Abstract
The title compound, C12H15NO, a degradation product of molindone hydrochloride, was prepared by the reaction of molindone with methyl iodide and subsequent reaction of the resulting quaternary ammonium salt with 2N aqueous sodium hydroxide. The newly formed double bond is exocyclic in nature and the carbonyl group is conjugated with the π-electrons of the pyrrole ring. The six-membered ring is in the half-chair conformation. The H atom attached to the N atom is involved in an intermolecular hydrogen bond with the O atom of a screw-related molecule, thus forming a continuous chain.
Related literature
For related literature, see: Dudzinski et al. (1973 ▶).
Experimental
Crystal data
C12H15NO
M r = 189.25
Monoclinic,
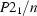
a = 9.0451 (3) Å
b = 8.5840 (3) Å
c = 14.3557 (5) Å
β = 107.355 (1)°
V = 1063.88 (6) Å3
Z = 4
Cu Kα radiation
μ = 0.59 mm−1
T = 90.0 (2) K
0.15 × 0.12 × 0.10 mm
Data collection
Bruker X8 Proteum diffractometer
Absorption correction: multi-scan (SADABS in APEX2; Bruker, 2006 ▶) T min = 0.837, T max = 0.944
15160 measured reflections
1972 independent reflections
1893 reflections with I > 2σ(I)
R int = 0.039
Refinement
R[F 2 > 2σ(F 2)] = 0.038
wR(F 2) = 0.097
S = 1.05
1972 reflections
129 parameters
H-atom parameters constrained
Δρmax = 0.24 e Å−3
Δρmin = −0.20 e Å−3
Data collection: APEX2 (Bruker, 2006 ▶); cell refinement: APEX2; data reduction: APEX2; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 1995 ▶); software used to prepare material for publication: SHELX97 and local procedures.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536807063076/om2194sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807063076/om2194Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1i | 0.88 | 1.91 | 2.7749 (12) | 169 |
Symmetry code: (i) 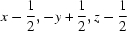 .
.
supplementary crystallographic information
Comment
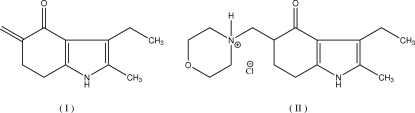
The Mannich condensation reaction is frequently used in the synthesis of pharmaceutical compounds. One such example is the synthesis of molindone, an antipsychotic agent. During stability studies and development of an assay for molindone hydrochloride, a degradation product was identified as 3-ethyl-2-methyl-5-methylene-6,7-dihydro-5H-indol-4-one (Dudzinski et al., 1973). Molindone has UV absorption peaks at 255 nm and 299 nm; these UV wavelengths can be used quantitatively for quantifying the drug substance. However, preliminary studies indicated that chemical degradation (as evidenced by color and precipitate formation) was not accompanied by a decrease in UV absorption, suggesting that the degradation product had a similar chromophore to molindone. The title compound was prepared by the reaction of molindone free base with methyl iodide and subsequent reaction of the resulting quaternary ammonium salt with 2 N aqueous sodium hydroxide. The structure of the resulting compound, 3-ethyl-2-methyl-5-methylene-6,7-dihydro-5H-indol-4-one, was initially characterized by NMR spectroscopy and shown to be identical to the degradation product of molindone hydrochloride. To confirm the exocyclic nature of newly formed double bond and to identify chromophoric group in the molecule responsible for its UV absorption profile, its crystal structure was determined by X-ray analysis.
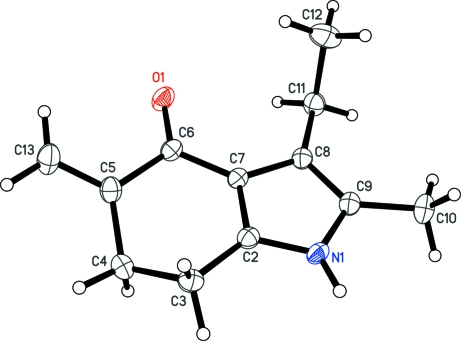
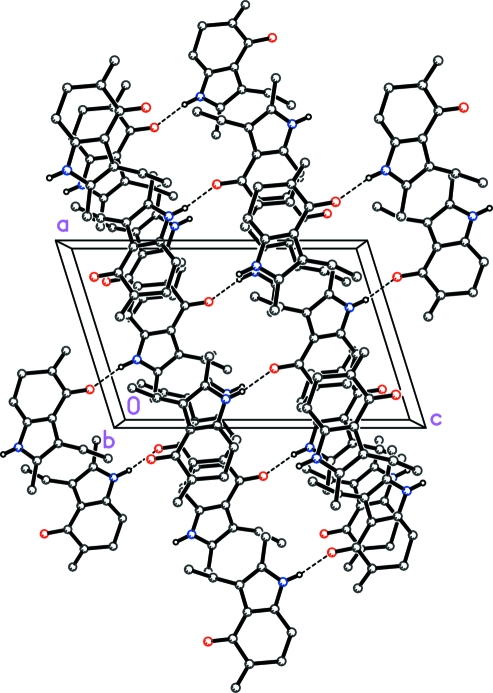
The molecular structure and the atom-numbering scheme are shown in Fig. 1. The bond length C4—C5 [1.5115 (16) Å] indicates that the newly formed double bond is exocyclic in nature. Further, it is evident from the bond lengths of C5—C6 and C6—C7 [1.5058 (15) and 1.4327 (15) Å, respectively] that the carbonyl group is conjugated with the π-electrons of pyrrole ring and not π-electrons of the exocyclic double. This explains why molindone and its degradation product, the title compound exhibit similar UV absorption. The mode of packing along the b direction is illustrated in Fig. 2. The H atom attached to atom N1 is involved in an intermolecular hydrogen bond [2.7749 (12) Å] with atom O1 of an inversion-related molecule, thus forming a continuous chain.
Experimental
A mixture of molindone (0.276 g, 1 mmol) and excess methyl iodide (2 ml) was stirred at ambient temperature. After completion of the reaction, unreacted methyl iodide was evaporated, and the crude quaternary ammonium salt was then mixed with 2 N aqueous sodium hydroxide (10 ml) and stirred for 1 h at ambient temperature. The resulting precipitate was collected by filtration and washed with water. Recrystallization from ethanol afforded the title compound as colorless crystalline product, which was suitable for X-ray analysis. Compound I: 1H NMR (400 MHz, CDCl3, p.p.m): δ 1.15 (t, J = 7.6 Hz, 3H), 2.16 (s, 3H), 2.70 (q, J = 7.6 Hz, 2H), 2.83 (s, 4H), 5.29 (d, J = 1.6 Hz, 1H), 6.03 (d, J = 1.6 Hz, 1H), 8.43 (sb, 1H); 13C NMR (75 MHz, CDCl3, p.p.m.): δ 10.68, 15.83, 18.41, 23.65, 32.13, 118.66, 119.16, 121.58, 124.74, 142.28, 144.84, 184.13.
Refinement
All H atoms were found in difference Fourier maps and but were subsequently placed in idealized positions with constrained distances of 0.98 Å (RCH3), 0.99 Å (R2CH2), 0.95 Å (RCsp2H2) and 0.88 Å (NH). Uiso(H) values were set to either 1.2Ueq or 1.5Ueq (RCH3 only) of the attached atom.
Figures
Fig. 1.
A view of the title compound I showing atom numbering scheme. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
A packing diagram viewed down the b axis, showing hydrogen bonding interactions (dashed lines). For clarity, only those H atoms involved in hydrogen bonding are shown.
Fig. 3.
Compounds (I) and (II).
Crystal data
| C12H15NO | F000 = 408 |
| Mr = 189.25 | Dx = 1.182 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation λ = 1.54178 Å |
| Hall symbol: -P 2yn | Cell parameters from 9990 reflections |
| a = 9.0451 (3) Å | θ = 3.2–69.4º |
| b = 8.5840 (3) Å | µ = 0.59 mm−1 |
| c = 14.3557 (5) Å | T = 90.0 (2) K |
| β = 107.355 (1)º | Block, colourless |
| V = 1063.88 (6) Å3 | 0.15 × 0.12 × 0.10 mm |
| Z = 4 |
Data collection
| Bruker X8 Proteum diffractometer | 1972 independent reflections |
| Radiation source: fine-focus rotating anode | 1893 reflections with I > 2σ(I) |
| Monochromator: graded multilayer optics | Rint = 0.039 |
| Detector resolution: 18 pixels mm-1 | θmax = 69.4º |
| T = 90.0(2) K | θmin = 5.2º |
| φ and ω scans | h = −10→10 |
| Absorption correction: multi-scan(SADABS in APEX2; Bruker, 2006) | k = −10→10 |
| Tmin = 0.837, Tmax = 0.944 | l = −17→17 |
| 15160 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.038 | H-atom parameters constrained |
| wR(F2) = 0.097 | w = 1/[σ2(Fo2) + (0.0464P)2 + 0.432P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 1972 reflections | Δρmax = 0.24 e Å−3 |
| 129 parameters | Δρmin = −0.20 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.68573 (9) | 0.20035 (10) | 0.42456 (6) | 0.0247 (2) | |
| N1 | 0.35484 (10) | 0.23361 (11) | 0.11591 (6) | 0.0186 (2) | |
| H1 | 0.3128 | 0.2575 | 0.0540 | 0.022* | |
| C2 | 0.49707 (13) | 0.27649 (13) | 0.17035 (8) | 0.0181 (2) | |
| C3 | 0.60968 (13) | 0.37109 (14) | 0.13708 (8) | 0.0232 (3) | |
| H3A | 0.5914 | 0.4835 | 0.1445 | 0.028* | |
| H3B | 0.5977 | 0.3499 | 0.0674 | 0.028* | |
| C4 | 0.77279 (13) | 0.32599 (14) | 0.20002 (8) | 0.0239 (3) | |
| H4A | 0.7983 | 0.2212 | 0.1805 | 0.029* | |
| H4B | 0.8484 | 0.4006 | 0.1879 | 0.029* | |
| C5 | 0.78799 (13) | 0.32500 (13) | 0.30777 (8) | 0.0212 (3) | |
| C6 | 0.66379 (13) | 0.24233 (12) | 0.33901 (8) | 0.0184 (2) | |
| C7 | 0.52110 (12) | 0.21662 (12) | 0.26362 (8) | 0.0168 (2) | |
| C8 | 0.38323 (12) | 0.13283 (12) | 0.26393 (8) | 0.0179 (3) | |
| C9 | 0.28372 (13) | 0.14590 (12) | 0.17184 (8) | 0.0190 (3) | |
| C10 | 0.12335 (13) | 0.08583 (15) | 0.12770 (9) | 0.0270 (3) | |
| H10A | 0.1004 | 0.0069 | 0.1708 | 0.041* | |
| H10B | 0.1149 | 0.0391 | 0.0640 | 0.041* | |
| H10C | 0.0494 | 0.1719 | 0.1193 | 0.041* | |
| C11 | 0.35382 (13) | 0.04614 (13) | 0.34712 (8) | 0.0224 (3) | |
| H11A | 0.4509 | −0.0057 | 0.3850 | 0.027* | |
| H11B | 0.2761 | −0.0362 | 0.3205 | 0.027* | |
| C12 | 0.29655 (17) | 0.14824 (17) | 0.41558 (10) | 0.0342 (3) | |
| H12A | 0.3746 | 0.2276 | 0.4444 | 0.051* | |
| H12B | 0.2784 | 0.0839 | 0.4675 | 0.051* | |
| H12C | 0.1996 | 0.1991 | 0.3790 | 0.051* | |
| C13 | 0.90486 (14) | 0.39164 (15) | 0.37417 (9) | 0.0283 (3) | |
| H13A | 0.9101 | 0.3872 | 0.4412 | 0.034* | |
| H13B | 0.9835 | 0.4439 | 0.3548 | 0.034* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0246 (4) | 0.0296 (5) | 0.0160 (4) | −0.0050 (3) | 0.0003 (3) | 0.0018 (3) |
| N1 | 0.0188 (5) | 0.0208 (5) | 0.0137 (4) | 0.0021 (4) | 0.0010 (4) | −0.0001 (3) |
| C2 | 0.0186 (5) | 0.0177 (5) | 0.0177 (5) | 0.0014 (4) | 0.0046 (4) | −0.0012 (4) |
| C3 | 0.0249 (6) | 0.0265 (6) | 0.0184 (5) | −0.0025 (5) | 0.0070 (5) | 0.0024 (4) |
| C4 | 0.0204 (6) | 0.0266 (6) | 0.0260 (6) | −0.0032 (5) | 0.0091 (5) | 0.0003 (5) |
| C5 | 0.0180 (5) | 0.0193 (5) | 0.0245 (6) | 0.0003 (4) | 0.0040 (4) | 0.0025 (4) |
| C6 | 0.0198 (6) | 0.0166 (5) | 0.0174 (5) | 0.0004 (4) | 0.0033 (4) | −0.0008 (4) |
| C7 | 0.0172 (5) | 0.0165 (5) | 0.0158 (5) | 0.0003 (4) | 0.0037 (4) | −0.0009 (4) |
| C8 | 0.0176 (5) | 0.0163 (5) | 0.0194 (5) | 0.0001 (4) | 0.0049 (4) | −0.0012 (4) |
| C9 | 0.0178 (5) | 0.0169 (5) | 0.0212 (6) | 0.0005 (4) | 0.0043 (4) | −0.0018 (4) |
| C10 | 0.0189 (6) | 0.0272 (6) | 0.0304 (6) | −0.0025 (5) | 0.0002 (5) | −0.0010 (5) |
| C11 | 0.0221 (6) | 0.0218 (6) | 0.0233 (6) | −0.0024 (4) | 0.0067 (4) | 0.0028 (4) |
| C12 | 0.0415 (8) | 0.0368 (7) | 0.0303 (7) | 0.0054 (6) | 0.0200 (6) | 0.0059 (5) |
| C13 | 0.0245 (6) | 0.0282 (6) | 0.0280 (6) | −0.0066 (5) | 0.0011 (5) | 0.0058 (5) |
Geometric parameters (Å, °)
| O1—C6 | 1.2377 (14) | C7—C8 | 1.4407 (15) |
| N1—C2 | 1.3423 (14) | C8—C9 | 1.3640 (15) |
| N1—C9 | 1.3905 (14) | C8—C11 | 1.4968 (15) |
| N1—H1 | 0.8800 | C9—C10 | 1.4911 (15) |
| C2—C7 | 1.3894 (15) | C10—H10A | 0.9800 |
| C2—C3 | 1.4883 (15) | C10—H10B | 0.9800 |
| C3—C4 | 1.5322 (16) | C10—H10C | 0.9800 |
| C3—H3A | 0.9900 | C11—C12 | 1.5185 (17) |
| C3—H3B | 0.9900 | C11—H11A | 0.9900 |
| C4—C5 | 1.5115 (16) | C11—H11B | 0.9900 |
| C4—H4A | 0.9900 | C12—H12A | 0.9800 |
| C4—H4B | 0.9900 | C12—H12B | 0.9800 |
| C5—C13 | 1.3237 (17) | C12—H12C | 0.9800 |
| C5—C6 | 1.5058 (15) | C13—H13A | 0.9500 |
| C6—C7 | 1.4327 (15) | C13—H13B | 0.9500 |
| C2—N1—C9 | 109.90 (9) | C9—C8—C7 | 106.11 (9) |
| C2—N1—H1 | 125.1 | C9—C8—C11 | 126.25 (10) |
| C9—N1—H1 | 125.1 | C7—C8—C11 | 127.64 (10) |
| N1—C2—C7 | 107.88 (10) | C8—C9—N1 | 108.61 (9) |
| N1—C2—C3 | 126.27 (10) | C8—C9—C10 | 131.34 (11) |
| C7—C2—C3 | 125.85 (10) | N1—C9—C10 | 120.05 (10) |
| C2—C3—C4 | 107.68 (9) | C9—C10—H10A | 109.5 |
| C2—C3—H3A | 110.2 | C9—C10—H10B | 109.5 |
| C4—C3—H3A | 110.2 | H10A—C10—H10B | 109.5 |
| C2—C3—H3B | 110.2 | C9—C10—H10C | 109.5 |
| C4—C3—H3B | 110.2 | H10A—C10—H10C | 109.5 |
| H3A—C3—H3B | 108.5 | H10B—C10—H10C | 109.5 |
| C5—C4—C3 | 112.49 (9) | C8—C11—C12 | 113.92 (10) |
| C5—C4—H4A | 109.1 | C8—C11—H11A | 108.8 |
| C3—C4—H4A | 109.1 | C12—C11—H11A | 108.8 |
| C5—C4—H4B | 109.1 | C8—C11—H11B | 108.8 |
| C3—C4—H4B | 109.1 | C12—C11—H11B | 108.8 |
| H4A—C4—H4B | 107.8 | H11A—C11—H11B | 107.7 |
| C13—C5—C6 | 119.71 (11) | C11—C12—H12A | 109.5 |
| C13—C5—C4 | 122.95 (11) | C11—C12—H12B | 109.5 |
| C6—C5—C4 | 117.34 (10) | H12A—C12—H12B | 109.5 |
| O1—C6—C7 | 123.12 (10) | C11—C12—H12C | 109.5 |
| O1—C6—C5 | 121.31 (10) | H12A—C12—H12C | 109.5 |
| C7—C6—C5 | 115.57 (9) | H12B—C12—H12C | 109.5 |
| C2—C7—C6 | 121.18 (10) | C5—C13—H13A | 120.0 |
| C2—C7—C8 | 107.51 (9) | C5—C13—H13B | 120.0 |
| C6—C7—C8 | 131.31 (10) | H13A—C13—H13B | 120.0 |
| C9—N1—C2—C7 | 0.04 (12) | C5—C6—C7—C2 | −5.13 (15) |
| C9—N1—C2—C3 | 179.92 (10) | O1—C6—C7—C8 | −5.42 (19) |
| N1—C2—C3—C4 | −152.52 (11) | C5—C6—C7—C8 | 174.62 (10) |
| C7—C2—C3—C4 | 27.34 (15) | C2—C7—C8—C9 | 0.04 (12) |
| C2—C3—C4—C5 | −48.30 (13) | C6—C7—C8—C9 | −179.74 (11) |
| C3—C4—C5—C13 | −133.03 (12) | C2—C7—C8—C11 | 179.14 (10) |
| C3—C4—C5—C6 | 47.57 (14) | C6—C7—C8—C11 | −0.63 (19) |
| C13—C5—C6—O1 | −18.54 (17) | C7—C8—C9—N1 | −0.02 (12) |
| C4—C5—C6—O1 | 160.88 (10) | C11—C8—C9—N1 | −179.14 (10) |
| C13—C5—C6—C7 | 161.42 (11) | C7—C8—C9—C10 | −179.29 (11) |
| C4—C5—C6—C7 | −19.16 (14) | C11—C8—C9—C10 | 1.6 (2) |
| N1—C2—C7—C6 | 179.76 (9) | C2—N1—C9—C8 | −0.01 (12) |
| C3—C2—C7—C6 | −0.13 (17) | C2—N1—C9—C10 | 179.35 (10) |
| N1—C2—C7—C8 | −0.05 (12) | C9—C8—C11—C12 | −96.71 (14) |
| C3—C2—C7—C8 | −179.93 (10) | C7—C8—C11—C12 | 84.36 (14) |
| O1—C6—C7—C2 | 174.83 (10) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1i | 0.88 | 1.91 | 2.7749 (12) | 169 |
Symmetry codes: (i) x−1/2, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: OM2194).
References
- Bruker (2006). APEX2 Bruker AXS Inc., Madison, Wisconcin, USA.
- Dudzinski, J., Lachman, L., Shami, E. & Tingstad, J. (1973). J. Pharm. Sci.62, 622–624. [DOI] [PubMed]
- Sheldrick, G. M. (1995). XP in SHELXTL/PC Siemens Analytical Instruments Inc., Madison, Wisconsin, USA.
- Sheldrick, G. M. (1997). SHELXS97 and SHELXL97 University of Göttingen, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536807063076/om2194sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807063076/om2194Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report