Abstract
In the molecule of the title compound, C8H6BrI, the H atoms of methyl groups are disordered; site-occupation factors were fixed at 0.50. The non-H atoms all lie on a crystallographic mirror plane. Weak intramolecular C—H⋯Br hydrogen bonds result in the formation of two non-planar five-membered rings.
Related literature
For bond-length data, see: Allen et al. (1987 ▶).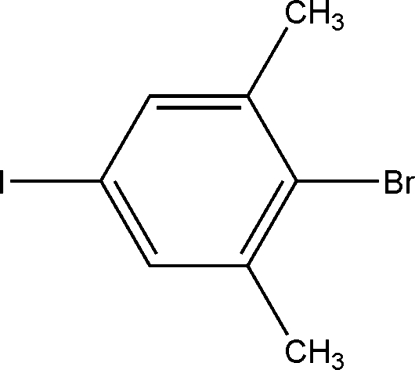
Experimental
Crystal data
C8H8BrI
M r = 310.94
Orthorhombic,
a = 16.686 (3) Å
b = 7.0640 (14) Å
c = 8.2130 (16) Å
V = 968.1 (3) Å3
Z = 4
Mo Kα radiation
μ = 7.37 mm−1
T = 294 (2) K
0.40 × 0.20 × 0.10 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Absorption correction: ψ scan (North et al., 1968 ▶) T min = 0.157, T max = 0.479
1030 measured reflections
1030 independent reflections
659 reflections with I > 2σ(I)
3 standard reflections frequency: 120 min intensity decay: none
Refinement
R[F 2 > 2σ(F 2)] = 0.056
wR(F 2) = 0.137
S = 1.10
1030 reflections
63 parameters
H-atom parameters constrained
Δρmax = 0.62 e Å−3
Δρmin = −0.85 e Å−3
Data collection: CAD-4 Software (Enraf–Nonius, 1985 ▶); cell refinement: CAD-4 Software; data reduction: XCAD4 (Harms & Wocadlo, 1995 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997 ▶); molecular graphics: PLATON (Spek, 2003 ▶); software used to prepare material for publication: SHELXTL (Bruker, 2000 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536807065415/hk2261sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807065415/hk2261Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C7—H7C⋯Br2 | 0.96 | 2.74 | 3.156 (6) | 107 |
| C8—H8C⋯Br2 | 0.96 | 2.77 | 3.115 (5) | 102 |
Acknowledgments
The authors thank the Center of Testing and Analysis, Nanjing University, for support.
supplementary crystallographic information
Comment
The title compound, (I), is a fine organic intermediate, which can be utilized to construct practical functional molecules. We herein report its crystal structure.
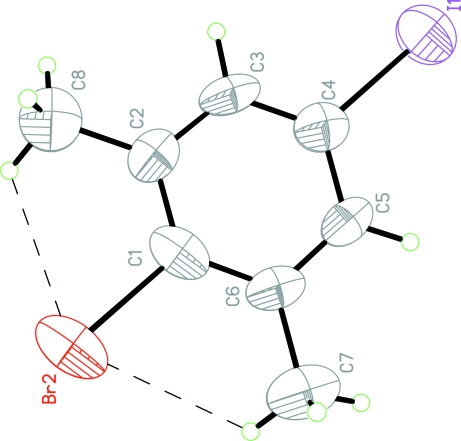
In the molecule of (I), (Fig. 1), the bond lengths and angles are within normal ranges (Allen et al., 1987). When the crystal structure was solved, H atoms of methyl groups were found to be disordered.
The atoms Br2, I1, C7 and C8 lie in the benzene ring plane. The weak intra- molecular C—H···Br hydrogen bonds (Table 1) result in the formations of two non-planar five-membered rings; B (Br2/C1/C6/C7/H7C) and C (Br2/C1/C2/C8/H8C). Ring A (C1—C6) is, of course, planar.
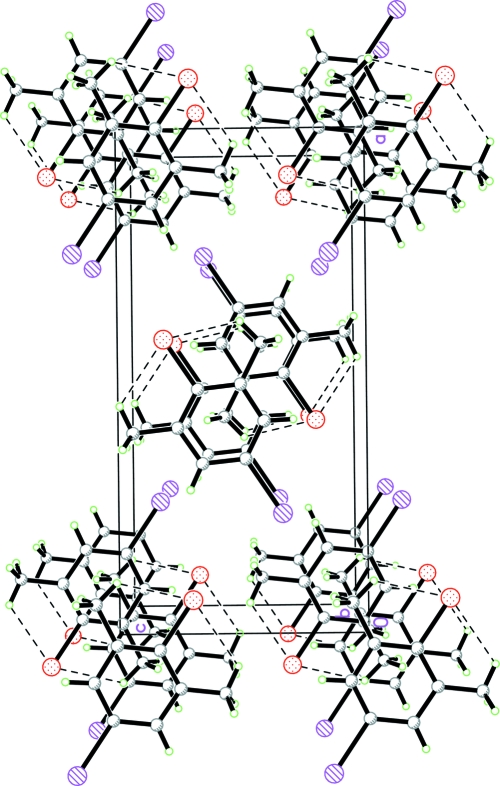
As can be seen from the packing diagram, (Fig. 2), the molecules are stacked along the b axis.
Experimental
For the preparation of the title compound, 4-iodo-2,6-dimethylaniline (5.0 g, 20 mmol), concentrated sulfuric acid (40 mmol, 2.23 ml) and water (100 ml) were stirred in an ice bath. When the temperature was below 278 K, the solution of sodium nitrite (1.44 g, 21 mmol) in water (100 ml) was added dropwise. Then, the mixture was added to a solution of CuBr (2.86 g, 20 mmol) and hydrobromic acid (20 mmol, 2.71 ml) with stirring. The solid residue was extracted with boiling hexane (40 ml) and hexane was distilled off. Crystals suitable for X-ray analysis were obtained by slow evaporation of ethanol at room temperature for about 20 d.
Refinement
When the crystal structure was solved, the H atoms of methyl groups were found to be disordered over two mirror image sites of the symmetry plane passing through the benzene ring. The occupancies of disordered H atoms were kept fixed as 0.50. H atoms were positioned geometrically, with C—H = 0.93 and 0.96 Å for aromatic and methyl H, respectively, and constrained to ride on their parent atoms, with Uiso(H) = xUeq(C), where x = 1.2 for aromatic H, and x = 1.5 for methyl H atoms.
Figures
Fig. 1.
The molecular structure of the title molecule with the atom-numbering scheme. Displacement ellipsoids are drawn at the 50% probability level. Hydrogen bonds are shown as dashed lines.
Fig. 2.
A packing diagram of (I). Hydrogen bonds are shown as dashed lines.
Crystal data
| C8H8BrI | Dx = 2.133 Mg m−3 |
| Mr = 310.94 | Melting point: 307 K |
| Orthorhombic, Pnma | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ac 2n | Cell parameters from 25 reflections |
| a = 16.686 (3) Å | θ = 10–13º |
| b = 7.0640 (14) Å | µ = 7.37 mm−1 |
| c = 8.2130 (16) Å | T = 294 (2) K |
| V = 968.1 (3) Å3 | Needle, colorless |
| Z = 4 | 0.40 × 0.20 × 0.10 mm |
| F000 = 576.0 |
Data collection
| Enraf–Nonius CAD-4 diffractometer | Rint = 0.0000 |
| Radiation source: fine-focus sealed tube | θmax = 26.0º |
| Monochromator: graphite | θmin = 2.4º |
| T = 294(2) K | h = 0→20 |
| ω/2θ scans | k = 0→8 |
| Absorption correction: ψ scan(North et al., 1968) | l = 0→10 |
| Tmin = 0.157, Tmax = 0.479 | 3 standard reflections |
| 1030 measured reflections | every 120 min |
| 1030 independent reflections | intensity decay: none |
| 659 reflections with I > 2σ(I) |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.056 | H-atom parameters constrained |
| wR(F2) = 0.137 | w = 1/[σ2(Fo2) + (0.0602P)2 + 1.4567P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.10 | (Δ/σ)max < 0.001 |
| 1030 reflections | Δρmax = 0.62 e Å−3 |
| 63 parameters | Δρmin = −0.85 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| I1 | 0.25441 (7) | 0.2500 | 1.15579 (9) | 0.0902 (5) | |
| Br2 | −0.08058 (5) | 0.2500 | 0.69651 (7) | 0.0800 (5) | |
| C1 | 0.0160 (4) | 0.2500 | 0.8282 (5) | 0.055 (3) | |
| C2 | 0.0066 (4) | 0.2500 | 0.9955 (6) | 0.051 (3) | |
| C3 | 0.0768 (4) | 0.2500 | 1.0850 (6) | 0.054 (3) | |
| H3 | 0.0740 | 0.2500 | 1.1981 | 0.065* | |
| C4 | 0.1500 (5) | 0.2500 | 1.0117 (6) | 0.057 (3) | |
| C5 | 0.1580 (5) | 0.2500 | 0.8449 (5) | 0.055 (3) | |
| H5 | 0.2085 | 0.2500 | 0.7972 | 0.066* | |
| C6 | 0.0897 (4) | 0.2500 | 0.7489 (5) | 0.050 (3) | |
| C7 | 0.0984 (4) | 0.2500 | 0.5721 (4) | 0.093 (5) | |
| H7A | 0.0947 | 0.1226 | 0.5322 | 0.139* | 0.50 |
| H7B | 0.1496 | 0.3022 | 0.5434 | 0.139* | 0.50 |
| H7C | 0.0567 | 0.3252 | 0.5244 | 0.139* | 0.50 |
| C8 | −0.0730 (4) | 0.2500 | 1.0755 (6) | 0.077 (4) | |
| H8A | −0.0672 | 0.2878 | 1.1871 | 0.116* | 0.50 |
| H8B | −0.0955 | 0.1251 | 1.0709 | 0.116* | 0.50 |
| H8C | −0.1078 | 0.3371 | 1.0203 | 0.116* | 0.50 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| I1 | 0.0675 (6) | 0.1300 (10) | 0.0731 (6) | 0.000 | −0.0170 (5) | 0.000 |
| Br2 | 0.0850 (9) | 0.0714 (8) | 0.0837 (9) | 0.000 | −0.0394 (8) | 0.000 |
| C1 | 0.065 (7) | 0.038 (6) | 0.063 (8) | 0.000 | −0.022 (6) | 0.000 |
| C6 | 0.074 (7) | 0.049 (5) | 0.047 (6) | 0.000 | 0.006 (5) | 0.000 |
| C4 | 0.068 (7) | 0.057 (7) | 0.046 (7) | 0.000 | 0.002 (6) | 0.000 |
| C3 | 0.079 (8) | 0.053 (6) | 0.031 (5) | 0.000 | 0.012 (6) | 0.000 |
| C2 | 0.054 (5) | 0.046 (6) | 0.052 (7) | 0.000 | 0.010 (5) | 0.000 |
| C5 | 0.060 (6) | 0.059 (7) | 0.045 (6) | 0.000 | 0.017 (5) | 0.000 |
| C7 | 0.108 (13) | 0.098 (10) | 0.083 (6) | 0.000 | 0.012 (7) | 0.000 |
| C8 | 0.071 (8) | 0.079 (8) | 0.082 (9) | 0.000 | 0.003 (7) | 0.000 |
Geometric parameters (Å, °)
| I1—C4 | 2.105 (7) | C3—H3 | 0.9300 |
| Br2—C1 | 1.941 (6) | C2—C8 | 1.481 (6) |
| C1—C2 | 1.383 (6) | C5—H5 | 0.9300 |
| C1—C6 | 1.392 (6) | C7—H7A | 0.9600 |
| C6—C5 | 1.385 (6) | C7—H7B | 0.9600 |
| C6—C7 | 1.460 (5) | C7—H7C | 0.9600 |
| C4—C3 | 1.362 (6) | C8—H8A | 0.9600 |
| C4—C5 | 1.377 (6) | C8—H8B | 0.9600 |
| C3—C2 | 1.384 (6) | C8—H8C | 0.9600 |
| C2—C1—C6 | 124.4 (5) | C4—C5—C6 | 119.2 (5) |
| C2—C1—Br2 | 117.4 (5) | C4—C5—H5 | 120.4 |
| C6—C1—Br2 | 118.2 (3) | C6—C5—H5 | 120.4 |
| C5—C6—C1 | 117.4 (4) | C6—C7—H7A | 109.5 |
| C5—C6—C7 | 118.9 (5) | C6—C7—H7B | 109.5 |
| C1—C6—C7 | 123.6 (5) | H7A—C7—H7B | 109.5 |
| C3—C4—C5 | 121.7 (4) | C6—C7—H7C | 109.5 |
| C3—C4—I1 | 119.6 (4) | H7A—C7—H7C | 109.5 |
| C5—C4—I1 | 118.7 (4) | H7B—C7—H7C | 109.5 |
| C4—C3—C2 | 121.6 (5) | C2—C8—H8A | 109.5 |
| C4—C3—H3 | 119.2 | C2—C8—H8B | 109.5 |
| C2—C3—H3 | 119.2 | H8A—C8—H8B | 109.5 |
| C1—C2—C3 | 115.6 (5) | C2—C8—H8C | 109.5 |
| C1—C2—C8 | 122.8 (6) | H8A—C8—H8C | 109.5 |
| C3—C2—C8 | 121.5 (5) | H8B—C8—H8C | 109.5 |
| C2—C1—C6—C5 | 0.000 (3) | C6—C1—C2—C8 | 180.000 (3) |
| Br2—C1—C6—C5 | 180.000 (2) | Br2—C1—C2—C8 | 0.000 (3) |
| C2—C1—C6—C7 | 180.000 (2) | C4—C3—C2—C1 | 0.000 (3) |
| Br2—C1—C6—C7 | 0.000 (2) | C4—C3—C2—C8 | 180.000 (3) |
| C5—C4—C3—C2 | 0.000 (3) | C3—C4—C5—C6 | 0.000 (3) |
| I1—C4—C3—C2 | 180.000 (3) | I1—C4—C5—C6 | 180.000 (2) |
| C6—C1—C2—C3 | 0.000 (3) | C1—C6—C5—C4 | 0.000 (3) |
| Br2—C1—C2—C3 | 180.000 (2) | C7—C6—C5—C4 | 180.000 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C7—H7C···Br2 | 0.96 | 2.74 | 3.156 (6) | 107 |
| C8—H8C···Br2 | 0.96 | 2.77 | 3.115 (5) | 102 |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HK2261).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bruker (2000). SHELXTL Bruker AXS Inc., Madison, Wisconsin, USA.
- Enraf–Nonius (1985). CAD-4 Software Version 5.0. Enraf–Nonius, Delft, The Netherlands.
- Harms, K. & Wocadlo, S. (1995). XCAD4 University of Marburg, Germany.
- North, A. C. T., Phillips, D. C. & Mathews, F. S. (1968). Acta Cryst. A24, 351–359.
- Sheldrick, G. M. (1997). SHELXS97 and SHELXL97 University of Göttingen, Germany.
- Spek, A. L. (2003). J. Appl. Cryst.36, 7–13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536807065415/hk2261sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807065415/hk2261Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report