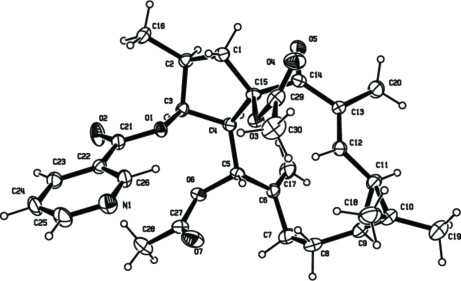
Abstract
The title compound [systematic name: (2S*,3S*,4R*,5R*,9S*,11S*,15R*)-5,15-diacetoxy-3-nicotinoyloxy-14-oxolathyra-6(17),12(E)-diene], C30H37NO7, was isolated from the seeds of Euphorbia lathyris. The tricyclic diterpenoid molecule contains an 11-membered ring, a five-membered ring exhibiting an envelope conformation and a three-membered ring. The 11-membered ring is cis-fused with the three-membered ring and trans-fused with the five-membered ring.
Related literature
For related literature, see: Appendino et al. (1999 ▶); Fujiwara et al. (1996 ▶); Kupchan et al. (1976 ▶); the Pharmacopoeia Commission of the People’s Republic of China (2005 ▶).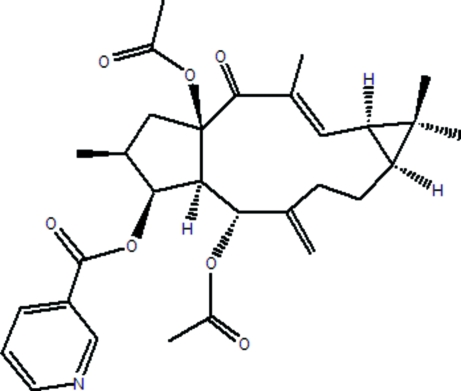
Experimental
Crystal data
C30H37NO7
M r = 523.61
Orthorhombic,
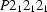
a = 10.162 (6) Å
b = 15.249 (5) Å
c = 18.802 (9) Å
V = 2914 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.08 mm−1
T = 298 (2) K
0.36 × 0.34 × 0.25 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Absorption correction: none
3444 measured reflections
3065 independent reflections
1462 reflections with I > 2σ(I)
R int = 0.003
3 standard reflections every 300 reflections intensity decay: 0.3%
Refinement
R[F 2 > 2σ(F 2)] = 0.049
wR(F 2) = 0.118
S = 0.93
3065 reflections
354 parameters
H-atom parameters constrained
Δρmax = 0.18 e Å−3
Δρmin = −0.24 e Å−3
Data collection: DIFRAC (Gabe & White, 1993 ▶); cell refinement: DIFRAC; data reduction: NRCVAX (Gabe et al., 1989 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536807066901/ww2099sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807066901/ww2099Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
Support by the ‘Western Light’ Joint Research Program of the Chinese Academy of Sciences is acknowledged. The authors are also grateful to the staff of the analytical group of Chengdu Institute of Biology, Chinese Academy of Sciences, for the NMR spectroscopic data.
supplementary crystallographic information
Comment
The seed of Euphorbia lathyris is a traditional Chinese medicine which has been used for the treatment of hydropsy, ascites, amenorrhea, scabies (Pharmacopoeia Commission of the People's Republic of China, 2005). Several constituents in this plant proved to have significant activity (Kupchan et al., 1976; Fujiwara et al., 1996) and this medicine has been used to treat tumors and cancer in many countries. In our current investigation, Euphorbia Factor L8 (I) was isolated from the seeds of this plant. The structure of (I) was elucidated by comprehensive spectroscopic analysis, and was confirmed by single-crystal X-ray diffraction analysis reported here (Fig. 1). The title compound shows the tricyclic terpenoid skeleton of lathyrane, consisting of fused five-, eleven- and three-membered rings (A: C1–C4/C15, B: C4–C9/C11–C15, C: C9–C11). Rings A and B are trans-joined (torsion angle H4–C4–C15–O3 = -152.8°), while rings B and C are cis-joined (H9–C9–C11–H11 = 0.99°). Ring A adopts an envelope conformation, with atom C3 0.64 Å out of the plane defined by atoms C1/C2/C4/C15.
Experimental
The seeds of E. lathyris (10 kg) were collected in Sichuan province, People's Republic of China and extracted with 95% EtOH at room temperature. The extract was concentrated in vacuo and filtered. The filtrate was partitioned between EtOAc and H2O. The EtOAc soluble materials (1 kg) were subjected to silica-gel column chromatography (160–200 mesh, 4 kg) with petrol-EtOAc stepwise elution. The column chromatographic fractions (500 ml each) were combined into 12 fractions according to thin-layer chromatography monitoring analysis. Fraction 5 (7.5 g) was applied to a RP-18 silica-gel column and eluted with MeOH/H2O (7:3) to yield five fractions. Fraction 5.2 (1.4 g) was subjected to silica-gel column chromatography (200–300 mesh, 50 g) and eluted with petrol-EtOAc (5:1) to afford the compound (I). The isolated product was recrystallized at room temperature from acetone to afford the block crytals. 13C NMR (150 MHz, CDCl3, δ, p.p.m.): 48.6(C1), 37.7(C2), 81.6(C3), 52.3(C4), 65.5(C5), 144.4(C6), 34.9(C7), 21.0(C8), 35.4(C9), 25.3(C10), 28.5(C11), 146.6(C12), 134.3(C13), 196.6(C14), 92.5(C15), 14.2(C16), 115.5(C17), 29.0(C18), 16.8(C19), 12.4(C20), 164.9(C21), 126.0(C22), 137.0(C23), 123.3(C24), 153.5(C25), 151.0(C26), 170.2(C27), 21.6(C28), 169.7(C29), 22.1(C30).
Refinement
All hydrogen atoms were located geometrically with C—H distances of 0.93–0.98 Å, and refined using a riding model. The absolute configuration could not be determined from the X-ray analysis, owing to the absence of strong anomalous scatterers, and Friedel pairs were averaged. However, the absolute configuration can be suggested on a biogenetic basis (Appendino et al., 1999).
Figures
Fig. 1.
ORTEP plot of compound (I) showing the atom-numbering scheme and displacement ellipsoids drawn at the 50% probability level.
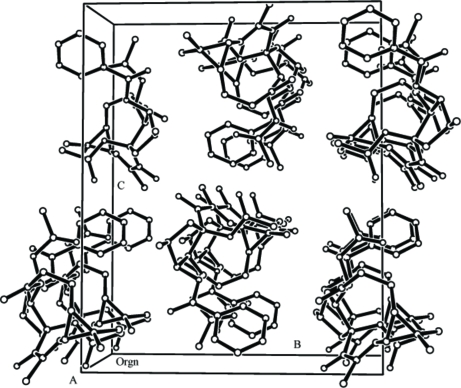
Fig. 2.
The crystal packing of (I), viewed down the a axis. H-atoms omitted for clarity.
Crystal data
| C30H37NO7 | Dx = 1.194 Mg m−3 |
| Mr = 523.61 | Melting point: 469(1) K |
| Orthorhombic, P212121 | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 24 reflections |
| a = 10.162 (6) Å | θ = 4.5–5.5º |
| b = 15.249 (5) Å | µ = 0.08 mm−1 |
| c = 18.802 (9) Å | T = 298 (2) K |
| V = 2914 (2) Å3 | Block, colourless |
| Z = 4 | 0.36 × 0.34 × 0.25 mm |
| F000 = 1120 |
Data collection
| Enraf–Nonius CAD-4 diffractometer | Rint = 0.003 |
| Radiation source: fine-focus sealed tube | θmax = 25.5º |
| Monochromator: graphite | θmin = 1.7º |
| T = 298(2) K | h = −1→12 |
| ω/2θ scans | k = −3→18 |
| Absorption correction: none | l = −1→22 |
| 3444 measured reflections | 3 standard reflections |
| 3065 independent reflections | every 300 reflections |
| 1462 reflections with I > 2σ(I) | intensity decay: 0.3% |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.049 | H-atom parameters constrained |
| wR(F2) = 0.118 | w = 1/[σ2(Fo2) + (0.0541P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 0.93 | (Δ/σ)max < 0.001 |
| 3065 reflections | Δρmax = 0.18 e Å−3 |
| 354 parameters | Δρmin = −0.24 e Å−3 |
| Primary atom site location: structure-invariant direct methods | Extinction correction: none |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.4410 (3) | 0.60363 (17) | 0.75057 (16) | 0.0422 (8) | |
| O2 | 0.3383 (4) | 0.6889 (2) | 0.67028 (18) | 0.0745 (12) | |
| O3 | 0.6027 (3) | 0.50915 (17) | 0.86366 (15) | 0.0442 (8) | |
| O4 | 0.5648 (4) | 0.4774 (2) | 0.97859 (19) | 0.0725 (11) | |
| O5 | 0.6858 (4) | 0.6902 (2) | 0.97413 (16) | 0.0600 (10) | |
| O6 | 0.6894 (4) | 0.63864 (19) | 0.68532 (16) | 0.0539 (9) | |
| O7 | 0.6709 (5) | 0.4992 (3) | 0.6477 (2) | 0.0887 (14) | |
| N1 | 0.3198 (5) | 0.3782 (2) | 0.6543 (2) | 0.0653 (13) | |
| C1 | 0.4680 (5) | 0.6336 (3) | 0.9021 (2) | 0.0463 (13) | |
| H1A | 0.4056 | 0.5857 | 0.8985 | 0.071 (7)* | |
| H1B | 0.4675 | 0.6552 | 0.9506 | 0.071 (7)* | |
| C2 | 0.4308 (5) | 0.7065 (3) | 0.8509 (2) | 0.0465 (12) | |
| H2 | 0.4703 | 0.7610 | 0.8684 | 0.045 (5)* | |
| C3 | 0.5024 (5) | 0.6806 (3) | 0.7833 (2) | 0.0401 (12) | |
| H3 | 0.5082 | 0.7297 | 0.7498 | 0.045 (5)* | |
| C4 | 0.6377 (5) | 0.6534 (3) | 0.8103 (2) | 0.0405 (12) | |
| H4 | 0.6821 | 0.7076 | 0.8246 | 0.045 (5)* | |
| C5 | 0.7290 (5) | 0.6088 (3) | 0.7565 (2) | 0.0441 (12) | |
| H5 | 0.7175 | 0.5451 | 0.7596 | 0.045 (5)* | |
| C6 | 0.8749 (5) | 0.6304 (3) | 0.7607 (3) | 0.0491 (14) | |
| C7 | 0.9639 (6) | 0.5689 (4) | 0.7203 (3) | 0.0763 (18) | |
| H7A | 1.0510 | 0.5950 | 0.7184 | 0.071 (7)* | |
| H7B | 0.9318 | 0.5648 | 0.6718 | 0.071 (7)* | |
| C8 | 0.9773 (6) | 0.4764 (4) | 0.7498 (3) | 0.0732 (17) | |
| H8A | 0.8901 | 0.4547 | 0.7612 | 0.071 (7)* | |
| H8B | 1.0129 | 0.4391 | 0.7127 | 0.071 (7)* | |
| C9 | 1.0623 (6) | 0.4677 (4) | 0.8146 (3) | 0.0686 (15) | |
| H9 | 1.1538 | 0.4847 | 0.8055 | 0.045 (5)* | |
| C10 | 1.0512 (6) | 0.3977 (4) | 0.8693 (3) | 0.0697 (17) | |
| C11 | 1.0173 (5) | 0.4919 (3) | 0.8887 (3) | 0.0586 (16) | |
| H11 | 1.0834 | 0.5220 | 0.9178 | 0.045 (5)* | |
| C12 | 0.8824 (5) | 0.5235 (3) | 0.8982 (2) | 0.0454 (12) | |
| H12 | 0.8184 | 0.4984 | 0.8693 | 0.064 (7)* | |
| C13 | 0.8419 (5) | 0.5850 (3) | 0.9442 (2) | 0.0446 (12) | |
| C14 | 0.7110 (5) | 0.6257 (3) | 0.9385 (2) | 0.0443 (12) | |
| C15 | 0.6070 (5) | 0.6020 (3) | 0.8815 (2) | 0.0410 (12) | |
| C16 | 0.2825 (5) | 0.7213 (3) | 0.8442 (3) | 0.0612 (15) | |
| H16A | 0.2414 | 0.6687 | 0.8271 | 0.129 (6)* | |
| H16B | 0.2661 | 0.7683 | 0.8115 | 0.129 (6)* | |
| H16C | 0.2468 | 0.7361 | 0.8899 | 0.129 (6)* | |
| C17 | 0.9201 (6) | 0.7006 (4) | 0.7918 (3) | 0.0720 (16) | |
| H17A | 1.0093 | 0.7138 | 0.7894 | 0.089 (15)* | |
| H17B | 0.8631 | 0.7374 | 0.8164 | 0.089 (15)* | |
| C18 | 0.9437 (7) | 0.3300 (4) | 0.8661 (3) | 0.090 (2) | |
| H18A | 0.9760 | 0.2784 | 0.8426 | 0.129 (6)* | |
| H18B | 0.8702 | 0.3532 | 0.8401 | 0.129 (6)* | |
| H18C | 0.9164 | 0.3153 | 0.9134 | 0.129 (6)* | |
| C19 | 1.1778 (7) | 0.3627 (5) | 0.9013 (4) | 0.112 (3) | |
| H19A | 1.2163 | 0.3207 | 0.8694 | 0.129 (6)* | |
| H19B | 1.1591 | 0.3350 | 0.9460 | 0.129 (6)* | |
| H19C | 1.2382 | 0.4102 | 0.9086 | 0.129 (6)* | |
| C20 | 0.9313 (6) | 0.6255 (3) | 0.9990 (3) | 0.0698 (16) | |
| H20A | 0.9596 | 0.6822 | 0.9829 | 0.129 (6)* | |
| H20B | 1.0067 | 0.5885 | 1.0059 | 0.129 (6)* | |
| H20C | 0.8847 | 0.6316 | 1.0431 | 0.129 (6)* | |
| C21 | 0.3664 (5) | 0.6179 (3) | 0.6929 (3) | 0.0453 (12) | |
| C22 | 0.3204 (5) | 0.5350 (3) | 0.6607 (2) | 0.0411 (12) | |
| C23 | 0.2381 (5) | 0.5386 (3) | 0.6026 (3) | 0.0531 (13) | |
| H23 | 0.2099 | 0.5925 | 0.5852 | 0.064 (7)* | |
| C24 | 0.1981 (6) | 0.4628 (4) | 0.5708 (3) | 0.0655 (15) | |
| H24 | 0.1428 | 0.4639 | 0.5314 | 0.064 (7)* | |
| C25 | 0.2414 (6) | 0.3849 (4) | 0.5984 (3) | 0.0697 (17) | |
| H25 | 0.2141 | 0.3334 | 0.5764 | 0.064 (7)* | |
| C26 | 0.3572 (5) | 0.4531 (3) | 0.6841 (3) | 0.0511 (13) | |
| H26 | 0.4121 | 0.4500 | 0.7236 | 0.064 (7)* | |
| C27 | 0.6664 (6) | 0.5762 (4) | 0.6360 (3) | 0.0630 (16) | |
| C28 | 0.6346 (7) | 0.6157 (4) | 0.5654 (2) | 0.089 (2) | |
| H28A | 0.6076 | 0.5703 | 0.5332 | 0.129 (6)* | |
| H28B | 0.7111 | 0.6447 | 0.5469 | 0.129 (6)* | |
| H28C | 0.5646 | 0.6575 | 0.5709 | 0.129 (6)* | |
| C29 | 0.5746 (5) | 0.4539 (3) | 0.9179 (3) | 0.0548 (13) | |
| C30 | 0.5620 (7) | 0.3613 (3) | 0.8920 (4) | 0.085 (2) | |
| H30A | 0.5472 | 0.3230 | 0.9318 | 0.129 (6)* | |
| H30B | 0.6414 | 0.3444 | 0.8681 | 0.129 (6)* | |
| H30C | 0.4891 | 0.3572 | 0.8597 | 0.129 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.043 (2) | 0.0396 (17) | 0.0435 (17) | −0.0049 (17) | −0.0102 (19) | −0.0047 (16) |
| O2 | 0.093 (3) | 0.0434 (18) | 0.088 (3) | −0.003 (2) | −0.046 (3) | 0.0091 (19) |
| O3 | 0.046 (2) | 0.0370 (17) | 0.0495 (18) | −0.0018 (16) | 0.0047 (19) | −0.0033 (15) |
| O4 | 0.087 (3) | 0.067 (2) | 0.063 (2) | 0.003 (2) | 0.026 (3) | 0.013 (2) |
| O5 | 0.070 (3) | 0.0543 (18) | 0.0553 (18) | 0.013 (2) | −0.012 (2) | −0.0226 (17) |
| O6 | 0.059 (2) | 0.065 (2) | 0.0376 (17) | 0.0056 (19) | 0.004 (2) | −0.0063 (17) |
| O7 | 0.119 (4) | 0.076 (3) | 0.072 (2) | 0.003 (3) | 0.004 (3) | −0.026 (2) |
| N1 | 0.080 (4) | 0.044 (2) | 0.072 (3) | 0.000 (3) | −0.014 (3) | −0.010 (2) |
| C1 | 0.043 (3) | 0.046 (3) | 0.050 (3) | −0.001 (2) | 0.006 (3) | −0.006 (2) |
| C2 | 0.042 (3) | 0.044 (3) | 0.054 (3) | 0.001 (3) | 0.000 (3) | −0.010 (2) |
| C3 | 0.032 (3) | 0.040 (3) | 0.049 (3) | −0.003 (2) | −0.002 (3) | −0.001 (2) |
| C4 | 0.037 (3) | 0.044 (2) | 0.040 (2) | −0.007 (2) | 0.001 (3) | −0.005 (2) |
| C5 | 0.046 (3) | 0.052 (3) | 0.034 (3) | 0.002 (3) | 0.007 (3) | 0.001 (2) |
| C6 | 0.036 (3) | 0.068 (3) | 0.044 (3) | 0.003 (3) | 0.004 (3) | 0.008 (3) |
| C7 | 0.055 (4) | 0.107 (5) | 0.067 (4) | 0.016 (4) | 0.019 (4) | 0.004 (4) |
| C8 | 0.061 (4) | 0.094 (4) | 0.065 (3) | 0.024 (4) | 0.013 (4) | −0.018 (3) |
| C9 | 0.038 (3) | 0.096 (4) | 0.072 (4) | 0.016 (4) | 0.006 (3) | −0.008 (4) |
| C10 | 0.056 (4) | 0.079 (4) | 0.074 (4) | 0.026 (4) | 0.002 (4) | −0.009 (3) |
| C11 | 0.042 (4) | 0.067 (4) | 0.067 (3) | 0.011 (3) | −0.005 (3) | −0.016 (3) |
| C12 | 0.039 (3) | 0.052 (3) | 0.046 (3) | −0.004 (3) | −0.003 (3) | −0.005 (2) |
| C13 | 0.042 (3) | 0.050 (3) | 0.042 (3) | 0.008 (3) | 0.001 (3) | −0.004 (2) |
| C14 | 0.052 (4) | 0.043 (3) | 0.037 (3) | 0.001 (3) | 0.004 (3) | −0.002 (3) |
| C15 | 0.046 (3) | 0.035 (2) | 0.042 (3) | 0.000 (2) | 0.004 (3) | −0.007 (2) |
| C16 | 0.037 (3) | 0.075 (3) | 0.072 (4) | 0.010 (3) | −0.002 (3) | −0.007 (3) |
| C17 | 0.036 (4) | 0.097 (4) | 0.082 (4) | −0.013 (4) | 0.006 (4) | 0.011 (4) |
| C18 | 0.098 (5) | 0.074 (4) | 0.097 (4) | 0.020 (4) | 0.012 (5) | −0.023 (3) |
| C19 | 0.092 (6) | 0.132 (6) | 0.111 (5) | 0.060 (5) | −0.011 (5) | −0.021 (5) |
| C20 | 0.061 (4) | 0.073 (3) | 0.075 (3) | 0.006 (3) | −0.017 (4) | −0.026 (3) |
| C21 | 0.046 (3) | 0.042 (3) | 0.048 (3) | 0.002 (3) | −0.008 (3) | −0.002 (3) |
| C22 | 0.039 (3) | 0.039 (2) | 0.046 (3) | 0.002 (3) | −0.002 (3) | −0.002 (2) |
| C23 | 0.053 (4) | 0.053 (3) | 0.053 (3) | 0.004 (3) | −0.006 (3) | 0.000 (3) |
| C24 | 0.073 (4) | 0.071 (3) | 0.053 (3) | 0.002 (4) | −0.029 (3) | −0.014 (3) |
| C25 | 0.082 (5) | 0.058 (4) | 0.069 (3) | −0.015 (4) | −0.016 (4) | −0.025 (3) |
| C26 | 0.051 (3) | 0.050 (3) | 0.052 (3) | 0.000 (3) | −0.014 (3) | −0.002 (2) |
| C27 | 0.058 (4) | 0.079 (4) | 0.052 (3) | 0.009 (4) | 0.007 (3) | −0.016 (3) |
| C28 | 0.104 (5) | 0.117 (5) | 0.045 (3) | 0.003 (5) | −0.004 (4) | −0.006 (3) |
| C29 | 0.044 (3) | 0.044 (3) | 0.077 (4) | −0.004 (3) | 0.007 (3) | 0.007 (3) |
| C30 | 0.085 (5) | 0.045 (3) | 0.126 (5) | −0.011 (4) | 0.011 (5) | −0.004 (3) |
Geometric parameters (Å, °)
| O1—C21 | 1.340 (5) | C10—C11 | 1.522 (7) |
| O1—C3 | 1.465 (5) | C11—C12 | 1.463 (7) |
| O2—C21 | 1.198 (5) | C11—H11 | 0.9800 |
| O3—C29 | 1.354 (6) | C12—C13 | 1.341 (6) |
| O3—C15 | 1.457 (5) | C12—H12 | 0.9300 |
| O4—C29 | 1.199 (6) | C13—C14 | 1.471 (7) |
| O5—C14 | 1.217 (5) | C13—C20 | 1.507 (7) |
| O6—C27 | 1.349 (6) | C14—C15 | 1.547 (6) |
| O6—C5 | 1.470 (5) | C16—H16A | 0.9600 |
| O7—C27 | 1.196 (6) | C16—H16B | 0.9600 |
| N1—C25 | 1.323 (6) | C16—H16C | 0.9600 |
| N1—C26 | 1.328 (5) | C17—H17A | 0.9300 |
| C1—C2 | 1.518 (6) | C17—H17B | 0.9300 |
| C1—C15 | 1.542 (6) | C18—H18A | 0.9600 |
| C1—H1A | 0.9700 | C18—H18B | 0.9600 |
| C1—H1B | 0.9700 | C18—H18C | 0.9600 |
| C2—C3 | 1.516 (6) | C19—H19A | 0.9600 |
| C2—C16 | 1.529 (7) | C19—H19B | 0.9600 |
| C2—H2 | 0.9800 | C19—H19C | 0.9600 |
| C3—C4 | 1.523 (6) | C20—H20A | 0.9600 |
| C3—H3 | 0.9800 | C20—H20B | 0.9600 |
| C4—C5 | 1.532 (6) | C20—H20C | 0.9600 |
| C4—C15 | 1.582 (6) | C21—C22 | 1.478 (6) |
| C4—H4 | 0.9800 | C22—C26 | 1.375 (6) |
| C5—C6 | 1.521 (7) | C22—C23 | 1.377 (6) |
| C5—H5 | 0.9800 | C23—C24 | 1.364 (6) |
| C6—C17 | 1.303 (7) | C23—H23 | 0.9300 |
| C6—C7 | 1.508 (7) | C24—C25 | 1.368 (7) |
| C7—C8 | 1.521 (7) | C24—H24 | 0.9300 |
| C7—H7A | 0.9700 | C25—H25 | 0.9300 |
| C7—H7B | 0.9700 | C26—H26 | 0.9300 |
| C8—C9 | 1.500 (7) | C27—C28 | 1.492 (7) |
| C8—H8A | 0.9700 | C28—H28A | 0.9600 |
| C8—H8B | 0.9700 | C28—H28B | 0.9600 |
| C9—C10 | 1.487 (7) | C28—H28C | 0.9600 |
| C9—C11 | 1.513 (7) | C29—C30 | 1.499 (7) |
| C9—H9 | 0.9800 | C30—H30A | 0.9600 |
| C10—C18 | 1.504 (8) | C30—H30B | 0.9600 |
| C10—C19 | 1.518 (8) | C30—H30C | 0.9600 |
| C21—O1—C3 | 116.8 (3) | O5—C14—C13 | 119.4 (5) |
| C29—O3—C15 | 115.9 (3) | O5—C14—C15 | 115.2 (4) |
| C27—O6—C5 | 117.0 (4) | C13—C14—C15 | 124.7 (4) |
| C25—N1—C26 | 116.1 (4) | O3—C15—C1 | 109.5 (4) |
| C2—C1—C15 | 107.4 (4) | O3—C15—C14 | 114.0 (4) |
| C2—C1—H1A | 110.2 | C1—C15—C14 | 112.4 (4) |
| C15—C1—H1A | 110.2 | O3—C15—C4 | 106.9 (3) |
| C2—C1—H1B | 110.2 | C1—C15—C4 | 103.7 (4) |
| C15—C1—H1B | 110.2 | C14—C15—C4 | 109.6 (4) |
| H1A—C1—H1B | 108.5 | C2—C16—H16A | 109.5 |
| C3—C2—C1 | 102.8 (3) | C2—C16—H16B | 109.5 |
| C3—C2—C16 | 116.3 (4) | H16A—C16—H16B | 109.5 |
| C1—C2—C16 | 113.9 (4) | C2—C16—H16C | 109.5 |
| C3—C2—H2 | 107.8 | H16A—C16—H16C | 109.5 |
| C1—C2—H2 | 107.8 | H16B—C16—H16C | 109.5 |
| C16—C2—H2 | 107.8 | C6—C17—H17A | 120.0 |
| O1—C3—C2 | 110.9 (4) | C6—C17—H17B | 120.0 |
| O1—C3—C4 | 107.8 (3) | H17A—C17—H17B | 120.0 |
| C2—C3—C4 | 103.0 (4) | C10—C18—H18A | 109.5 |
| O1—C3—H3 | 111.6 | C10—C18—H18B | 109.5 |
| C2—C3—H3 | 111.6 | H18A—C18—H18B | 109.5 |
| C4—C3—H3 | 111.6 | C10—C18—H18C | 109.5 |
| C3—C4—C5 | 116.6 (4) | H18A—C18—H18C | 109.5 |
| C3—C4—C15 | 103.8 (4) | H18B—C18—H18C | 109.5 |
| C5—C4—C15 | 117.3 (4) | C10—C19—H19A | 109.5 |
| C3—C4—H4 | 106.1 | C10—C19—H19B | 109.5 |
| C5—C4—H4 | 106.1 | H19A—C19—H19B | 109.5 |
| C15—C4—H4 | 106.1 | C10—C19—H19C | 109.5 |
| O6—C5—C6 | 104.4 (4) | H19A—C19—H19C | 109.5 |
| O6—C5—C4 | 107.3 (4) | H19B—C19—H19C | 109.5 |
| C6—C5—C4 | 117.4 (4) | C13—C20—H20A | 109.5 |
| O6—C5—H5 | 109.2 | C13—C20—H20B | 109.5 |
| C6—C5—H5 | 109.2 | H20A—C20—H20B | 109.5 |
| C4—C5—H5 | 109.2 | C13—C20—H20C | 109.5 |
| C17—C6—C7 | 121.7 (5) | H20A—C20—H20C | 109.5 |
| C17—C6—C5 | 123.0 (5) | H20B—C20—H20C | 109.5 |
| C7—C6—C5 | 115.1 (5) | O2—C21—O1 | 124.7 (4) |
| C6—C7—C8 | 116.5 (4) | O2—C21—C22 | 123.5 (4) |
| C6—C7—H7A | 108.2 | O1—C21—C22 | 111.8 (4) |
| C8—C7—H7A | 108.2 | C26—C22—C23 | 117.1 (4) |
| C6—C7—H7B | 108.2 | C26—C22—C21 | 124.0 (4) |
| C8—C7—H7B | 108.2 | C23—C22—C21 | 118.8 (4) |
| H7A—C7—H7B | 107.3 | C24—C23—C22 | 119.6 (5) |
| C9—C8—C7 | 115.4 (5) | C24—C23—H23 | 120.2 |
| C9—C8—H8A | 108.4 | C22—C23—H23 | 120.2 |
| C7—C8—H8A | 108.4 | C23—C24—C25 | 118.3 (5) |
| C9—C8—H8B | 108.4 | C23—C24—H24 | 120.9 |
| C7—C8—H8B | 108.4 | C25—C24—H24 | 120.9 |
| H8A—C8—H8B | 107.5 | N1—C25—C24 | 124.2 (5) |
| C10—C9—C8 | 125.6 (5) | N1—C25—H25 | 117.9 |
| C10—C9—C11 | 61.0 (3) | C24—C25—H25 | 117.9 |
| C8—C9—C11 | 123.5 (5) | N1—C26—C22 | 124.6 (4) |
| C10—C9—H9 | 112.5 | N1—C26—H26 | 117.7 |
| C8—C9—H9 | 112.5 | C22—C26—H26 | 117.7 |
| C11—C9—H9 | 112.5 | O7—C27—O6 | 124.0 (5) |
| C9—C10—C18 | 121.3 (5) | O7—C27—C28 | 124.7 (5) |
| C9—C10—C19 | 117.5 (6) | O6—C27—C28 | 111.4 (5) |
| C18—C10—C19 | 113.0 (5) | C27—C28—H28A | 109.5 |
| C9—C10—C11 | 60.4 (4) | C27—C28—H28B | 109.5 |
| C18—C10—C11 | 119.6 (5) | H28A—C28—H28B | 109.5 |
| C19—C10—C11 | 115.4 (5) | C27—C28—H28C | 109.5 |
| C12—C11—C9 | 118.4 (5) | H28A—C28—H28C | 109.5 |
| C12—C11—C10 | 123.5 (5) | H28B—C28—H28C | 109.5 |
| C9—C11—C10 | 58.7 (3) | O4—C29—O3 | 123.3 (4) |
| C12—C11—H11 | 114.9 | O4—C29—C30 | 125.7 (5) |
| C9—C11—H11 | 114.9 | O3—C29—C30 | 111.0 (5) |
| C10—C11—H11 | 114.9 | C29—C30—H30A | 109.5 |
| C13—C12—C11 | 126.6 (5) | C29—C30—H30B | 109.5 |
| C13—C12—H12 | 116.7 | H30A—C30—H30B | 109.5 |
| C11—C12—H12 | 116.7 | C29—C30—H30C | 109.5 |
| C12—C13—C14 | 121.7 (4) | H30A—C30—H30C | 109.5 |
| C12—C13—C20 | 122.9 (5) | H30B—C30—H30C | 109.5 |
| C14—C13—C20 | 115.0 (4) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: WW2099).
References
- Appendino, G., Tron, G. C., Cravotto, G., Palmisano, G. & Jakupovic, J. (1999). J. Nat. Prod.62, 76–79. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Fujiwara, M., Ijichi, K., Tokuhisa, K., Katsuura, K., Shigeta, S., Konno, K., Wang, G.-Y.-S., Uemura, D., Yokota, T. & Baba, M. (1996). Antimicrob. Agents Chemother.40, 271–273. [DOI] [PMC free article] [PubMed]
- Gabe, E. J., Le Page, Y., Charland, J.-P., Lee, F. L. & White, P. S. (1989). J. Appl. Cryst.22, 384–387.
- Gabe, E. J. & White, P. S. (1993). DIFRAC Am. Crystallogr. Assoc. Meet., Pittsburgh, Abstract PA 104.
- Kupchan, S. M., Uchida, I., Branfman, A. R., Dailey, R. G. Jr & Fel, B. Y. (1976). Science, 191, 571–572. [DOI] [PubMed]
- Pharmacopoeia Commission of the People’s Republic of China (2005). Pharmacopoeia of the People’s Republic of China 2005, p. 24. Beijing: Chemical Industry Press.
- Sheldrick, G. M. (1997). SHELXS97 and SHELXL97 University of Göttingen, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536807066901/ww2099sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536807066901/ww2099Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report