 178.82 (13) | C11—C16—N13—O17 | −48.03 (19) |
| C13—C14—C15—C16 | −1.4 (2) | C15—C16—N13—O16 | −44.61 (19) |
| N12—C14—C15—C16 | 179.93 (13) | C11—C16—N13—O16 | 133.57 (14) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···N6i | 0.87 (2) | 2.09 (2) | 2.958 (2) | 177.0 (18) |
| N1—H1B···O11 | 0.91 (2) | 1.97 (2) | 2.7577 (19) | 143.7 (18) |
| N1—H1B···O17 | 0.91 (2) | 2.50 (2) | 3.2488 (18) | 140.0 (17) |
| N2—H2···O11 | 0.90 (2) | 1.84 (2) | 2.6501 (16) | 148.6 (19) |
| N2—H2···O12 | 0.90 (2) | 2.31 (2) | 2.9792 (18) | 131.6 (17) |
Symmetry codes: (i) −x+2, −y+1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: AT2509).
References
- Barraclough, P. & Smith, S. (1995). J. Chem. Res. pp. 56–59.
- Etter, M. C., Adsmond, D. A. & Britton, D. (1990). Acta Cryst. C46, 933–934.
- Fischer, A., Yathirajan, H. S., Mithun, A., Bindya, S. & Narayana, B. (2007). Acta Cryst. E63, o1224–o1225.
- Goswami, S., Mukherjee, R., Ghosh, K., Razak, I. A., Shanmuga Sundara Raj, S. & Fun, H.-K. (2000). Acta Cryst. C56, 477–478. [DOI] [PubMed]
- Gueiffier, A., Lhassani, M., Elhakmaoui, A., Snoeck, R., Andrei, G., Chavignon, O., Teulade, J.-C., Kerbal, A., Essassi, E. M., Debouzy, J.-C., Witvrouw, M., Blache, Y., De Balzarini, J., Clercq, E. & Chapat, J.-P. (1996). J. Med. Chem.39, 2856–2859. [DOI] [PubMed]
- Katritzky, A. R., Xu, Y.-J. & Tu, H. (2003). J. Org. Chem.68, 4935–3937. [DOI] [PubMed]
- Rival, Y., Grassy, G., Taudou, A. & Ecalle, R. (1991). Eur. J. Med. Chem.26, 13–18.
- Sanfilippo, P. J., Urbanski, M., Press, J. B., Dubinsky, B. & Moore, J. B. Jr (1988). J. Med. Chem.31, 2221–2227. [DOI] [PubMed]
- Scheinbeim, J. & Schempp, E. (1976). Acta Cryst. B32, 607–609.
- Schlueter, J. A., Funk, R. J. & Geiser, U. (2006). Acta Cryst. E62, o339–o341.
- Sheldrick, G. M. (1990). Acta Cryst. A46, 467–473.
- Sheldrick, G. M. (1997). SHELXL97 University of Göttingen, Germany.
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
- Stoe & Cie (2001). X-AREA Stoe & Cie, Darmstadt, Germany.
- Tully, W. R., Gardner, C. R., Gillespie, R. J. & Westwood, R. (1991). J. Med. Chem.34, 2060–2067. [DOI] [PubMed]
- Yathirajan, H. S., Bindya, S., Sarojini, B. K., Narayana, B. & Bolte, M. (2007a). Acta Cryst. E63, o2566.
- Yathirajan, H. S., Bindya, S., Sarojini, B. K., Narayana, B. & Bolte, M. (2007b). Acta Cryst. E63, o2718.
- Yathirajan, H. S., Mayekar, A. N., Sarojini, B. K., Narayana, B. & Bolte, M. (2007). Acta Cryst. E63, o1395–o1397.
- Yathirajan, H. S., Narayana, B., Ashalatha, B. V., Sarojini, B. K. & Bolte, M. (2007). Acta Cryst. E63, o923–o924.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536807062599/at2509sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807062599/at2509Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report