Abstract
In the title compound, C20H27BrN2O2, molecules are linked into one-dimensional chains through (amide)N—H⋯O=C(amide) intermolecular hydrogen bonds.
Related literature
For related literature, see: Bohne et al. (2005 ▶); Bondy et al. (2004 ▶); Bruker (2000 ▶); Ślebioda (1995 ▶). For literature on related crystal structures, see: Ball et al. (1990 ▶); Chérioux et al. (2002 ▶); Gallagher et al. (1999 ▶); Govindasamy & Subramanian (1997 ▶); Toniolo et al. (1990 ▶); Wu et al. (2006 ▶).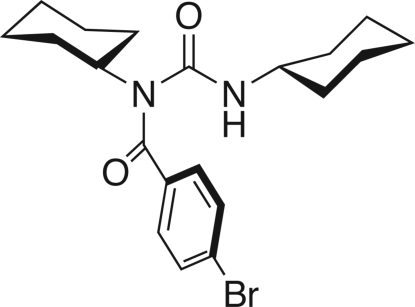
Experimental
Crystal data
C20H27BrN2O2
M r = 407.35
Monoclinic,
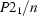
a = 13.501 (2) Å
b = 9.5621 (10) Å
c = 16.306 (2) Å
β = 114.443 (6)°
V = 1916.3 (4) Å3
Z = 4
Mo Kα radiation
μ = 2.16 mm−1
T = 113 (2) K
0.38 × 0.16 × 0.14 mm
Data collection
Rigaku Saturn CCD diffractometer
Absorption correction: multi-scan (REQABS; Jacobson, 1998 ▶) T min = 0.484, T max = 0.739
17461 measured reflections
4526 independent reflections
3651 reflections with I > 2σ(I)
R int = 0.044
Refinement
R[F 2 > 2σ(F 2)] = 0.036
wR(F 2) = 0.078
S = 1.06
4526 reflections
231 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.45 e Å−3
Δρmin = −0.44 e Å−3
Data collection: CrystalClear (Jacobson, 1999 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: CrystalStructure (Rigaku, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536807064756/pk2071sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807064756/pk2071Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2A⋯O2i | 0.898 (10) | 2.072 (11) | 2.961 (2) | 170 (2) |
Symmetry code: (i) 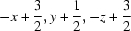 .
.
Acknowledgments
The authors acknowledge financial support from the Research Fund for New Faculty at the State Key Laboratory of Applied Organic Chemistry.
supplementary crystallographic information
Comment
The first conscious total synthesis of a natural product was that of urea in 1828 by Wohler, which marks the beginnings of organic synthesis. Since then, many urea derivatives have been prepared and have demonstrated a wide range of uses, including fluorescence probes (Bohne et al., 2005) and anion receptors (Bondy et al., 2004).
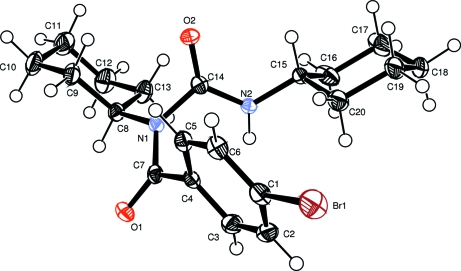
The title compound, an N-acylurea derivative, can be conveniently prepared from dicyclohexylcarbodiimide (DCC) and p-bromobenzoic acid according to reported methods (Ślebioda, 1995). The molecular structure is shown in Fig.1. Each cyclohexyl group adopts the chair conformation, as is required for energy minimization. The two carbonyl groups are twisted substantially at the central atom, N1, with a dihedral angle of 66.43 (10)° between the O1/C7/N1 and O2/C14/N2 planes, which increases the distance between atoms O1 and N2. As a result, no intramolecular N2–H2A···O1 hydrogen bond is formed. However, molecules are linked into chains through (amide) N–H···O=C (amide) intermolecular hydrogen bonds, reinforced by C–H···O=C interactions. Surprisingly, this supramolecular arrangement is not observed in a closely related X-ray structure (Gallagher et al., 1999).
Experimental
p-bromobenzoic acid (201 mg, 1 mmol) was dissolved in CHCl3 (5 ml) and DCC (206 mg, 1 mmol) and N,N-dimethylpyridin-4-amine (122 mg, 1 mmol) were added to the solution. The resulting mixture was stirred for 1 h at 298 K. After evaporation of the solvent, a colorless solid was isolated. Single crystals suitable for X-ray structure determination were obtained by slow evaporation of a EtOAc solution over a period of several days.
Refinement
The H atom bonded to N2 was found in a difference map and refined freely to obtain an unbiased geometry for the hydrogen bonding scheme. The H atoms bonded to C were placed geometrically (C—H values were set to 1.00, 0.99 and 0.95 A° for atoms CH2 and CH, respectively) and refined with a riding model, with Uiso(H) = 1.2 times Ueq(C).
Figures
Fig. 1.
An ellipsoid plot of the title compound, showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 50% probability level.
Crystal data
| C20H27BrN2O2 | F000 = 848 |
| Mr = 407.35 | Dx = 1.412 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation λ = 0.71070 Å |
| Hall symbol: -P 2yn | Cell parameters from 4164 reflections |
| a = 13.501 (2) Å | θ = 1.7–27.9º |
| b = 9.5621 (10) Å | µ = 2.16 mm−1 |
| c = 16.306 (2) Å | T = 113 (2) K |
| β = 114.443 (6)º | Prism, colorless |
| V = 1916.3 (4) Å3 | 0.38 × 0.16 × 0.14 mm |
| Z = 4 |
Data collection
| Rigaku Saturn CCD diffractometer | 4526 independent reflections |
| Radiation source: Rotating anode | 3651 reflections with I > 2σ(I) |
| Monochromator: confocal | Rint = 0.044 |
| Detector resolution: 7.31 pixels mm-1 | θmax = 27.9º |
| T = 113(2) K | θmin = 1.7º |
| ω scans | h = −17→17 |
| Absorption correction: multi-scan(REQABS; Jacobson, 1998) | k = −12→12 |
| Tmin = 0.484, Tmax = 0.739 | l = −21→20 |
| 17461 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.036 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.078 | w = 1/[σ2(Fo2) + (0.0381P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.003 |
| 4526 reflections | Δρmax = 0.45 e Å−3 |
| 231 parameters | Δρmin = −0.44 e Å−3 |
| 1 restraint | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.615319 (17) | 0.39160 (2) | 1.092233 (14) | 0.03591 (9) | |
| N1 | 0.86443 (11) | 0.53780 (15) | 0.80694 (9) | 0.0172 (3) | |
| N2 | 0.67615 (11) | 0.53992 (16) | 0.73657 (10) | 0.0180 (3) | |
| O1 | 0.95485 (9) | 0.68643 (13) | 0.92419 (8) | 0.0226 (3) | |
| O2 | 0.76754 (10) | 0.34042 (13) | 0.73292 (8) | 0.0234 (3) | |
| C1 | 0.69576 (14) | 0.4583 (2) | 1.02792 (12) | 0.0233 (4) | |
| C2 | 0.69278 (15) | 0.5987 (2) | 1.00682 (13) | 0.0267 (4) | |
| H2 | 0.6508 | 0.6624 | 1.0240 | 0.032* | |
| C3 | 0.75215 (15) | 0.6450 (2) | 0.96011 (13) | 0.0246 (4) | |
| H3 | 0.7519 | 0.7414 | 0.9460 | 0.029* | |
| C4 | 0.81235 (13) | 0.55047 (19) | 0.93373 (11) | 0.0176 (4) | |
| C5 | 0.81418 (14) | 0.40955 (19) | 0.95585 (12) | 0.0205 (4) | |
| H5 | 0.8551 | 0.3450 | 0.9381 | 0.025* | |
| C6 | 0.75616 (14) | 0.3634 (2) | 1.00396 (12) | 0.0223 (4) | |
| H6 | 0.7581 | 0.2677 | 1.0202 | 0.027* | |
| C7 | 0.88221 (13) | 0.60122 (18) | 0.88777 (12) | 0.0179 (4) | |
| C8 | 0.94261 (14) | 0.55666 (19) | 0.76516 (12) | 0.0199 (4) | |
| H8 | 0.9907 | 0.6372 | 0.7963 | 0.024* | |
| C9 | 1.01578 (15) | 0.4287 (2) | 0.77921 (14) | 0.0259 (4) | |
| H9A | 0.9702 | 0.3452 | 0.7531 | 0.031* | |
| H9B | 1.0574 | 0.4122 | 0.8445 | 0.031* | |
| C10 | 1.09504 (16) | 0.4493 (2) | 0.73482 (14) | 0.0331 (5) | |
| H10A | 1.1462 | 0.5260 | 0.7656 | 0.040* | |
| H10B | 1.1380 | 0.3628 | 0.7415 | 0.040* | |
| C11 | 1.03445 (17) | 0.4842 (2) | 0.63510 (14) | 0.0361 (5) | |
| H11A | 0.9878 | 0.4043 | 0.6032 | 0.043* | |
| H11B | 1.0874 | 0.5007 | 0.6086 | 0.043* | |
| C12 | 0.96424 (17) | 0.6143 (2) | 0.62288 (14) | 0.0341 (5) | |
| H12A | 0.9238 | 0.6338 | 0.5578 | 0.041* | |
| H12B | 1.0117 | 0.6956 | 0.6507 | 0.041* | |
| C13 | 0.88348 (15) | 0.5955 (2) | 0.66570 (13) | 0.0268 (4) | |
| H13A | 0.8424 | 0.6834 | 0.6599 | 0.032* | |
| H13B | 0.8308 | 0.5210 | 0.6335 | 0.032* | |
| C14 | 0.76568 (14) | 0.4618 (2) | 0.75616 (11) | 0.0180 (4) | |
| C15 | 0.56540 (13) | 0.48403 (19) | 0.69141 (12) | 0.0180 (4) | |
| H15 | 0.5702 | 0.3853 | 0.6733 | 0.022* | |
| C16 | 0.50196 (16) | 0.5682 (2) | 0.60708 (13) | 0.0313 (5) | |
| H16A | 0.5392 | 0.5639 | 0.5661 | 0.038* | |
| H16B | 0.4987 | 0.6673 | 0.6233 | 0.038* | |
| C17 | 0.38642 (16) | 0.5099 (2) | 0.55931 (15) | 0.0399 (6) | |
| H17A | 0.3452 | 0.5668 | 0.5051 | 0.048* | |
| H17B | 0.3899 | 0.4128 | 0.5396 | 0.048* | |
| C18 | 0.32755 (16) | 0.5114 (2) | 0.62076 (18) | 0.0509 (7) | |
| H18A | 0.3189 | 0.6091 | 0.6368 | 0.061* | |
| H18B | 0.2542 | 0.4702 | 0.5890 | 0.061* | |
| C19 | 0.39104 (16) | 0.4288 (2) | 0.70547 (16) | 0.0403 (6) | |
| H19A | 0.3934 | 0.3293 | 0.6896 | 0.048* | |
| H19B | 0.3535 | 0.4344 | 0.7462 | 0.048* | |
| C20 | 0.50804 (16) | 0.4842 (2) | 0.75473 (14) | 0.0296 (5) | |
| H20A | 0.5063 | 0.5805 | 0.7764 | 0.036* | |
| H20B | 0.5487 | 0.4245 | 0.8077 | 0.036* | |
| H2A | 0.6851 (15) | 0.6326 (10) | 0.7456 (12) | 0.023 (5)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.03720 (13) | 0.04748 (17) | 0.03229 (13) | −0.01255 (10) | 0.02362 (10) | −0.00410 (10) |
| N1 | 0.0147 (7) | 0.0164 (8) | 0.0206 (8) | −0.0024 (6) | 0.0075 (6) | −0.0004 (6) |
| N2 | 0.0154 (7) | 0.0136 (9) | 0.0238 (8) | −0.0013 (6) | 0.0070 (6) | −0.0024 (7) |
| O1 | 0.0190 (6) | 0.0223 (7) | 0.0233 (7) | −0.0054 (6) | 0.0056 (5) | −0.0015 (6) |
| O2 | 0.0213 (6) | 0.0140 (7) | 0.0322 (8) | 0.0006 (5) | 0.0084 (6) | −0.0032 (6) |
| C1 | 0.0217 (9) | 0.0320 (12) | 0.0174 (9) | −0.0071 (8) | 0.0093 (8) | −0.0027 (9) |
| C2 | 0.0257 (10) | 0.0269 (12) | 0.0320 (11) | −0.0048 (8) | 0.0165 (9) | −0.0102 (9) |
| C3 | 0.0259 (10) | 0.0198 (11) | 0.0297 (10) | −0.0034 (8) | 0.0132 (8) | −0.0030 (9) |
| C4 | 0.0141 (8) | 0.0212 (10) | 0.0151 (8) | −0.0030 (7) | 0.0037 (7) | −0.0019 (8) |
| C5 | 0.0164 (8) | 0.0239 (11) | 0.0194 (9) | 0.0003 (8) | 0.0057 (7) | 0.0005 (8) |
| C6 | 0.0218 (9) | 0.0244 (11) | 0.0191 (9) | −0.0012 (8) | 0.0067 (8) | 0.0040 (8) |
| C7 | 0.0145 (8) | 0.0166 (10) | 0.0198 (9) | 0.0026 (7) | 0.0045 (7) | 0.0037 (8) |
| C8 | 0.0171 (9) | 0.0203 (10) | 0.0252 (10) | −0.0019 (7) | 0.0117 (8) | −0.0001 (8) |
| C9 | 0.0188 (9) | 0.0267 (11) | 0.0306 (11) | 0.0044 (8) | 0.0087 (8) | 0.0030 (9) |
| C10 | 0.0221 (10) | 0.0396 (13) | 0.0405 (12) | 0.0087 (9) | 0.0159 (9) | 0.0002 (11) |
| C11 | 0.0321 (11) | 0.0479 (14) | 0.0351 (12) | 0.0045 (10) | 0.0209 (10) | −0.0050 (11) |
| C12 | 0.0348 (11) | 0.0463 (14) | 0.0290 (11) | 0.0046 (10) | 0.0210 (10) | 0.0048 (10) |
| C13 | 0.0245 (10) | 0.0332 (12) | 0.0264 (10) | 0.0058 (9) | 0.0144 (8) | 0.0065 (9) |
| C14 | 0.0176 (9) | 0.0184 (10) | 0.0179 (9) | −0.0019 (7) | 0.0074 (7) | 0.0019 (8) |
| C15 | 0.0141 (8) | 0.0162 (10) | 0.0219 (9) | −0.0008 (7) | 0.0055 (7) | −0.0019 (8) |
| C16 | 0.0314 (11) | 0.0229 (11) | 0.0270 (11) | −0.0040 (9) | −0.0006 (9) | 0.0023 (9) |
| C17 | 0.0268 (11) | 0.0284 (13) | 0.0408 (13) | 0.0028 (9) | −0.0096 (10) | 0.0015 (10) |
| C18 | 0.0150 (10) | 0.0391 (14) | 0.0841 (19) | 0.0034 (10) | 0.0060 (12) | −0.0301 (14) |
| C19 | 0.0277 (11) | 0.0528 (15) | 0.0518 (15) | −0.0179 (10) | 0.0279 (11) | −0.0276 (12) |
| C20 | 0.0262 (10) | 0.0367 (13) | 0.0304 (11) | −0.0088 (9) | 0.0163 (9) | −0.0101 (10) |
Geometric parameters (Å, °)
| Br1—C1 | 1.9048 (18) | C10—H10A | 0.9900 |
| N1—C7 | 1.379 (2) | C10—H10B | 0.9900 |
| N1—C14 | 1.441 (2) | C11—C12 | 1.526 (3) |
| N1—C8 | 1.485 (2) | C11—H11A | 0.9900 |
| N2—C14 | 1.342 (2) | C11—H11B | 0.9900 |
| N2—C15 | 1.467 (2) | C12—C13 | 1.529 (2) |
| N2—H2A | 0.898 (9) | C12—H12A | 0.9900 |
| O1—C7 | 1.223 (2) | C12—H12B | 0.9900 |
| O2—C14 | 1.225 (2) | C13—H13A | 0.9900 |
| C1—C6 | 1.380 (3) | C13—H13B | 0.9900 |
| C1—C2 | 1.382 (3) | C15—C16 | 1.516 (2) |
| C2—C3 | 1.387 (3) | C15—C20 | 1.526 (2) |
| C2—H2 | 0.9500 | C15—H15 | 1.0000 |
| C3—C4 | 1.397 (3) | C16—C17 | 1.531 (3) |
| C3—H3 | 0.9500 | C16—H16A | 0.9900 |
| C4—C5 | 1.393 (3) | C16—H16B | 0.9900 |
| C4—C7 | 1.507 (2) | C17—C18 | 1.515 (3) |
| C5—C6 | 1.390 (2) | C17—H17A | 0.9900 |
| C5—H5 | 0.9500 | C17—H17B | 0.9900 |
| C6—H6 | 0.9500 | C18—C19 | 1.512 (3) |
| C8—C13 | 1.528 (2) | C18—H18A | 0.9900 |
| C8—C9 | 1.529 (3) | C18—H18B | 0.9900 |
| C8—H8 | 1.0000 | C19—C20 | 1.540 (3) |
| C9—C10 | 1.532 (3) | C19—H19A | 0.9900 |
| C9—H9A | 0.9900 | C19—H19B | 0.9900 |
| C9—H9B | 0.9900 | C20—H20A | 0.9900 |
| C10—C11 | 1.524 (3) | C20—H20B | 0.9900 |
| C7—N1—C14 | 121.65 (14) | C11—C12—C13 | 111.49 (17) |
| C7—N1—C8 | 120.63 (14) | C11—C12—H12A | 109.3 |
| C14—N1—C8 | 117.50 (14) | C13—C12—H12A | 109.3 |
| C14—N2—C15 | 123.18 (15) | C11—C12—H12B | 109.3 |
| C14—N2—H2A | 117.5 (12) | C13—C12—H12B | 109.3 |
| C15—N2—H2A | 118.8 (12) | H12A—C12—H12B | 108.0 |
| C6—C1—C2 | 121.85 (17) | C8—C13—C12 | 110.81 (16) |
| C6—C1—Br1 | 118.30 (15) | C8—C13—H13A | 109.5 |
| C2—C1—Br1 | 119.85 (14) | C12—C13—H13A | 109.5 |
| C1—C2—C3 | 118.86 (18) | C8—C13—H13B | 109.5 |
| C1—C2—H2 | 120.6 | C12—C13—H13B | 109.5 |
| C3—C2—H2 | 120.6 | H13A—C13—H13B | 108.1 |
| C2—C3—C4 | 120.33 (18) | O2—C14—N2 | 125.83 (16) |
| C2—C3—H3 | 119.8 | O2—C14—N1 | 121.54 (16) |
| C4—C3—H3 | 119.8 | N2—C14—N1 | 112.60 (16) |
| C5—C4—C3 | 119.75 (17) | N2—C15—C16 | 109.75 (14) |
| C5—C4—C7 | 119.52 (16) | N2—C15—C20 | 110.93 (14) |
| C3—C4—C7 | 120.56 (17) | C16—C15—C20 | 111.03 (16) |
| C6—C5—C4 | 119.98 (17) | N2—C15—H15 | 108.3 |
| C6—C5—H5 | 120.0 | C16—C15—H15 | 108.3 |
| C4—C5—H5 | 120.0 | C20—C15—H15 | 108.3 |
| C1—C6—C5 | 119.22 (18) | C15—C16—C17 | 110.02 (16) |
| C1—C6—H6 | 120.4 | C15—C16—H16A | 109.7 |
| C5—C6—H6 | 120.4 | C17—C16—H16A | 109.7 |
| O1—C7—N1 | 122.99 (16) | C15—C16—H16B | 109.7 |
| O1—C7—C4 | 120.93 (16) | C17—C16—H16B | 109.7 |
| N1—C7—C4 | 115.88 (15) | H16A—C16—H16B | 108.2 |
| N1—C8—C13 | 110.98 (14) | C18—C17—C16 | 111.28 (18) |
| N1—C8—C9 | 111.83 (15) | C18—C17—H17A | 109.4 |
| C13—C8—C9 | 112.02 (16) | C16—C17—H17A | 109.4 |
| N1—C8—H8 | 107.2 | C18—C17—H17B | 109.4 |
| C13—C8—H8 | 107.2 | C16—C17—H17B | 109.4 |
| C9—C8—H8 | 107.2 | H17A—C17—H17B | 108.0 |
| C8—C9—C10 | 111.13 (16) | C19—C18—C17 | 110.18 (17) |
| C8—C9—H9A | 109.4 | C19—C18—H18A | 109.6 |
| C10—C9—H9A | 109.4 | C17—C18—H18A | 109.6 |
| C8—C9—H9B | 109.4 | C19—C18—H18B | 109.6 |
| C10—C9—H9B | 109.4 | C17—C18—H18B | 109.6 |
| H9A—C9—H9B | 108.0 | H18A—C18—H18B | 108.1 |
| C11—C10—C9 | 111.13 (16) | C18—C19—C20 | 111.59 (18) |
| C11—C10—H10A | 109.4 | C18—C19—H19A | 109.3 |
| C9—C10—H10A | 109.4 | C20—C19—H19A | 109.3 |
| C11—C10—H10B | 109.4 | C18—C19—H19B | 109.3 |
| C9—C10—H10B | 109.4 | C20—C19—H19B | 109.3 |
| H10A—C10—H10B | 108.0 | H19A—C19—H19B | 108.0 |
| C10—C11—C12 | 110.37 (17) | C15—C20—C19 | 110.18 (16) |
| C10—C11—H11A | 109.6 | C15—C20—H20A | 109.6 |
| C12—C11—H11A | 109.6 | C19—C20—H20A | 109.6 |
| C10—C11—H11B | 109.6 | C15—C20—H20B | 109.6 |
| C12—C11—H11B | 109.6 | C19—C20—H20B | 109.6 |
| H11A—C11—H11B | 108.1 | H20A—C20—H20B | 108.1 |
| C6—C1—C2—C3 | 0.1 (3) | C13—C8—C9—C10 | 54.1 (2) |
| Br1—C1—C2—C3 | 179.88 (14) | C8—C9—C10—C11 | −55.5 (2) |
| C1—C2—C3—C4 | 1.0 (3) | C9—C10—C11—C12 | 57.0 (2) |
| C2—C3—C4—C5 | −1.1 (3) | C10—C11—C12—C13 | −57.3 (2) |
| C2—C3—C4—C7 | −176.41 (17) | N1—C8—C13—C12 | −179.79 (15) |
| C3—C4—C5—C6 | 0.0 (3) | C9—C8—C13—C12 | −54.0 (2) |
| C7—C4—C5—C6 | 175.39 (15) | C11—C12—C13—C8 | 55.7 (2) |
| C2—C1—C6—C5 | −1.2 (3) | C15—N2—C14—O2 | 6.6 (3) |
| Br1—C1—C6—C5 | 179.06 (13) | C15—N2—C14—N1 | −175.26 (14) |
| C4—C5—C6—C1 | 1.1 (3) | C7—N1—C14—O2 | −125.42 (18) |
| C14—N1—C7—O1 | −166.44 (16) | C8—N1—C14—O2 | 60.0 (2) |
| C8—N1—C7—O1 | 7.9 (2) | C7—N1—C14—N2 | 56.4 (2) |
| C14—N1—C7—C4 | 18.6 (2) | C8—N1—C14—N2 | −118.19 (17) |
| C8—N1—C7—C4 | −167.06 (14) | C14—N2—C15—C16 | −124.11 (18) |
| C5—C4—C7—O1 | −117.18 (19) | C14—N2—C15—C20 | 112.83 (19) |
| C3—C4—C7—O1 | 58.1 (2) | N2—C15—C16—C17 | 179.70 (16) |
| C5—C4—C7—N1 | 57.9 (2) | C20—C15—C16—C17 | −57.3 (2) |
| C3—C4—C7—N1 | −126.75 (18) | C15—C16—C17—C18 | 57.9 (2) |
| C7—N1—C8—C13 | −131.60 (17) | C16—C17—C18—C19 | −57.3 (2) |
| C14—N1—C8—C13 | 43.0 (2) | C17—C18—C19—C20 | 56.3 (2) |
| C7—N1—C8—C9 | 102.52 (19) | N2—C15—C20—C19 | 178.56 (17) |
| C14—N1—C8—C9 | −82.87 (19) | C16—C15—C20—C19 | 56.2 (2) |
| N1—C8—C9—C10 | 179.38 (15) | C18—C19—C20—C15 | −55.8 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2A···O2i | 0.898 (10) | 2.072 (11) | 2.961 (2) | 170 (2) |
Symmetry codes: (i) −x+3/2, y+1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: PK2071).
References
- Ball, R. G., Brown, R. S. & Bennet, A. J. (1990). Acta Cryst. C46, 2491–2493.
- Bohne, C., Ihmels, H., Waidelich, M. & Yihwa, C. (2005). J. Am. Chem. Soc.127, 17158–17159. [DOI] [PubMed]
- Bondy, C. R., Gale, P. A. & Loeb, S. J. (2004). J. Am. Chem. Soc.126, 5030–5031. [DOI] [PubMed]
- Bruker (2000). SHELXTL Version 6.10. Bruker AXS Inc., Madison, Wisconsin, USA.
- Chérioux, F., Therrien, B., Stoeckli-Evans, H. & Süss-Fink, G. (2002). Acta Cryst. E58, o27–o29.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565
- Gallagher, J. F., Kenny, P. T. M. & Sheehy, M. J. (1999). Acta Cryst. C55, 1607–1610.
- Govindasamy, L. & Subramanian, E. (1997). Acta Cryst. C53, 927–928.
- Jacobson, R. A. (1998). REQABS. Version 1.1. MSC, The Woodlands, Texas, USA.
- Jacobson, R. A. (1999). CrystalClear Rigaku Corporation, The Woodlands, Texas, USA.
- Rigaku (1999). CrystalStructure Version 3.7.0. Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (1997). SHELXS97 and SHELXL97 University of Göttingen, Germany.
- Ślebioda, M. (1995). Tetrahedron, 51, 7829–7834.
- Toniolo, C., Valle, G., Crisma, M., Moretto, V., Izdebski, J., Pelka, J. & Schneider, C. H. (1990). Helv. Chim. Acta, 73, 626–634.
- Wu, L., Liu, H.-M., Zhao, W.-T. & Zhang, W.-Q. (2006). Acta Cryst. C62, o435–o437. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536807064756/pk2071sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807064756/pk2071Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report