Abstract
Purpose
About 65% to 70% of melanomas harbor a mutation in v-raf murine sarcoma viral oncogene homolog B1 (BRAF) that causes the steady-state activation of extracellular signal-regulated kinase (ERK). We sought to investigate the efficacy of PLX4032 (BRAF inhibitor) to identify patterns/predictors of response/resistance and to study the effects of BRAF in melanoma.
Experimental Design
Well-characterized melanoma cell lines, including several with acquired drug resistance, were exposed to PLX4032. Growth inhibition, phosphosignaling, cell cycle, apoptosis, and gene expression analyses were performed before and after exposure to drug.
Results
Using a growth-adjusted inhibitory concentration of 50% cutoff of 1 µM, 13 of 35 cell lines were sensitive to PLX4032, 16 resistant, and 6 intermediate (37%, 46%, and 17% respectively). PLX4032 caused growth inhibition, G0/G1 arrest, and restored apoptosis in the sensitive cell lines. A BRAF mutation predicted for but did not guarantee a response, whereas a neuroblastoma RAS viral oncogene homolog mutation or wild-type BRAF conferred resistance. Cells with concurrent BRAF mutations and melanocortin 1 receptor germ line variants and/or a more differentiated melanocyte genotype had a preferential response. Acquired PLX4032 resistance reestablishes ERK signaling, promotes a nonmelanocytic genotype, and is associated with an increase in the gene expression of certain metallothioneins and mediators of angiogenesis.
Conclusions
PLX4032 has robust activity in BRAF mutated melanoma. The preclinical use of this molecule identifies criteria for its proper clinical application, describes patterns of and reasons for response/resistance, and affords insight into the role of a BRAF mutation in melanoma.
Introduction
Malignant melanoma (MM) has long been considered a single histologic entity with heterogeneous clinical phenotypes. Evaluations of sun damage, mutations (v-raf murine sarcoma viral oncogene homolog B1 [BRAF], neuroblastoma RAS viral (v-ras) oncogene homolog [NRAS], and v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog [CKIT]), and gene polymorphisms (melanocortin 1 receptor [MC1R]) indicate that melanoma is actually composed of distinct clinical and molecular entities driven by various oncogenic events [1–3]. The identification of these molecular alterations permits the development of targeted therapies tailored to the specific pathway lesions of individual tumors. The presence of these aberrations, however, has not necessarily predicted for or conferred expected clinical responses [4,5]. This emphasizes the diverse nature of melanoma, questions its dependence on single “oncogenic” events, demonstrates the complexity inherent to and of targeting molecular pathways, and highlights the need to identify the proper application of novel inhibitors through preclinical models.
The mitogen-activated protein kinase (MAPK) pathway is implicated in the pathogenesis and propagation of melanoma because of mutations in transmembrane receptor tyrosine kinases (RTKs) (CKIT 3%–5%), membrane-associated guanine nucleotide binding proteins (NRAS 15%–20%), and cytoplasmic serine/threonine kinases (BRAF 60%–75%). Collectively, the effect of these aberrant signaling mediators is the steady-state activation of extracellular signal-regulated kinase (ERK) observed in 90% of melanomas [6,7]. Regardless of the causative upstream event, ERK overexpression promotes the differentiation, malignant transformation, proliferation, and survival of MM [7]. Specific to ERK activation in BRAF mutated melanoma, as opposed to RTK mediated ERK activation, is its resistance to negative feedback inhibition from the dual specific phosphatases (DUSP) and sprouty family (SPRY) of RAF binding proteins [8]. Also, the presence of a BRAF mutation (BRAFm) seems to evoke downstream transcriptional activity of ERK through MYC, FOS-like antigen 1 (FOSL1), and the ETS family of transcription factors [8]. Activation of the phosphoinositide-3-kinase (PI3K) pathway is documented in more than 70% of MM [9]. Synergistic activity of BRAF and v-akt murine thymoma viral oncogene homolog (AKT) has been implicated in the pathogenesis and malignant transformation of melanoma [10,11].
In BRAF-mutated melanomas, a simple amino acid transversion (exon 15, activation loop) confers a 500-fold increase in its kinase activity [12]. This, along with its role in the pathogenesis and propagation of MM, makes it an attractive clinical target. PLX4032 (RO5185426; Plexxikon/Roche, Berkeley, CA) inhibits oncogenic BRAF mutated at residue 600 with high affinity (IC50 44 nM) [13]. Early reporting of a phase 1 clinical trial of PLX4032 has yielded promising results in MM [14,15]. Clinical responses seem to be restricted to tumors with a BRAFm; however, the presence of a BRAFm did not unconditionally confer a response. Stratifying patients in the context of defined pathway lesions is a critical step in applying targeted therapies and in determining predictors of response. It is feasible to scrutinize the efficacy of PLX4032 in preclinical models to determine the effects of BRAF inhibition in melanoma and the molecular role of a BRAFm in melanogenesis and to gain an understanding as to which patient should be treated with MAPK pathway inhibitors. We investigated these questions by evaluating the inhibitory/molecular effects of PLX4032 in a well-characterized panel of MM cell lines.
Materials and Methods
Cell Lines, Culture, and Reagents
Cell lines are described in Figure 1A. SKMEL2, SKMEL28, and WM2664 were cultured with Eagle minimal essential medium (American Type Culture Collection [ATCC], Manassas, VA). G361 and SKMEL3 were cultured with McCoy's-5A modified medium with l-glutamine (ATCC). All others were cultured in RPMI-1640 (ATCC). All media were supplemented with 10% heat-inactivated FBS (Omega Scientific, Inc, Tarzana, CA) and 1% penicillin and streptomycin (Irvine Scientific, Santa Ana, CA).
Figure 1.
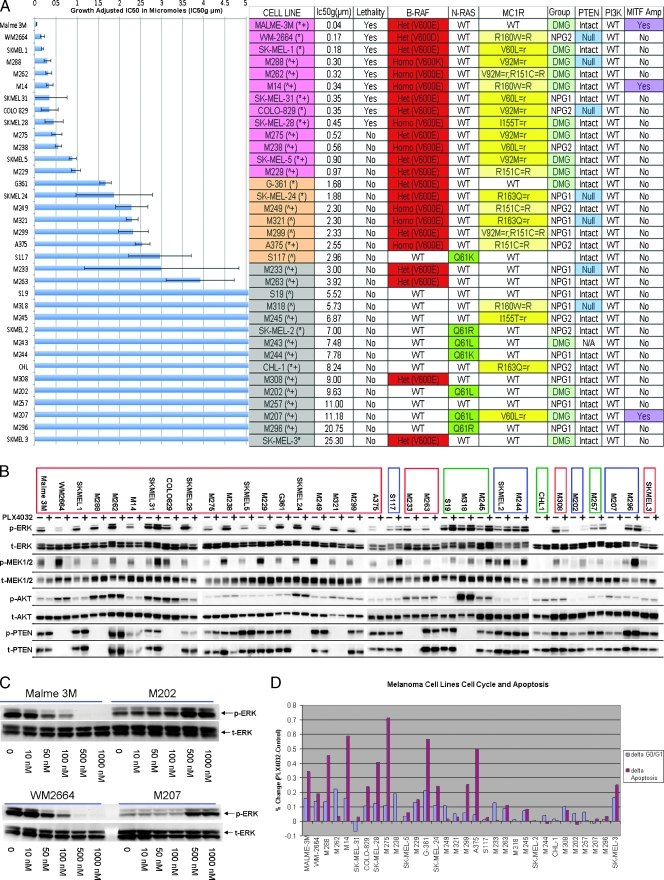
Functional activity of PLX4032. (A) Growth inhibition assay of MM cell lines (n = 35) after exposure to PLX4032. The concentration of drug in micromolars that achieves a growth adjusted inhibitory concentration of 50% (IC50g µM) (x-axis), melanoma cell line (y-axis). SE is depicted. Table outlines characteristics of the 35 MM cell lines. Order of cell lines corresponds to the growth inhibition chart. Cell lines are color coordinated based on response (pink—clear sensitivity, orange—intermediate sensitivity, gray—resistant). In parenthesis next to the cell line: *cells were purchased from ATCC or ^developed from tumor samples at UCLA (University California, Los Angeles), +cell lines that were included in the complementary RNA mixed reference pool for the microarray analysis. Lethality = cell death in addition to growth inhibition. BRAF and NRAS status with specific amino acid substitutions; MC1R germ line status/gene polymorphism; R versus r describes the functional effect of MC1R variant/relation to skin pigmentation [3]. Gene expression groupings: DMG = differentiated melanocyte group, NPG = neuronal precursor group. PTEN and PI3K status: presence of MITF amplification as determined by focal in situ hybridization (>2 genes per chromosome 3 centromeres). (B) Phosphoprotein signaling of MM cell lines through Western blot analysis before and after exposure to PLX4032. Cell lines are listed in order of sensitivity. Composite arrangement of multiple Western blots is depicted by dividing lines. BRAFm cell lines are outlined in a red box; NRASm, blue; and BRAFwt/NRASwt, green. (C) Western blot analysis of two BRAFm (Malme3M and WM2664) and two NRASm (M202 and M207) MM cell lines after exposure to increasing concentrations of PLX4032. (D) Cell cycle and apoptosis assays as done by flow cytometry. Chart depicts percent change in the number of cells in G0/1 (blue bar) and the number of cells in apoptosis (maroon bar) of the treated cell line versus the untreated control.
Polymerase Chain Reaction and Sequencing
DNA was extracted with DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). Exons were amplified by polymerase chain reaction (HotMaster Taq Polymerase; 5 Prime, Hamburg, Germany). Products were purified (PCR Purification Kit; Qiagen) and sequenced (BigDye Terminator Kit, 3730XL Instrument; Applied Biosystems, Inc, Foster City, CA). Primer sets are shown in the Supplemental Material.
Proliferation Assays
Proliferation assays were performed as previously described [16]. Serial dilutions of PLX4032 started at 10 µM. Growth inhibition/lethality was calculated as previously described [16] (RAD001 [Selleck Chemicals, Houston, TX] and LY294002 [Sigma-Aldrich, St Louis, MO]).
Western Blot
Western blot analysis was performed as previously described [16]. Cells were treated with 3 µM PLX4032 for 30 minutes. Antibodies (anti-AKT1, anti-AKT2, anti-AKT3, anti-AKT [C67E7], anti-phospho-p70 S6 kinase [Ser371], anti-p70 S6 kinase, anti-phospho-MAPK/ERK kinase [MEK1/2; Ser217/221], anti-MEK1/2, anti-phospho-A-Raf [Ser299], anti-A-Raf, anti-phospho-B-Raf [Ser445], anti-Braf [55C6], anti-phospho-C-Raf [Ser529], and anti-C-Raf) were purchased from Cell Signaling Technology, Inc (Danvers, MA).
Cell Cycle and Apoptosis
Cell cycle and apoptosis assays were performed as previously described [16]. Cells were treated with 3 µM PLX4032. For the cell cycle analysis, cells were harvested after 3 days. For the apoptosis assay, cells were harvested after 5 days.
Acquired PLX4032 Resistance
Cell lines were separated into two T-75 flasks and cultured in parallel; one was treated at their growth-adjusted inhibitory concentration of 50% (IC50g), the other with DMSO. Concentrations of PLX4032 were increased as resistance developed. Cell line identity was confirmed by mitochondrial DNA comparative analysis to the highly variable regions 1/2 of the modified Cambridge sequence. Growth rate of M14R, M288R, and SKMEL28R was determined as follows: M14R, M288R, and SKMEL28R were grown in two sets of flasks, one in standard culture medium, the other in culture medium supplemented with 3 µM of PLX4032. On day 7, cells from each flask were plated in duplicate in 12-well plates (10,000 cells/2 ml/well). PLX4032 (3 µM) was added to the medium of cells initially grown in the presence of PLX4032 (W/PLX4032); of the cells initially grown without PLX4032, one set was supplemented with the BRAF inhibitor (W/O → W/PLX4032), the other was continued to be grown in the absence of drug (W/O PLX4032). Cell counts were performed on days 0, 3, 4, 5, 6, 7, 10, 11, 12, 13, and 14 (Z1 Particle Counter; Beckman Coulter, Brea, CA).
Ras Activation Assay
Ras activation assay was performed as previously described [17]. The cells were cultured to approximately 85% to 90% confluence and were harvested after being treated with 3 µM PLX4032 or DMSO for 30 minutes. Equal amounts of cell lysates were used for the activation assay (Ras Activation Assay Kit; Millipore, Billerica, MA).
Microarray Analyses
Baseline characterization. RNA was extracted, quantified, and hybridized as previously described [16]. A total of 35 melanoma cell lines were hybridized to Agilent Human 1A-oligo arrayV1 chips (Agilent Technologies, Santa Clara, CA).Melanoma cell lines used in the mixed reference complementary RNA pool are listed (Figure 1A).
Before/After PLX4032. Cells were grown to midlog phase and treated with 3 µM PLX4032 or DMSO for 24 hours. Cell harvesting/RNA extraction was previously described [16]. Microarray hybridizations were performed using the Agilent Human 4x44K array chip (Agilent Technologies).
Microarray slides were read as previously described [16]. Rosetta Resolver analysis of variance (ANOVA) was performed using the Benjamini-Hochberg false discovery rate multiple test correction. Two-dimensional cluster analysis was performed using an agglomerate hierarchical clustering algorithm based on the cosine correlation similarity metric.
Results
Mutational Status/Response PLX4032
A panel of 35 melanoma cell lines was exposed to PLX4032 through a 7-day growth inhibition assay. A differential response pattern was established (Figure 1A). Using an IC50g cutoff of 1 µM as clearly sensitive and 3 µM as resistant, 13 cell lines were sensitive, 16 were resistant, and 6 were intermediate. Mutational analysis revealed that a V600 BRAFm conferred sensitivity (clear and intermediate) regardless of zygosity or amino acid substitution (E, D, K). Sensitive cell lines (clear and intermediate) harbored a BRAFm, whereas only four BRAF mutated cell lines were resistant. An NRAS mutation (NRASm) or the absence of either mutation (BRAFwt/NRASwt) conferred resistance (7/7 NRASm and 5/5 BRAFwt/NRASwt resistant). These data show the specificity of PLX4032 and suggest that its clinical application should focus on V600 BRAF mutated melanomas regardless of zygosity or specific amino acid substitution. However, as in the phase 1 clinical trial, a BRAFm did not necessarily confer a response to treatment [14,15].
Evaluation of MC1R status showed a predilection for MC1R variants (MC1Rv) in the sensitive cell lines but did not supersede the presence of a BRAFm (Figure 1A). Moreover, 12 of 13 sensitive and 5 of 6 intermediate lines had a MC1Rv. All of these cell lines also had a concurrent BRAFm. Only 4 of 16 resistant cell lines had a MC1Rv. None of these had a concurrent BRAFm. It has been reported that MC1Rv are more prevalent in BRAF mutated samples [3,18]. Our data confirm this and suggest that melanomas with a MC1Rv may be more dependent on selective ERK signaling associated with a BRAFm as opposed to an aberrant RTK or a NRASm. No correlation was observed in the BRAFm/MC1Rv cell lines regarding the specific MC1R polymorphism or its proposed functional effect (r vs R).
BRAFm cooperates with phosphatase and tensin homolog (PTEN) loss in MM development [11]. Of 35 cell lines, 7 (20%) had a PTEN mutation (PTENm) (Figure 1A). PTENm were mutually exclusive with NRASm as previously reported [19]. Of the seven PTENm samples, six also had concurrent BRAFm; therefore, a trend was observed between PTEN status and response to PLX4032. As most of the sensitive cell lines were BRAFm/PTENwt, it is apparent that the BRAFm, regardless of PTEN status, drives the response to PLX4032. These data suggest that although up-regulation of the PI3K/AKT/mechanistic target of rapamycin (mTOR) pathway through PTEN loss may potentiate melanoma formation in BRAF mutated nevi, once established, an MM sample depends preferentially on aberrant ERK signaling under the influence of BRAFm. The role of PTEN loss through mutations or epigenetic modifications may become more prominent in the BRAF mutated samples if ERK signaling is perturbed. The PI3K/AKT pathway may then regulate the MAPK pathway similar to its activity in early melanocytogenesis [10].
Phosphosignaling
Western blot analysis was performed before and after exposure to PLX4032 (Figure 1B). p-ERK was downregulated after treatment in all BRAF mutated samples including the resistant ones. SKMEL31, sensitive, was an exception to this. A discrepancy exists in the literature regarding the BRAF status of SKMEL31 [8,20–23]. In this analysis, SKMEL31 was confirmed through repeated sequencing as having a heterozygous BRAFm. However, evaluation of its chromatogram suggests that it may have an allelic imbalance in which the percentage of the wild-type allele is increased compared to the mutant [24]. A decrease in p-ERK was not noted in NRAS mutated or BRAFwt/NRASwt samples (Figure 1B). Rather, after treatment, these samples had an initial increase in p-ERK signaling. This was time- (data not shown) and dose-dependent (Figure 1C). Evaluation of total and p-MEK, a direct readout of RAF signaling, mirrored p-ERK signaling (Figure 1B). This demonstrates that no additional cellular input occurs in between RAF and ERK signaling and that the evaluation p-ERK signaling gives an accurate assessment of RAF activity after exposure to PLX4032. No significant changes were seen in p-AKT in any of the samples (Figure 1B).
Cell Cycle and Apoptosis
ERK activity in BRAF mutated melanoma cells is capable of driving cellular proliferation through dysregulation of the cell cycle. Aberrant MAPK activity can also suppress apoptosis by inactivating proapoptotic members of the Bcl-2 family (BH3 and BH1/BH3 domain containing proteins) and by increasing antiapoptotic members (Bcl-2, Mcl-1, and Bcl-XL). Cell cycle and apoptosis assays were performed before and after exposure to PLX4032 (Figure 1D). An accumulation of cells in the G0/G1 phase of the cell cycle was noted in the majority of sensitive but not resistant lines. Evaluation of apoptosis showed that PLX4032 caused an increase in apoptosis in the majority of the sensitive as opposed to the resistant samples. Together, these data show that inhibiting the mutated BRAF kinase can abrogate the proliferative effects and survival advantage of aberrant ERK signaling.
Baseline Gene Expression
We evaluated the gene expression profiles of the cell lines before exposure to PLX4032. Unsupervised clustering revealed two distinct molecular groups (Figure 2A). This is consistent with others who have also shown heterogeneity in the gene expression profiles of melanoma samples [25–27]. An ANOVA was performed to identify and evaluate the individual transcriptomes of each group.
Figure 2.
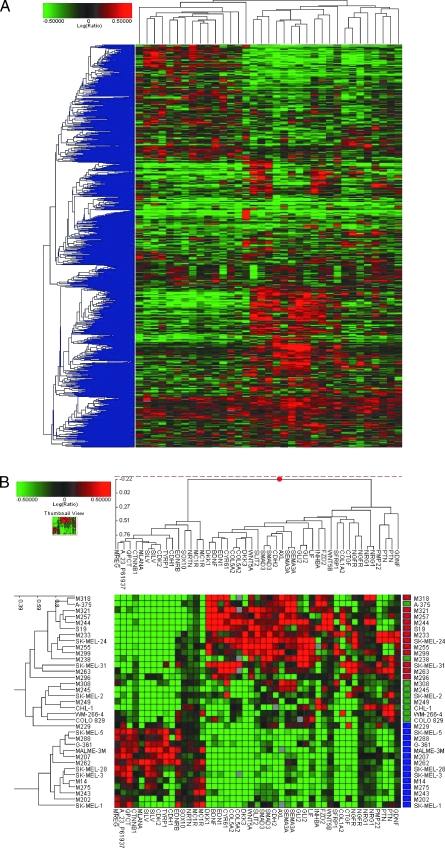
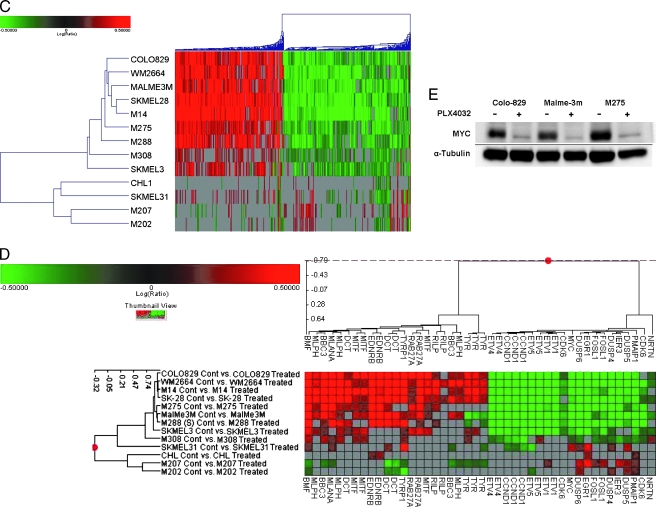
Gene expression analysis of the 35 MM cell lines. (A) Global view of unsupervised two-dimensional clustering of the 35 cell lines using a statistical cutoff for genes with a two-fold change in at least four experiments (3997 resulting genes). Cell lines (top), individual genes (left). Red increased expression; green, decreased expression. Color intensity correlates with degree of expression compared with the complementary RNA mixed reference pool. (B) Supervised clustering: differential gene expression patterns of melanoma cell lines generated by an ANOVA. Two-dimensional clustering of 35 melanoma cell lines based on the expression pattern of selective genes associated with MITF (A_23_P61937) and its downstream signaling pathway, melanocyte function, melanin synthesis, noncanonical Wnt and Tgfβ pathways, and neuronal mediators of NCC development. Length of dendogram arm depicts degree of association of the cell lines based on the expression pattern of these 37 genes. MM subgroups are represented by color blocks on the right of the cluster diagram (blue—group A [DMG], red—group B1 [NPG1], green—group B2 [NPG2]). (C) Global clustering of eight sensitive and five resistant cell lines after their exposure to PLX4032. Expression signatures depict the fold change of the posttreatment versus pretreatment samples. A statistical cutoff for genes with at least a two-fold change in at least four experiments was performed, resulting in 1927 genes that had a P < .01. (D) Two-dimensional clustering that displays the expression patterns of genes associated with aberrant ERK signaling and melanocyte differentiation and function. Each row displays the difference in the expression of genes in the post treatment line compared with the pretreatment line. (E) Western blot analysis of C-MYC expression in selected cell lines before and after exposure to PLX4032.
Group A is dominated by melanoma markers and genes associated with melanin production and melanocyte function (Table W1A). We therefore designated group A as the differentiated melanocyte group (DMG). The gene expression profile of the DMG includes genes upregulated through the activity of the microphthalmia-associated transcription factor (MITF) and its downstream signaling pathway. As an illustration, differential clustering highlights the link between and shows the expression pattern of melanocyte-specific genes MITF, β-catenin (CTNNB1), and cyclin-dependent kinase 2 (CDK2; Figure 2B). CTNNB1, through activity of the canonical Wnt pathway, is known to increase the expression of MITF [28]. CDK2 is a major cell cycle regulator linked to melanoma growth through MITF [29].
Comparatively, group B is defined by the lack of expression of melanocyte-specific genes. A significant decrease in MITF and β-catenin is noted compared with the DMG (Figure 2B). Correlative changes were noted in the activity of the Wnt receptor signaling pathways, suggesting activity of the noncanonical Wnt pathway in group B with resulting inhibition of the canonical pathway [30–32]. Wingless-type MMTV integration site family 5a (Wnt5a), upregulated in group B, increases the motility and metastatic potential of melanoma [33,34], and consistent with this analysis, Wnt5a has also been shown to suppress melanoma associated antigens such as gp100 [35].
CTNNB1 and MITF play a crucial role in melanoblast differentiation from the neural crest (NC) [28,36]. The relative decreased expression of these genes suggests that cells of group B develop along a NC lineage distinct from typical melanocytes. Specific growth factors have been identified that regulate NC cell (NCC) differentiation and development. Endothelin 1, neuregulin, neurturin, and glial cell line-derived neurotrophic factor influence the development of NCC precursors along mesenchymal and neural lineages [37]. The differential expression of these ligands in the cell lines affords insight into their potential developmental patterns (Figure 2B; Table W1B).
A subset of melanocytes that are produced from skin-innervating nerve cells has been described [38]. These nerve cells develop from Schwann cell precursors (SCPs) that retain the developmental potential to form melanocytes. The melanocyte-forming SCPs migrate along the ventral aspect of the neural tube, opposed to the melanoblast-fated NCC that migrate from the dorsal aspect. This neuronal versus melanoblastic fate decision is determined by the dueling expression of H6 family homeobox 1 (HMX1) and SRY (sex-determining region Y)-box 10 (SOX10). HMX1 expression in the dorsal root ganglia differentiates NCC into neuronal phenotypes. Absence of HMX1 allows the influence of SOX10 to dominate development. SOX10+ cells express MITF and develop along the well-described melanocytic lineage [38].
We suspect that cell lines in the DMG follow typical patterns of melanocyte development from the dorsal aspect of the neural tube. This group is defined by the expression of MITF, CTNNB1, and other melanocyte specific NC mediators (endothelin receptor type B [EDNRB] and neurturin). Cells in group B lack the relative influences of MITF and CTNNB1 and express ligands typical of neuronal and mesenchymal NCC development (Figure 2B and Table W1B). This suggests that they may represent the versatility of less differentiated progenitor/precursor cells that develop into melanocytes under strong neuronal influences, possibly the SOX10- SCP cells. As such, melanoma group B is designated the neuronal precursor group (NPG). The NPG is composed of two subgroups (NPG1 and NPG2), as defined by their respective gene expression signature. NPG2 lacks the expression of genes that strongly differentiate the DMG from the NPG1 (Figure 2B).
Gene Expression Groupings/Response PLX4032
PLX4032 growth inhibition results were compared with the gene expression groupings that are listed above (Figure 1A). Of the 13 cell lines in the clearly sensitive group, 9 belong to the DMG, whereas 4 belong to the NPG (1 in NPG1 and 3 in NPG2). Of the intermediate group, one of six cell lines is in the DMG, whereas five are in the NPG (three in NPG1 and two NPG2). Of the 16 resistant cell lines, 4 are in the DMG and 12 are in the NPG (8 in NPG1, 3 in NPG2, and 1 N/A). Specifically, 23 cell lines have a BRAFm (11 in DMG and 12 in NPG). Of those in the DMG, nine are clearly sensitive, one is intermediate, and one is resistant. Of the BRAFm cell lines in the NPG, four are clearly sensitive (one in NPG1 and three in NPG2), five are intermediate (three in NPG1 and two in NPG2), and three are resistant (three in NPG1 and none in NPG2). Taken together, these observations suggest that melanoma cells become more dependent on downstream transcriptional effects of BRAF mutated ERK signaling as they become more differentiated, possibly because of the increased interactions with MITF.
Gene Expression Profile Before/After Exposure PLX4032
An ANOVA was performed comparing PLX4032-sensitive and -resistant cell lines. Distinct gene expression signals could not be identified apart from the patterns that define the DMG and the NPG. To overcome the expressional strength of each subgroup, gene expression profiling was performed on individual samples before and after PLX4032 exposure (Figure 2C and Table W2). Thirteen cell lines were chosen (eight were sensitive and five were resistant; ten with a BRAFm, two with a NRASm, and one WT; eight in DMG, two in NPG1, and three in NPG2). Three cell lines clustered differently than their growth inhibition patterns would have suggested. SKMEL31, BRAFm/sensitive, had few changes in its posttreatment array; M308 and SKMEL3, BRAFm/resistant, had an expression profile more similar to the sensitive lines. The cluster pattern of these 13 cell lines is very consistent with the patterns of p-ERK signaling that were noted on Western analysis after exposure to PLX4032. The expression profile of SKMEL31, sensitive, clusters with the resistant cell lines CHL1, M207, and M202; these four cell lines show continued p-ERK signaling on Western analysis after exposure to PLX4032. M308 and SKMEL3 show loss of p-ERK despite being resistant to PLX4032. These cell lines cluster with the seven PLX4032-sensitive cell lines, all of which had abrogation of p-ERK signaling. These observations suggest that the dominant genetic signatures generated by this analysis are due to the effect/lack of effect of PLX4032 on p-ERK signaling.
To investigate the individual genes and molecular processes associated with the application of PLX4032, a function-based gene analysis was performed on the before/after treatment data. Illustrative gene changes are depicted (Figure 2D).
Protein Kinase Activity
An assessment of protein kinase activity in the sensitive lines revealed a significant decrease in DUSP 4, 5, and 6 and in SPRY 2 and 4. DUSPs are highly inducible cytoplasmic and nuclear phosphatases that negatively regulate ERK activity. SPRYs are also negative regulators of MAPK signaling with target effects above ERK. These noted decreases support down-regulation of the MAPK pathway in BRAF mutated cells through target inhibition with PLX4032. Similar reciprocal events were seen in melanoma samples treated with selective MEK inhibitors [8,39].
Cellular Proliferation/Apoptosis
The effects of ERK signaling on cellular proliferation and apoptosis in cancer are well described [7,40]. Activity of ERK-dependent transcription factors, including MYC, EGR1, FOSL1, IER3, and the family of ETS variants (ETV1, ETV4, and ETV5), has been noted in BRAF mutated MM [8]. The treatment of BRAF mutated melanoma cells with PLX4032 caused a significant decrease in the expression of these transcription factors. The decreased expression of MYC was confirmed on the protein level (Figure 2E). Significant changes were noted in the downstream effectors of these factors including a decrease in selective markers of cellular proliferation (CCND1 and CDK6) and an increase in several BH3 domain-containing proapoptotic factors (BMF and PUMA/BBC3; Figure 2D).
Differentiation/Functional Genes
An interesting phenomenon noted with the application of selective MEK inhibitors in melanoma is an increase in markers of melanocyte differentiation. This was originally described with tyrosinase (TYR) and EDNRB [39,41]. TYR is an enzyme necessary for the conversion of tyrosine to melanin; EDNRB is a receptor for endothelin, a ligand that is essential for melanocyte development and the transcription of MITF. TYR and EDNRB were preferentially expressed in the DMG. Because of this observation, the significant disparity in the expression of melanocyte differentiation markers between the DMG and NPG, and the preferential response of the DMG to BRAF inhibition, we examined the patterns of differentiation that occur after exposure to PLX4032 (Figure 2D). PLX4032 caused an increase in the expression of melanocyte-specific genes in sensitive cell lines of both the DMG and NPG. Significant changes were seen in TYR, TYRP1, EDNRB, MLANA, DCT, and MITF. Genes associated with melanosome function, RAB27A, MYO5, MLPH, and RILP, were also upregulated. It seems that a BRAFm causes a differentiation arrest that inhibits typical melanocyte function as pertaining to melanin synthesis and transport. Prolonged p-ERK signaling has been shown to be a negative regulator of melanogenesis [42]. This prevents the overproduction of lethal amounts of melanin. The inhibition of aberrant ERK signaling may relieve the negative feedback inhibition on melanogenesis and explain why an increase in differentiation markers specific to melanin production and transport is noted after treatment with PLX4032.
Acquired PLX4032 Resistance
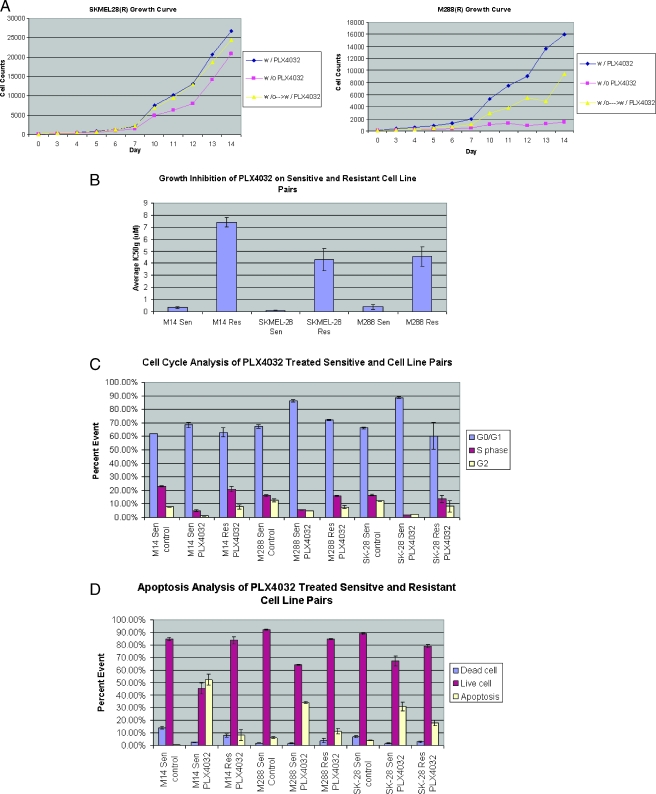
Six BRAFm/PLX4032-sensitive cell lines (M288, SKMEL28, M14, WM2664, COLO829, and M238) were grown in increasing concentrations of PLX4032. Three have acquired resistance (M288R, SKMEL28R, and M14R). The growth rate of these cell lines is, to a variable degree, dependent on the continued presence of PLX4032 (Figure 3A). Growth of M288R, PTEN null, seems fully dependent on the presence of PLX4032. This cell line did not grow in the absence of PLX4032 and remained quiescent until the starved serum was resupplemented with drug (Figure 3A). SKMEL28R and M14R grew slower in the absence of PLX4032; however, these cells were able to overcome their lag phase and enter a growth phase that was less than, but parallel to, the growth of the cell line that was maintained in the continued presence of drug. This is depicted in the growth assay of SKMEL28R (Figure 3A).
Figure 3.
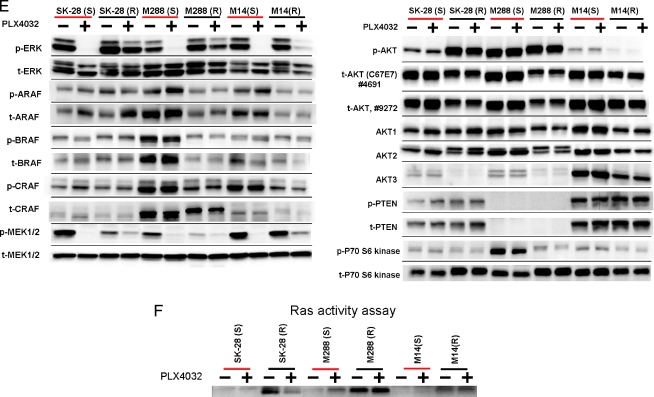
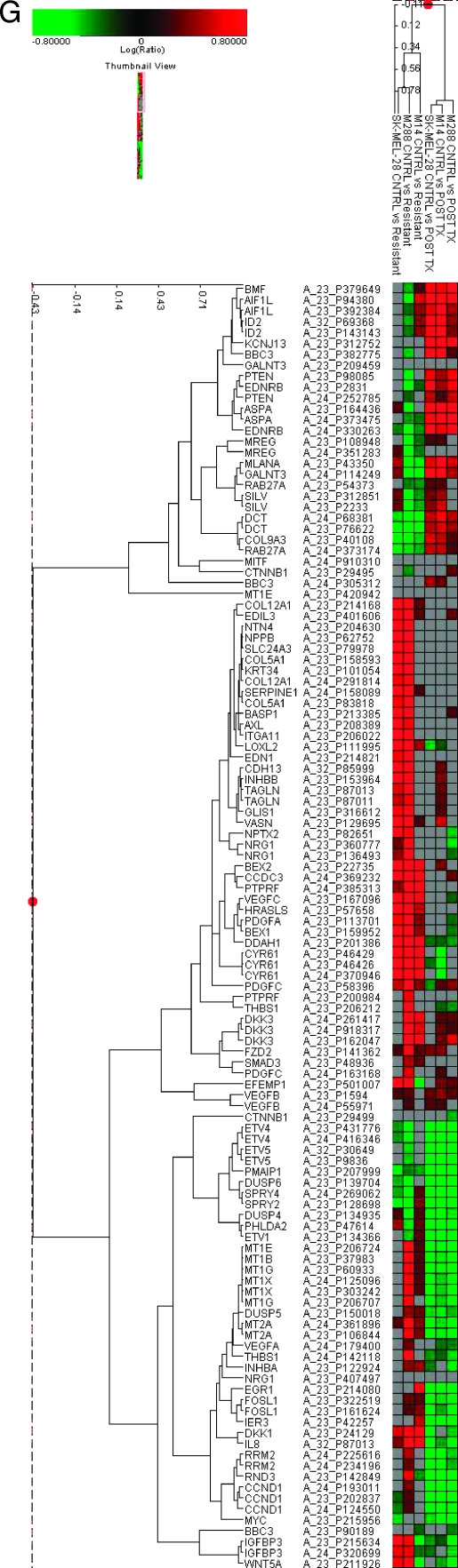
Analysis of resistant cell lines (M288R, SK-MEL-28R, and M14R) compared with their sensitive parental counterparts (M288, SK-MEL-28, and M14). (A) Growth curves of SKMEL28R and M288R. Blue lines—cells grown in the continued presence of PLX4032, pink—absence of PLX4032, yellow—grown in the absence and then the presence of PLX4032; y-axis—cell count, x-axis—day of count. (B) Growth inhibition assay (IC50g µM) in the presence of PLX4032. (C) Cell cycle and (D) apoptosis assays as done by flow cytometry. (E) Phosphoprotein signaling of MM cell lines before and after exposure to PLX4032. Two separate total AKT antibodies that recognize different epitopes were used. (F) RAS activation assay in sensitive and resistant counterparts before and after exposure to PLX4032. (G) A comparative gene expression analysis of the expression profiles of M288R, SKMEL28R, and M14R and M14, M288, and SKMEL28. Selected genes are listed. Columns depict fold change in the gene expression of M288R, SKMEL28R, and M14R compared with M14, M288, and SKMEL28 and the fold change in the gene expression in the post PLX4032 treatment samples of M14, SKMEL28, and M288 compared with pretreatment controls.
Of the six cell lines grown in the constant presence of PLX4032, M288, SKMEL28, and M14 have, whereas WM2664, COLO829, and M238 have not yet, acquired resistance. M288, SKMEL28, and M14 are all in the DMG. This is opposed to WM2664, COLO829, and M238 who are all in the NPG2. This suggests that, although cell lines of the DMG show a preferential response to PLX4032, their response may be more transient. Such an observation may provide, by evaluating genes specific to the DMG, molecular biomarkers that could be used clinically to identify patients who are at high risk for developing early PLX4032 resistance.
Resistance was confirmed with growth inhibition assays, lack of a cell cycle arrest, and absence of apoptosis (Figure 3, B–D). Western blot analysis shows that p-ERK signaling was maintained after exposure to PLX4032 in M288R, SK-MEL-28R, and, to a lesser degree, M14R (Figure 3E). These data suggest that acquired resistance develops in part by reestablishing MAPK signaling. The fact that only a partial restoration of p-ERK signaling was noted in M14R suggests that the acquisition of resistance may be multifactorial and mediated by pathways other than the MAPK. Evaluation of ARAF, BRAF, and CRAF signaling in SKMEL28R, M14R, and M288R did not show an increase in the phosphosignaling of these mediators compared with their parental counterparts (Figure 3E).
MEK signaling was also evaluated in the sensitive/resistant pairs (Figure 3E). p-MEK signaling was not as strong in SKMEL28R, M14R, and M288R compared with SKMEL28, M14, and M288. As expected, treatment with PLX4032 abrogated p-MEK signaling in SKMEL28, M14, and M288; however, p-MEK signaling was only partially reestablished in the resistant counterparts. The intensity of p-MEK did not correlate with the brisk p-ERK signal that was noted in SKMEL28R and M288R after exposure to PLX4032. This suggests that mediators other than RAF/MEK may activate ERK in the cell lines with acquired PLX4032 resistance.
RAS status was also evaluated in the three cell lines with acquired resistance. Exon 2 of NRAS was sequenced. As expected, sequences in M288R, SKMEL28R, M14R were identical to their sensitive parental cell lines (data not shown). As numerous additional mechanisms exist, which could cause RAS activation, we performed a RAS assay (Figure 3F). M288R and, to a lesser degree, SKMEL28R and M14R had an increase in RAS-GTP levels compared with their parental counterparts. These data suggest that RAS activity is upregulated in BRAFm MM samples that acquire resistance to PLX4032.
Evaluation of the AKT/mTOR pathway was also performed. Subtle increases in p-AKT were noted in SKMEL28R and M288R (PTEN null) but not in M14 (Figure 3E). The amount of total AKT seemed to be less in SKMEL28R, M14R, and M288R compared with SKMEL28, M14, and M288. Evaluation of the three AKT isoforms suggests that this is, in part, explained by decreasing levels of AKT 3.
M14, M288, and SKMEL28 (sensitive and resistant) were exposed to RAD001, an mTOR inhibitor, and to LY294002, a PI3K inhibitor. Both RAD001 and LY294002 had minimal effect as a single agent on SKMEL28 (IC50g 12 and 8 µM, respectively) and M14 (15 and 7 µM) or SKMEL28R (11 and 7 µM) and M14R (14 and 5 µM). M288 was resistant to LY294002 (IC50g 7 µM) and relatively resistant to RAD001 (IC50g 3 µM). Comparatively, M288R was more sensitive to both RAD001 and LY294002 (IC50g 1 and 2 µM, respectively). However, the number of generations observed in M288R in the growth inhibition assay was limited because the single-agent assays were done in the absence of PLX4032. In M288R, the combination of RAD001 and PLX4032 or LY294002 and PLX4032 did not improve the response to either drug alone (data not shown). Overall, these data do not offer strong support for the PI3K pathway contributing to the growth and survival of BRAFm MM cell lines that acquired PLX4032 resistance.
Because of the rapidity in which resistance developed, we favor the reestablishment of MAPK signaling, the activation of feedback mechanisms, and/or epigenetic modifications rather than secondary point mutations that prevent PLX4032 from interacting with the mutated BRAF. We sequenced the G loop and the activation domain, exons 11 and 15, respectively, of the BRAF kinase. Sequences in M288R, SKMEL28R, and M14R were identical to their sensitive parental cell lines (data not shown). This suggests that resistance does not occur through a change in ATP binding or phosphorylation capacity.
A gene expression analysis was conducted comparing M288R, SKMEL28R, and M14R to M14, M288, and SKMEL28 at baseline and after the parental strains were treated with PLX4032 (Figure 3G and Table W3). Such an analysis reveals the genetic changes that a cell undergoes as it acquires drug resistance and how resistance affects the expression signature that is generated by PLX4032.
The expression profile of M288R, SKMEL28R, and M14R revealed variable changes in the expression patterns of DUSP 4, 5, and 6 and SPRY 2 and 4, the nuclear transcription factors MYC, EGR1, FOSL1, IER3, ETV1, ETV4, and ETV5 as well as certain melanocytic genes (DCT, RAB27A, MLANA, and SILV). Genes that defined the NPG such as wnt5A, neuregulin, endothelin 1, DKK1, INHBB, AXL, and CYR61 were upregulated to variable degrees in M288R, SKMEL28R, and M14R compared with their parental counterparts. It seems that although MAPK signaling is reestablished when resistance develops, it is not specifically driven by a BRAFm because the characteristic gene expression signature of a BRAFm was not encountered. This is in accordance with the patterns of RAS activity and p-ERK/p-MEK signaling that was noticed in the cell lines with acquired resistance. In addition, M288R, SKMEL28R, and M14R belong to the DMG. The fact that an NPG expression signature is encountered in cells of the DMG suggests that the expression signature of the NPG represents a more primordial/default signature that develops in the absence of melanocytic differentiation or differentiation signals.
Finally, numerous genes were upregulated in M288R, SKMEL28R, and M14R compared to M14, M288, and SKMEL28 (Figure 3G and Table W3). This included platelet-derived growth factor A and certain mediators of angiogenesis (interleukin 8, VEGFC). Significant increases were also noted in the expression of certain metallothioneins (MTs). MTs are low-molecular weight cysteine-rich proteins that function in intracellular metal metabolism/detoxification. A role for these proteins is emerging in cancer, including melanoma in which their expression is an independent prognostic variable [43]. MTs also have antiapoptotic activity and are implicated in metal- and nonmetal-containing cytostatic drugs' resistance [44,45].
The increased expression of these mediators in PLX4032-acquired resistance suggests the activity of additional survival pathways that may be amenable to targeted therapy either in rescue of or in combination with BRAF inhibition. Also, genes such as interleukin 8 or the MTs may be useful biomarkers that signal when resistance is about to develop in patients undergoing treatment with PLX4032.
Discussion
MM, once thought to be a single entity, is now considered a molecular and developmentally diverse disease. This observation allows for the generation and application of novel therapies specifically targeted to the molecular pathway lesions of a patient's disease. Such an approach affords the opportunity to positively affect the historically low survival rates in melanoma and serves as a molecular tool to help dissect the oncogenic events that drive this disease.
The effect of aberrant ERK signaling in MM is well described [7]. Consequently, the clinical application of selective MAPK inhibitors is currently underway. PLX4032 is a potent inhibitor of the mutated form of BRAF. An early phase 1 trial of this compound in MM has been promising [14]. However, concurrent preclinical molecular research is required to supplement clinical findings and to help direct the proper clinical application of PLX4032, including how to predict and overcome the development of drug resistance.
We tested the efficacy of PLX4032 in a large panel of well-characterized MM cell lines. The presence of BRAFm predicted for, but did not guarantee, a response to therapy. In addition to a BRAFm, samples that had a concurrent MC1Rv and a more differentiated melanocytic genotype had a preferential response. This provides valuable information that could be used to screen and select future clinical trial participants and to retrospectively analyze tissue and outcome results from the completed phase 1 trial.
Phosphoprofiling of our samples before and after PLX4032 treatment confirmed the inhibition of p-ERK signaling in all cell lines with a BRAFm regardless of response. This confirms the specificity of PLX4032 and the dependency of most BRAFm MM cells on p-ERK signaling. SKMEL31, sensitive, was an exception to this. After exposure to PLX4032, SKMEL31 had an increase in p-ERK signaling, much like NRAS mutated or BRAFwt/NRASwt samples. It is possible that SKMEL31 has a reverse mutant allele-specific imbalance in which the wild-type allele is increased compared to the mutant [24]. The presence of the mutated allele must be sufficient to drive its malignant phenotype and confer sensitivity to PLX4032. However, the imbalance in favor of the wild-type allele could explain why its post-PLX4032 p-ERK signaling pattern is more consistent with a BRAFwt cell line. Of the 23 MM cell lines with a BRAFm, 15 are heterozygous. Evaluation of the chromatogram of these cell lines revealed SKMEL31 and SKMEL3 as the only two lines that have a higher peak of the wild-type allele compared with the mutant. Interestingly, SKMEL3 is the most resistant cell line in the panel. It is possible that this reverse allelic imbalance will explain the phosphosignaling pattern of SKMEL31 and provide a way to predict resistance in patients who are heterozygous for the BRAFm.
Abrogation of MAPK signaling in the PLX4032-resistant BRAFm cell lines suggests that a small population of MM samples exist that have aberrant ERK signaling but can survive in the absence of this pathway's functional input. These samples (BRAFm PLX4032-resistant) were heterozygous for the BRAFm, tended to be in the NPG, and were without a MC1Rv. Further validation of the unique qualities of this population may identify alternate signaling pathways, treatment options, and those patients with a BRAFm who would not benefit from single-agent RAF inhibition. Also, our analysis clearly suggests that patients with NRASm or NRASwt/BRAFwt tumors would not benefit from PLX4032. As expected, the application of PLX4032 in these cells did not abrogate p-ERK signaling. Instead, selective BRAF inhibition seemed to activate the MAPK pathway as previously described [46,47]. A correlation in response to PLX4032 was noted in the BRAF mutated samples that have a concurrent MC1R polymorphism. BRAFm/MC1R normal cell lines were resistant to the effects of PLX4032. This suggests that melanoma cells that have BRAFm/MC1Rv are more dependent on aberrant p-ERK signaling as driven by the BRAFm. The dominant effect of both pathways may converge on MITF signaling. This may explain why cell lines in the DMG that are BRAFm/MC1Rv seem to be more sensitive to PLX4032 than cell lines in the NPG that are BRAFm/MC1Rv.
The application of PLX4032 in the MM cell lines shows the downstream effect of ERK signaling in BRAF mutated samples. We performed cell cycle and apoptotic analyses before/after exposure to the drug. Interruption of ERK signaling decreased cellular proliferation and the ability to escape apoptosis. Our gene expression data suggest that BMF and PUMA mediate the restoration of apoptosis. Our expression analysis also confirmed that MAPK activity in BRAF mutated MM is resistant to negative feedback inhibition of DUSP and SPRY and that MYC, FOSL1, EGR1, and ETV 1, 4, and 5 are the main transcriptional activators of ERK as driven by a BRAFm.
An interesting observation noted with the disruption of ERK signaling is the reexpression of melanocyte-specific genes and markers of melanocyte differentiation. This includes TYR, MLANA, DCT, and MITF. It is uncertain why these differentiation markers are decreased in MM by ERK signaling. However, it has been suggested that prolonged ERK signaling will decrease the expression of MITF with subsequent down-regulation of melanogenesis to avoid the cytotoxic effects of excess melanin production [42]. Consistent with this is our observation that genes associated with melanosome function, RAB27A, MYO5, MLPH, and RILP, were reexpressed after MM cell lines with a BRAFm were treated with PLX4032. The inhibition of aberrant ERK signaling in BRAF mutated samples, especially ones that have a MC1Rv, may relieve the feedback inhibition of ERK and reestablish the innate drive for melanin production.
Adding to the complexity of the relationship between ERK signaling and melanocyte differentiation is the identification of a subset of melanoma samples whose genotype is more characteristic of neuronal precursors than that of typical melanocytes. These cells behave and function like melanocytes, develop into clinically indistinguishable melanomas, and are susceptible to common melanoma mutations. However, they have very distinct gene expression signatures that suggest activity of the noncanonical Wnt and TGFβ pathways at the expense of MITF, CTNNB1, and the canonical Wnt pathway. Interestingly, BRAF mutated MM samples of the NPG have an increase in melanocytic genes after PLX4032 exposure. Also, BRAF mutated MM cell lines of the DMG that develop resistance to PLX4032 have an increase in the expression of genes that are more characteristic of cells with in the NPG. These data suggest that plasticity does exist between the DMG and NPG and that cell lines of the NPG may be able to differentiate into a more melanocytic phenotype if they encounter the proper internal/external cues.
Our data show that the cells of DMG were more dependent on ERK signaling when driven by a BRAFm. Cell lines of the DMG were more sensitive to PLX4032 than were BRAFm cells in the NPG. Interestingly, those cell lines in the NPG that were clearly sensitive to PLX4032 were mostly in the NPG2, the group that was not defined by melanocytic or by neuronal genes. It is possible that this group represents a genetic bridge that links the DMG with the NPG1 and the plasticity inherent to NCC development.
We propose that cells of the NPG develop from the NC apart from the influence of SOX10 and MITF. In such, they never develop a strong melanocytic genotype and retain much of the neuronal influences of their precursors. A higher incidence of PTEN loss was noted in these cells. PTEN loss has been shown in conjunction with a BRAFm to facilitate melanoma development [10,11]. We noticed a predilection of the PTENm to be in the NPG regardless of BRAF status. This suggests that the less differentiated cells of the NPG may be more dependent on the cross talk between the MAPK and PI3K pathways that is thought to potentiate melanoma formation in BRAF mutated nevi.
As seen clinically, continual exposure of melanoma to PLX4032 can promote drug resistance especially in the more differentiated samples. Western blot analysis showed reactivation of p-ERK signaling. However, a gene expression analysis did not consistently reproduce the downstream signaling patterns of a fully penetrable BRAFm. These data suggest that other mediators may contribute to the acquisition of resistance. In the samples with PLX4032 acquired resistance, we noticed increase activity of RAS and a discordant signaling pattern between MEK and ERK. This may afford some insights into points of influence on MAPK pathway apart from a BRAFm. Changes in AKT and p-AKT signaling were noted in the resistant samples compared with their sensitive parental counterparts. However, further assessment, including treatment data with selective PI3K and mTOR inhibitors, did not make a strong argument for this pathways involvement in acquired PLX4032 resistance.
A gene expression analysis implicated several other mediators that may be involved in the acquisition of resistance to PLX4032; this includes members of the platelet derived growth factor family, increased expression of angiogenic factors, and increased activity of certain MTs. Such observations afford biomarkers to screen for the development of resistance in patients who are actively being treated with PLX4032 and possible clinical avenues to pursue when resistance occurs.
PLX4032 is a selective BRAF inhibitor that seems to have significant clinical activity in BRAF mutated MM. The development of this compound provides hope in a malignancy that has been notoriously refractory to standard therapies. The preclinical use of this molecule sheds light onto its proper clinical application and affords new insight into patterns of response and resistance. Such information furthers our understanding of this terrible malignancy and opens the door to new targeted therapies and the clinical means to properly apply them in the context of well-defined pathway lesions.
Supplementary Material
Acknowledgments
The authors thank UCLA Sequencing Core and Jim Economou and Antoni Ribas for providing the cell lines that were developed at UCLA.
Abbreviations
- AKT
v-akt murine thymoma viral oncogene homolog
- ANOVA
analysis of variance
- ATCC
American Type Culture Collection
- BRAF
v-raf murine sarcoma viral oncogene homolog B1
- BRAFm
BRAF mutation
- CKIT
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
- CTNNB1
β-catenin
- DMG
differentiated melanocyte group
- DUSP
dual specific phosphatase
- EDNRB
endothelin receptor type B
- ERK
extracellular signal-regulated kinase
- ETV
ets variant
- FOSL1
FOS-like antigen 1
- HMX
H6 family homeobox
- IC50g
growth-adjusted inhibitory concentration of 50%
- M14R
M14 with acquired PLX4032 resistance
- M288R
M288 with acquired PLX4032 resistance
- MM
malignant melanoma
- MAPK
mitogen-activated protein kinase
- MC1R
melanocortin 1 receptor
- MC1Rv
MC1R variant (gene polymorphism)
- MEK
MAPK/ERK kinase
- MITF
microphthalmia-associated transcription factor
- MT
metallothioneins
- MYC
v-myc myelocytomatosis viral oncogene homolog
- mTOR
mechanistic target of rapamycin
- NCC
neural crest cell
- NPG
neuronal precursor group
- NRAS
neuroblastoma RAS viral (v-ras) oncogene homolog
- NRASm
NRAS mutation
- PI3K
phosphoinositide-3-kinase
- PTEN
phosphatase and tensin homolog
- PTENm
PTEN mutation
- RTK
receptor tyrosine kinase
- SCP
Schwann cell precursor
- SOX
SRY (sex-determining region Y)-box
- SKMEL28R
SKMEL28 with acquired PLX4032 resistance
- SPRY
sprouty family of RAF binding proteins
- TYR
tyrosinase
Footnotes
This study was supported in part by the National Institutes of Health grant CA 120228-01 (R. Essner, P.I.).
This article refers to supplementary materials, which are designated by Tables W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 2.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 3.Landi MT, Bauer J, Pfeiffer RM, Elder DE, Hulley B, Minghetti P, Calista D, Kanetsky PA, Pinkel D, Bastian BC. MC1R germline variants confer risk for BRAF-mutant melanoma. Science. 2006;313:521–522. doi: 10.1126/science.1127515. [DOI] [PubMed] [Google Scholar]
- 4.Kim KB, Eton O, Davis DW, Frazier ML, McConkey DJ, Diwan AH, Papadopoulos NE, Bedikian AY, Camacho LH, Ross MI, et al. Phase II trial of imatinib mesylate in patients with metastatic melanoma. Br J Cancer. 2008;99:734–740. doi: 10.1038/sj.bjc.6604482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amaravadi RK, Schuchter LM, McDermott DF, Kramer A, Giles L, Gramlich K, Carberry M, Troxel AB, Letrero R, Nathanson KL, et al. Phase II trial of temozolomide and sorafenib in advanced melanoma patients with or without brain metastases. Clin Cancer Res. 2009;15:7711–7718. doi: 10.1158/1078-0432.CCR-09-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen C, Zavala-Pompa A, Sequeira JH, Shoji M, Sexton DG, Cotsonis G, Cerimele F, Govindarajan B, Macaron N, Arbiser JL. Mitogen-activated protein kinase activation is an early event in melanoma progression. Clin Cancer Res. 2002;8:3728–3733. [PubMed] [Google Scholar]
- 7.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 8.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kantrow SM, Boyd AS, Ellis DL, Nanney LB, Richmond A, Shyr Y, Robbins JB. Expression of activated Akt in benign nevi, Spitz nevi and melanomas. J Cutan Pathol. 2007;34:593–596. doi: 10.1111/j.1600-0560.2006.00675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung M, Sharma A, Madhunapantula SV, Robertson GP. Akt3 and mutant V600E B-Raf cooperate to promote early melanoma development. Cancer Res. 2008;68:3429–3439. doi: 10.1158/0008-5472.CAN-07-5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, You MJ, DePinho RA, McMahon M, Bosenberg M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray-Schopfer VC, Karasarides M, Hayward R, Marais R. Tumor necrosis factor-alpha blocks apoptosis in melanoma cells when BRAF signaling is inhibited. Cancer Res. 2007;67:122–129. doi: 10.1158/0008-5472.CAN-06-1880. [DOI] [PubMed] [Google Scholar]
- 13.Sala E, Mologni L, Truffa S, Gaetano C, Bollag GE, Gambacorti-Passerini C. BRAF silencing by short hairpin RNA or chemical blockade by PLX4032 leads to different responses in melanoma and thyroid carcinoma cells. Mol Cancer Res. 2008;6:751–759. doi: 10.1158/1541-7786.MCR-07-2001. [DOI] [PubMed] [Google Scholar]
- 14.Chapman P, Puzanov I, Sosman J, Kim KB, Ribas A, McArthur G, Lee R, Grippo J, Nolop K, Flaherty K. Early efficacy signal demonstrated in advanced melanoma in a phase I trial of the oncogenic BRAF-selective inhibitor PLX4032. Eur J Cancer. 2009;7(suppl 3):5. [Google Scholar]
- 15.Sondak VK, Smalley K. Targeting mutant BRAF and KIT in metastatic melanoma: ASCO 2009 meeting report. Pigment Cell Melanoma Res. 2009;22:386–387. doi: 10.1111/j.1755-148X.2009.00593.x. [DOI] [PubMed] [Google Scholar]
- 16.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamoutpour F, Bodempudi V, Park SE, Pan W, Mauzy MJ, Kratzke RA, Dudek A, Potter DA, Woo RA, O'Rourke DM, et al. Gene silencing for epidermal growth factor receptor variant III induces cell-specific cytotoxicity. Mol Cancer Ther. 2008;7:3586–3597. doi: 10.1158/1535-7163.MCT-08-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fargnoli MC, Pike K, Pfeiffer RM, Tsang S, Rozenblum E, Munroe DJ, Golubeva Y, Calista D, Seidenari S, Massi D, et al. MC1R variants increase risk of melanomas harboring BRAF mutations. J Invest Dermatol. 2008;128:2485–2490. doi: 10.1038/jid.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abi-Habib RJ, Urieto JO, Liu S, Leppla SH, Duesbery NS, Frankel AE. BRAF status and mitogen-activated protein/extracellular signal-regulated kinase kinase 1/2 activity indicate sensitivity of melanoma cells to anthrax lethal toxin. Mol Cancer Ther. 2005;4:1303–1310. doi: 10.1158/1535-7163.MCT-05-0145. [DOI] [PubMed] [Google Scholar]
- 22.Christensen C, Guldberg P. Growth factors rescue cutaneous melanoma cells from apoptosis induced by knockdown of mutated (V 600 E) B-RAF. Oncogene. 2005;24:6292–6302. doi: 10.1038/sj.onc.1208758. [DOI] [PubMed] [Google Scholar]
- 23.Hao H, Muniz-Medina VM, Mehta H, Thomas NE, Khazak V, Der CJ, Shields JM. Context-dependent roles of mutant B-Raf signaling in melanoma and colorectal carcinoma cell growth. Mol Cancer Ther. 2007;6:2220–2229. doi: 10.1158/1535-7163.MCT-06-0728. [DOI] [PubMed] [Google Scholar]
- 24.Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, Chari R, Shames DS, Tang X, MacAulay C, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009;4:e7464. doi: 10.1371/journal.pone.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 26.Shields JM, Thomas NE, Cregger M, Berger AJ, Leslie M, Torrice C, Hao H, Penland S, Arbiser J, Scott G, et al. Lack of extracellular signal-regulated kinase mitogen-activated protein kinase signaling shows a new type of melanoma. Cancer Res. 2007;67:1502–1512. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- 27.Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 28.Schepsky A, Bruser K, Gunnarsson GJ, Goodall J, Hallsson JH, Goding CR, Steingrimsson E, Hecht A. The microphthalmia-associated transcription factor Mitf interacts with β-catenin to determine target gene expression. Mol Cell Biol. 2006;26:8914–8927. doi: 10.1128/MCB.02299-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent β-catenin degradation. J Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedi P, Bafico A, Nieto Soria A, Burgess WH, Miki T, Bottaro DP, Kraus MH, Aaronson SA. Isolation and biochemical characterization of the human Dkk-1 homologue, a novel inhibitor of mammalian Wnt signaling. J Biol Chem. 1999;274:19465–19472. doi: 10.1074/jbc.274.27.19465. [DOI] [PubMed] [Google Scholar]
- 32.Finch PW, He X, Kelley MJ, Uren A, Schaudies RP, Popescu NC, Rudikoff S, Aaronson SA, Varmus HE, Rubin JS. Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci USA. 1997;94:6770–6775. doi: 10.1073/pnas.94.13.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, Trent JM. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 34.O'Connell MP, Fiori JL, Xu M, Carter AD, Frank BP, Camilli TC, French AD, Dissanayake SK, Indig FE, Bernier M, et al. The orphan tyrosine kinase receptor, ROR2, mediates Wnt5A signaling in metastatic melanoma. Oncogene. 2010;29(1):34–44. doi: 10.1038/onc.2009.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dissanayake SK, Olkhanud PB, O'Connell MP, Carter A, French AD, Camilli TC, Emeche CD, Hewitt KJ, Rosenthal DT, Leotlela PD, et al. Wnt5A regulates expression of tumor-associated antigens in melanoma via changes in signal transducers and activators of transcription 3 phosphorylation. Cancer Res. 2008;68:10205–10214. doi: 10.1158/0008-5472.CAN-08-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larue L, Kumasaka M, Goding CR. β-Catenin in the melanocyte lineage. Pigment Cell Res. 2003;16:312–317. doi: 10.1034/j.1600-0749.2003.00050.x. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Morales JR, Henrich T, Ramialison M, Wittbrodt J. New genes in the evolution of the neural crest differentiation program. Genome Biol. 2007;8:R36. doi: 10.1186/gb-2007-8-3-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adameyko I, Lallemend F, Aquino JB, Pereira JA, Topilko P, Muller T, Fritz N, Beljajeva A, Mochii M, Liste I, et al. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139:366–379. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 39.Ciuffreda L, Del Bufalo D, Desideri M, Di Sanza C, Stoppacciaro A, Ricciardi MR, Chiaretti S, Tavolaro S, Benassi B, Bellacosa A, et al. Growth-inhibitory and antiangiogenic activity of the MEK inhibitor PD0325901 in malignant melanoma with or without BRAF mutations. Neoplasia. 2009;11:720–731. doi: 10.1593/neo.09398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray-Schopfer VC, da Rocha Dias S, Marais R. The role of B-RAF in melanoma. Cancer Metastasis Rev. 2005;24:165–183. doi: 10.1007/s10555-005-5865-1. [DOI] [PubMed] [Google Scholar]
- 41.Solit DB, Santos E, Pratilas CA, Lobo J, Moroz M, Cai S, Blasberg R, Sebolt-Leopold J, Larson S, Rosen N. 3′-Deoxy-3′-[18F]fluorothymidine positron emission tomography is a sensitive method for imaging the response of BRAF-dependent tumors to MEK inhibition. Cancer Res. 2007;67:11463–11469. doi: 10.1158/0008-5472.CAN-07-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khaled M, Larribere L, Bille K, Aberdam E, Ortonne JP, Ballotti R, Bertolotto C. Glycogen synthase kinase 3β is activated by cAMP and plays an active role in the regulation of melanogenesis. J Biol Chem. 2002;277:33690–33697. doi: 10.1074/jbc.M202939200. [DOI] [PubMed] [Google Scholar]
- 43.Weinlich G, Eisendle K, Hassler E, Baltaci M, Fritsch PO, Zelger B. Metallothionein—overexpression as a highly significant prognostic factor in melanoma: a prospective study on 1270 patients. Br J Cancer. 2006;94:835–841. doi: 10.1038/sj.bjc.6603028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Endo T, Yoshikawa M, Ebara M, Kato K, Sunaga M, Fukuda H, Hayasaka A, Kondo F, Sugiura N, Saisho H. Immunohistochemical metallothionein expression in hepatocellular carcinoma: relation to tumor progression and chemoresistance to platinum agents. J Gastroenterol. 2004;39:1196–1201. doi: 10.1007/s00535-004-1471-1. [DOI] [PubMed] [Google Scholar]
- 45.Shimoda R, Achanzar WE, Qu W, Nagamine T, Takagi H, Mori M, Waalkes MP. Metallothionein is a potential negative regulator of apoptosis. Toxicol Sci. 2003;73:294–300. doi: 10.1093/toxsci/kfg095. [DOI] [PubMed] [Google Scholar]
- 46.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.