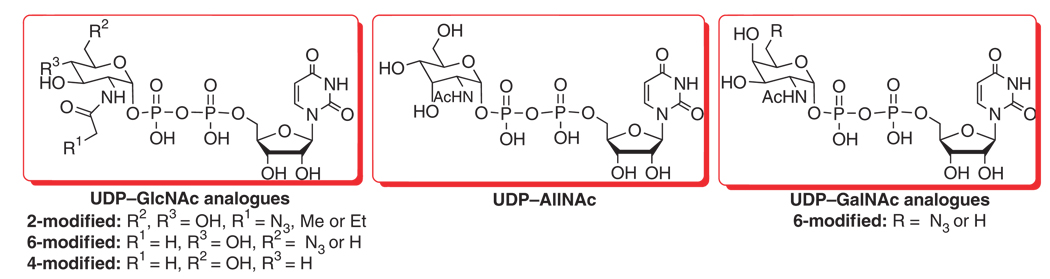
Abstract
Enzymatic synthesis using glycosyltransferases is a powerful approach to building polysaccharides with high efficiency and selectivity. Sugar nucleotides are fundamental donor molecules in enzymatic glycosylation reactions by Leloir-type glycosyltransferases. The applications of these donors are restricted by their limited availability. In this protocol, N-acetylglucosamine (GlcNAc)/N-acetylgalactosamine (GalNAc) are phosphorylated by N-acetylhexosamine 1-kinase (NahK) and subsequently pyrophosphorylated by N-acetylglucosamine uridyltransferase (GlmU) to give UDP–GlcNAc/GalNAc. Other UDP–GlcNAc/GalNAc analogues can also be prepared depending on the tolerance of these enzymes to the modified sugar substrates. Starting from l-fucose, GDP–fucose is constructed by one bifunctional enzyme l-fucose pyrophosphorylase (FKP) via two reactions.
INTRODUCTION
Carbohydrates have important structural and functional roles in numerous physiological processes, which include the development, growth, function and survival of an organism1,2. Many carbohydrates have greatly attracted the attention of scientists because of their potential medical applications, such as α-Gal epitope3, isoglobotrihexosylceramide derivatives4, Lewis X and other antigens from bacteria and their derivatives5–8.
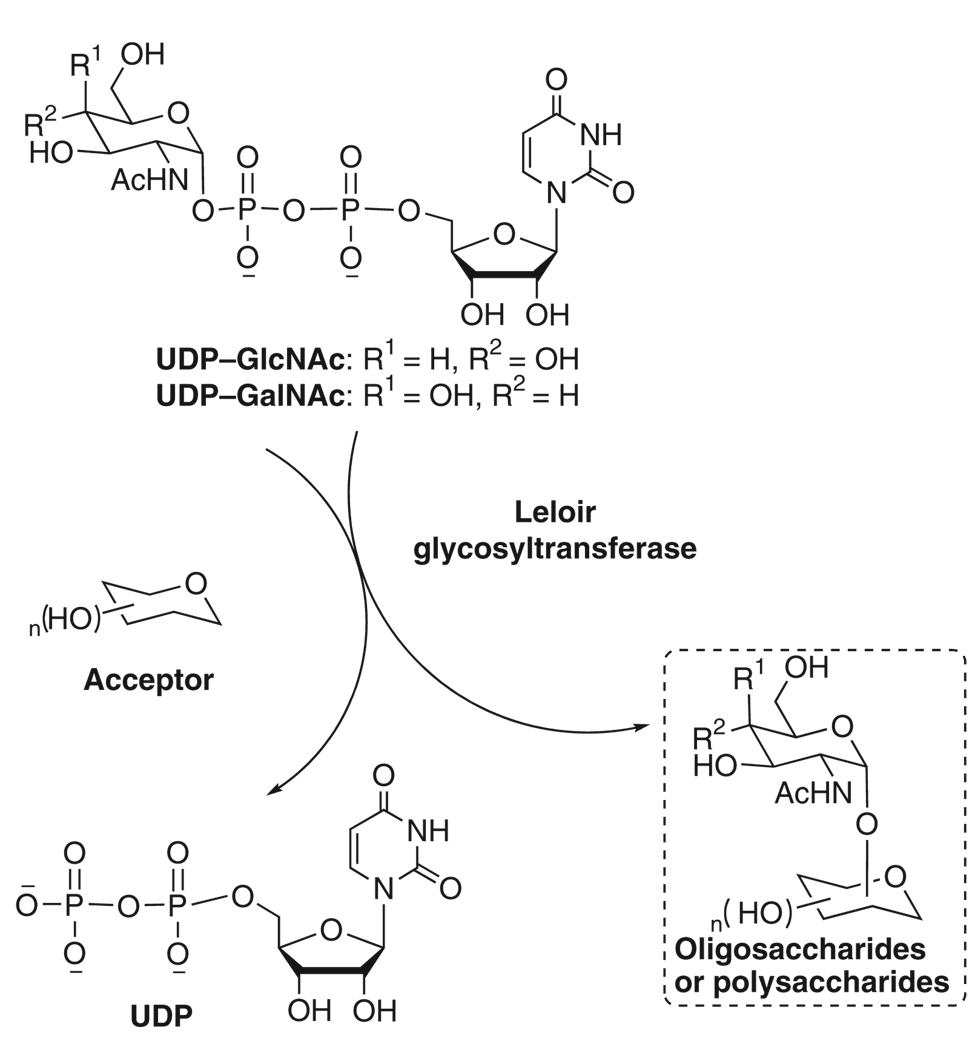
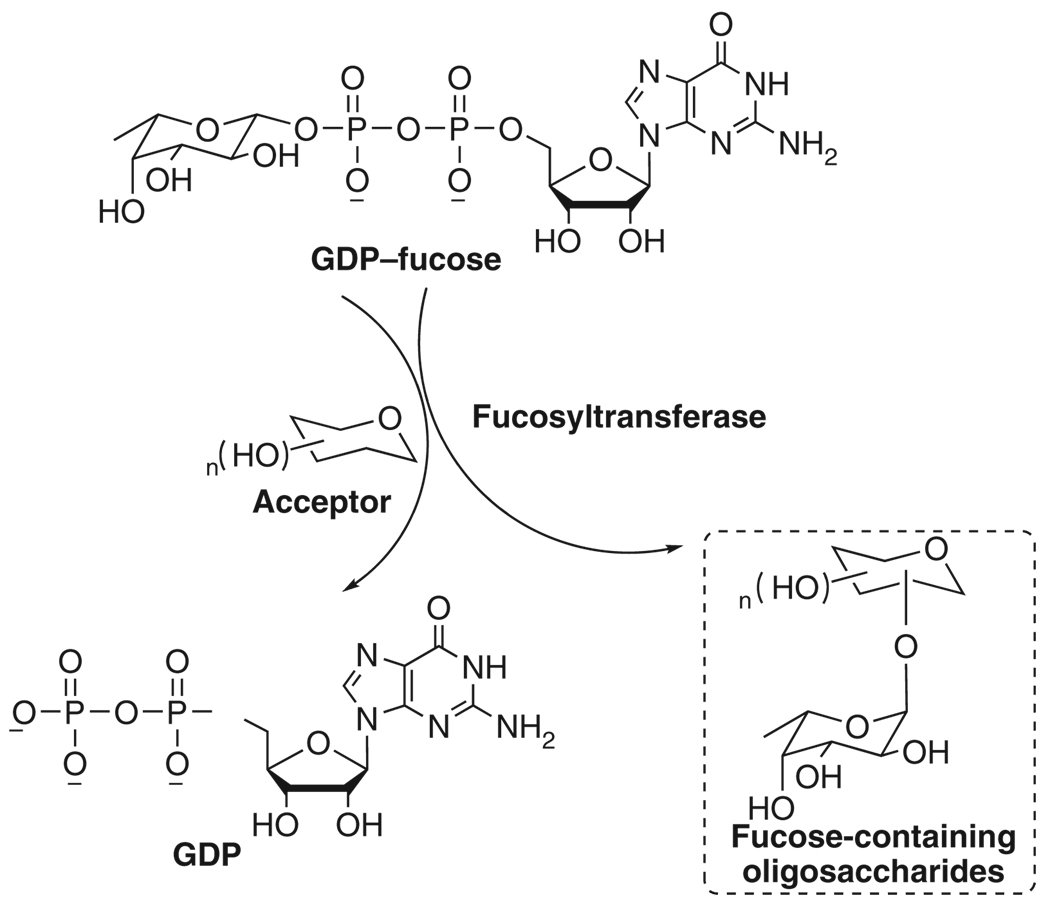
Glycosyltransferases have been used for the enzymatic synthesis of these significant carbohydrates, of which the main advantage compared with traditional chemical synthesis is the regio- and stereo-selectivity that can be achieved without protection of functional groups9. Sugar nucleotides are universal donors of enzymatic glycosylation reactions by Leloir pathway. They can be regarded as activated forms of sugar molecules, which will be added onto the nonreducing end of oligosaccharide by glycosyltransferases10. For example, N-acetylglucosamine (GlcNAc) and N-acetylgalactosamine (GalNAc) are incorporated into GlcNAc/GalNAc-containing polysaccharides by corresponding glycosyltransferases using UDP–GlcNAc/GalNAc (Fig. 1). GDP–fucose is the donor substrate of fucosyltransferases to synthesize fucosylated antigens such as sialyl Lewis X (Fig. 2). In eukaryotic cells, there are only ten sugar nucleotides which are the construction units for most glycobiological reactions11. In bacteria, the category of sugar nucleotides is diverse and many uncommon sugar nucleotides have been found.
Figure 1.
Incorporation of N-acetylglucosamine (GlcNAc)/N-acetylgalactosamine (GalNAc) into oligosaccharides or polysaccharides using UDP–GlcNAc/GalNAc as donor.
Figure 2.
Incorporation of fucose into target molecules using GDP–fucose as glycosylation donor.
However, the limited availability of sugar nucleotide donors and the key intermediate sugar-1-phosphates (sugar-1-Ps), as well, turns out to be the major bottleneck to the basic research and application of these components. Chemical synthesis, which involves multi-step synthetic manipulations and tedious purification processes, is laborious and time consuming. The yield is normally rather low as well12–15. Thus, an enzymatic route offers a more promising alternative, specifically if a monosaccharide can be directly 1-phosphorylated into sugar-1-P and followed by pyrophosphorylation to give a sugar nucleotide.
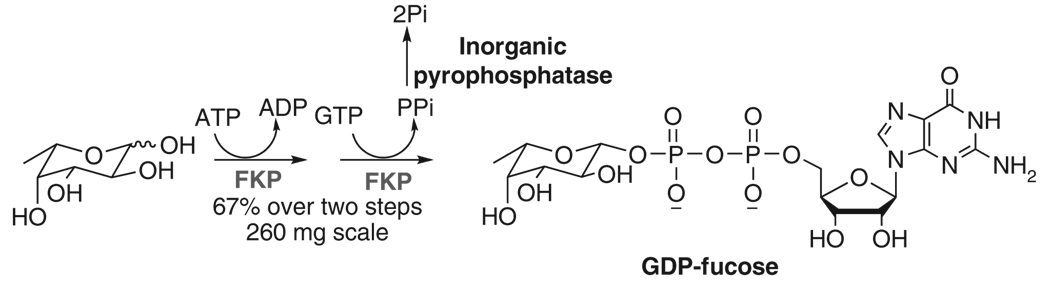
N-acetylhexosamine 1-kinase (NahK) is an N-acetylhexosamine kinase found in Bifidobacterium longum and is the first gluco-type 1-kinase reported16. One-step formation of GlcNAc/GalNAc-1-phosphate (GlcNAc/GalNAc-1-P) (and analogues) from GlcNAc/GalNAc (and analogues) and ATP can be accomplished with high efficiency17,18. N-acetylglucosamine uridyltransferase (GlmU), an N-acetyl-d-glucosamine 1-phosphate (GlcNAc-1-P) uridyltransferase (pyrophosphorylase) from Escherichia coli, could pyrophosphorylate GlcNAc/GalNAc-1-P (and analogues). UDP/GlcNAc–GalNAc (and analogues) can thus be produced at preparative scale by the function of these two enzymes19. Bacteroides fragilis l-fucokinase–GDP–l-fucose pyrophosphorylase (FKP) is a bifunctional enzyme with a kinase domain and a pyrophosphorylase domain in one peptide chain. It catalyzes the transformation of l-fucose to GDP–fucose in a two-step reaction (Box 1)20. We acquired 150–250 mg scale of GDP–fucose using this enzyme. FKP is a promiscuous enzyme with relaxed substrate specificity to modifications at the C-6 position of l-fucose in vitro21,22. The procedure reported in the protocol also could be used for the synthesis of GDP–fucose derivatives. Scaling up (or down) preparation can be realized by proportionally increasing (or decreasing) the amount of reagents for enzymatic reactions as well as the volume of chromatography resins used for purification.
PREPARATIVE-SCALE SYNTHESIS OF GDP–FUCOSE.
Prepare the mixture solution without enzyme: weigh out 300 mg (0.54 mmol) ATP, 300 mg (0.53 mmol) GTP. Dissolve them in 8 ml deionized water in a 15 ml centrifuge tube.
-
Check the pH value of the mixture solution. If pH is too low, adjust pH using 10 M NaOH solution (solid NaOH is also acceptable but must be used carefully to avoid excessive NaOH) to pH 7.5.
 pH value is important in this step. If pH is too low, the enzyme will precipitate.
pH value is important in this step. If pH is too low, the enzyme will precipitate. NaOH is a strong base and highly corrosive. Wear gloves and safety goggles while handling it.
NaOH is a strong base and highly corrosive. Wear gloves and safety goggles while handling it. Weigh out 150 mg (0.91 mmol) l-fucose and add it directly into the solution. Swirl the tube manually until l-fucose dissolves completely.
-
Add 0.5 ml 1 M Tris–HCl buffer (pH 7.5), 0.5 ml 1 M MgCl2 solution and 100 µl inorganic pyrophosphatase (PPA) (1 U µl−1) into the mixture.
 Keep ATP, GTP and PPA on ice. Store at − 20 °C after use. PPA should be stored in aliquots at − 20 °C. Avoid freezing and thawing repeatedly.
Keep ATP, GTP and PPA on ice. Store at − 20 °C after use. PPA should be stored in aliquots at − 20 °C. Avoid freezing and thawing repeatedly. -
Add 100 µl l-fucose pyrophosphorylase (FKP) (5 mg ml−1) to the reaction mixture. Add deionized water to bring the total volume of the reaction mixture to 10 ml.
 FKP should be stored in aliquots at − 20 °C. Avoid freezing and thawing repeatedly.
FKP should be stored in aliquots at − 20 °C. Avoid freezing and thawing repeatedly.
-
Leave the tube in a Fisher water bath at 25 °C and let the reaction proceed for 2 d.
 GDP–fucose is not stable at higher temperature.
GDP–fucose is not stable at higher temperature. When no further formation of the product is observed by TLC, stop the reaction by heating the reaction tube in boiling water for 5 min. Then immediately transfer the tube onto ice to cool the solution completely.
Centrifuge the tube at 5,000g at room temperature for 30 min to remove the protein precipitate. Transfer the supernatant to a new 15 ml centrifuge tube.
-
Add 100 U alkaline phosphatase into the mixture and mix gently by inversion. Continue to incubate the tube in the 25 °C water bath for one more day.
 Alkaline phosphatase should be stored at − 20 °C after use.
Alkaline phosphatase should be stored at − 20 °C after use. Heat the reaction mixture in boiling water bath for about 5 min until the protein precipitates completely. Then immediately transfer the tube onto ice to cool the solution completely. Centrifuge the tube at 5,000g for 30 min to remove the precipitate.
-
Transfer the supernatant to a new 15 ml centrifuge tube.
 The tube containing the reaction mixture could be capped and sealed by Parafilm and stored at − 20 °C until purification.
The tube containing the reaction mixture could be capped and sealed by Parafilm and stored at − 20 °C until purification.Steps 1–11: 3–4 d.
Purification and characterization of GDP–fucose
Steps 12–22: 2–3 d.
Thaw the solution from Step 11. Place the tube on ice for use.
-
Open the valve of a Bio-Gel P-2 column (2.5 cm × 120 cm). Let the water flow down to the top level of the gel bed.
 Increasing the length or diameter of the gel filtration column can improve the purification.
Increasing the length or diameter of the gel filtration column can improve the purification. Pipette 10 ml reaction mixture onto the top of the gel bed. Let the solution flow down to the top level of the gel bed. Then add 1–2 ml water onto the top of gel bed and let it flow down.
-
Pipette 5 ml degassed deionized water into the column without disturbing the top of the gel bed. Connect degassed water source pipe and let about 1,000 ml deionized water run through via gravity from a 4-liter flask. Collect 8 ml fractions in 10 ml test tubes using a fraction collector.
 Watch the fraction collector collect three test tubes before leaving to guarantee that it works well.
Watch the fraction collector collect three test tubes before leaving to guarantee that it works well.
Carry out the quick TLC screening procedure described in Step 20 in the PROCEDURE section and reference 25 to see which fractions contain sugar nucleotides. You will need squares for 36 spots starting from the 25th tube collected.
-
After identifying tubes containing GDP–fucose product, spot 1 µl of sample from product-containing tubes, five tubes before the first tube and after the last one onto a TLC plate (3 cm × 6 cm). Load ATP and l-fucose onto the plate as controls and use n-butanol:acetic acid:water = 2:1:1 (v/v/v) as the mobile phase.
 The developing solvents are irritants and should be handled in a fume hood with care. Wear gloves and safety goggles.
The developing solvents are irritants and should be handled in a fume hood with care. Wear gloves and safety goggles. Weigh a dry, clean 100 ml round-bottom flask (normally, 100 ml round-bottom flask should be appropriate for the reaction scale in Step 3). Record the weight in the notebook and/or on the wall of the flask.
Transfer and combine the fractions only containing GDP–fucose into the 100 ml round-bottom flask. Lyophilize the combined solution using a freeze-dryer to provide a white powder.
Weigh the flask containing the GDP–fucose dry white powder. Record the weight in the notebook. Calculate the yield.
Carefully transfer the dry white powder into a 1.7 ml microcentrifuge tube. Store the sample at − 20 °C.
-
Characterize the GDP–fucose using 1H NMR and mass spectrometry (negative mode).
Steps 1–11: 3–4 d
Steps 12–22: 2–3 d
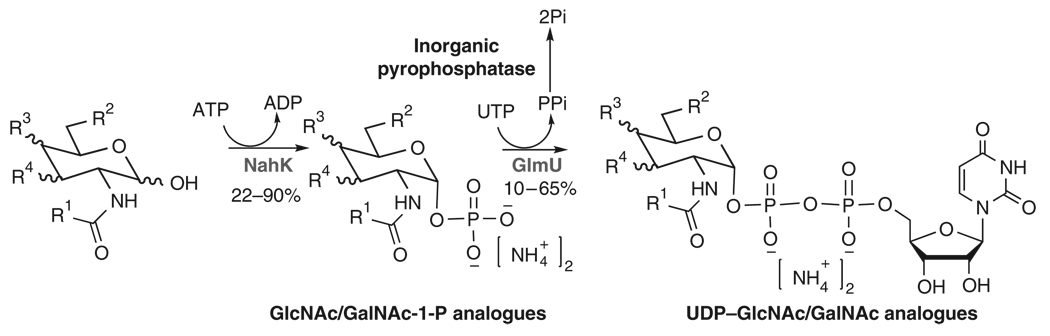
In this protocol, the B. longum NahK and E. coli GlmU are used as UDP–GlcNAc/GalNAc biosynthetic enzymes (Fig. 3). FKP is used as GDP–fucose biosynthetic enzyme (Fig. 4). Yeast inorganic pyrophosphatase (PPA) is added in pyrophosphorylation reactions to drive reaction forward by degrading the by-product pyrophosphate (PPi) because the accumulation of PPi may inhibit the enzymatic activity of GlmU or FKP. The detailed procedures for the preparation of UDP–GlcNAc/GalNAc and GDP–fucose from GlcNAc/GalNAc and l-fucose respectively are described here. As long as the GlcNAc/GalNAc derivatives can be tolerated by NahK and the corresponding modified GlcNAc/GalNAc-1-Ps can be tolerated by GlmU, a similar approach can be applied to the enzymatic synthesis of structurally modified UDP-GlcNAc/GalNAc analogues19 (Fig. 5, NMR and MS data and spectra for UDP–GlcNAc/GalNAc analogues; please see Supplementary Data). For the preparation of UDP–GlcNAc/GalNAc analogues starting from a GlcNAc/GalNAc derivative, initial small-scale test is recommended to check whether the new structure is tolerated by the enzymes. Some GlcNAc/GlaNAc-1-P analogues failed to be substrates of GlmU to produce corresponding UDP–GlcNAc/GalNAc analogues. Mutants of GlmU can be designed according to the crystal structure of this enzyme23 to accommodate more GlcNAc/GalNAc-1-P analogues. Other pyrophosphorylases, such as mammalian UDP–GalNAc pyrophosphorylase (AGX1)24, can also be considered under such circumstances.
Figure 3.
Enzymatic preparation of UDP–N-acetylglucosamine (GlcNAc)/N-acetylgalactosamine (GalNAc) and their analogues using N-acetylhexosamine 1-kinase (NahK) and N-acetylglucosamine uridyltransferase (GlmU).
Figure 4.
Enzymatic large-scale preparation of GDP–fucose using l-fucose pyrophosphorylase (FKP).
Figure 5.
Structure of UDP–N-acetylglucosamine (GlcNAc)/N-acetylgalactosamine (GalNAc) analogues synthesized.
MATERIALS
REAGENTS
Acetic acid (Fisher, cat. no. A38C-212)
GalNAc (Sigma-Aldrich, cat. no. A2795-5G)
GlcNAc (Sigma-Aldrich, cat. no. A4106-100G)
GlcNAc-1-P disodium salt (Sigma-Aldrich, cat. no. A2142-25MG)
ATP disodium salt hydrate (Sigma-Aldrich, cat. no. A26209-10G)
Alkaline phosphatase, calf intestinal (CIP; NEB, cat. no. M0290S)
Ammonium bicarbonate (Sigma-Aldrich, cat. no. A6141-1KG)
p-Anisaldehyde (Sigma-Aldrich, cat. no. A0519-1L)
Bio-Gel P-2 gel, fine (Bio-Rad, cat. no. 150-4115)
n-Butanol (J. T. Baker, cat. no. 9054)
DEAE-cellulose (Sigma-Aldrich, cat. no. D3764-500G)
Dichloromethane (EMD, cat. no. DX0835-5)
Ethanol 200 proof (Decon Labs, cat. no. 2701)
l-Fucose (V-LABS, cat. no. BF61)
GTP sodium salt hydrate (Sigma-Aldrich, cat. no. 51120-1G)
Hydrochloric acid (Fisher, cat. no. A144C-212)
Magnesium chloride (Sigma-Aldrich, cat. no. M8266-100G)
Methanol (EMD, cat. no. MX0485-5)
Pyrophosphatase, inorganic from baker’s yeast (Saccharomyces cerevisiae) (Sigma-Aldrich, cat. no. I1643)
Silica gel 60 (EMD, cat. no. 9385-9)
Sodium hydroxide, pellet (Mallinchrodt, cat. no. 7708-10)
Sulfuric acid (Fisher, cat. no. A300C-212)
Tris (Invitrogen, cat. no. 15504-020)
1 M Tris–HCl pH 7.5 (Invitrogen, cat. no. 15567-027)
UDP–GlcNAc sodium salt (Sigma-Aldrich, cat. no. U4375-100MG)
UTP trisodium salt hydrate (Sigma-Aldrich, cat. no. U6625-1G)
EQUIPMENT
Benchtop centrifuge (Eppendorf, cat. no. 5415 D)
Buchi rotary evaporator (Fisher, cat. no. 04-987-222)
Econo-column chromatography column (Bio-Rad, cat. no. 737-1091)
Freeze-dry systems (LABCONCO, cat. no. 7753000)
GeneMate 15 ml sterile centrifuge tubes (ISC Bioexpress, cat. no. C-3394-1)
GeneMate 50 ml sterile centrifuge tubes (ISC Bioexpress, cat. no. C-3394-3)
Hot plate/stirrer (Corning, cat. no. 6795-220)
Isotemp economy analog-control water bath Model 115 (Fisher, cat. no. 15-460-15Q)
Master heat gun (UL listed, cat. no. HG-501A)
Model 2110 fraction collector (Bio-Rad, cat. no. 731-8120)
‘Thermo’ 1.7 ml graduated microcentrifuge tubes for boiling applications (ISC Bioexpress, cat. no. C-3260-1)
TLC silica gel 60 F254 (EMD, cat. no. 5554-7)
REAGENT SETUP
Enzymes (preparation of UDP–GlcNAc/GalNAc)
The PPA is a commercially available enzyme. In the preparation of UDP–GlcNAc/GalNAc, solid PPA powder (500 U) is dissolved in 1 ml deionized water to prepare 0.5 U µl−1 storage solution. NahK and GlmU are recombinant proteins cloned and purified in our laboratory17,19. Briefly, E. coli BL21 (DE3) containing pET22b–nahK or pET15b–glmU recombinant plasmid is cultivated at 37 °C until OD600 reaches 0.7 and induces protein expression with 0.1 mM IPTG at 16 °C for 20 h. The cells are then collected and sonicated. After centrifugation at 16,000 r.p.m. for 30 min at 4 °C, the NahK or GlmU protein in the supernatant is purified by Ni-NTA and gel-filtration columns. All enzymes are stored at − 20 °C.
Enzymes (preparation of GDP–fucose)
The PPA and alkaline phosphatase are commercially available. In the preparation of GDP–fucose (Box 1), solid PPA powder (1,000 U) is dissolved in 1 ml deionized water to prepare 1 U µl−1 storage solution. FKP is a recombinant protein that was purified in our laboratory21. Briefly, E. coli BL21 (DE3) containing pMCSG7–fkp recombinant plasmid is incubated at 37 °C until OD600 reaches 0.7 and induces with 0.1 mM IPTG at 16 °C for 20 h. Then the cells are collected and sonicated. After centrifugation at 16,000 r.p.m. for 30 min at 4 °C, the FKP protein in the supernatant is purified by Ni-NTA and gel-filtration columns.
All enzymes are stored at − 20 °C.
p-Anisaldehyde sugar staining solution
Dissolve 18 ml of p-anisaldehyde in 540 ml 95% ethanol and cool the solution in an ice/water bath. Mix 30 ml of 97% H2SO4 and 6 ml of acetic acid. Cautiously add the acid mixture to the prechilled ethanol solution dropwise at 0 °C with vigorous stirring without splashing. Store the resulting colorless solution in a − 20 °C refrigerator before use.  p-Anisaldehyde is harmful. Sulfuric acid and acetic acid are highly corrosive. Handle the solution with care in a fume hood. Wear gloves and safety goggles.
p-Anisaldehyde is harmful. Sulfuric acid and acetic acid are highly corrosive. Handle the solution with care in a fume hood. Wear gloves and safety goggles.
EQUIPMENT SETUP
Packing a silica gel column
Mix 5–6 g silica gel with 20 ml CH2Cl2. Swirl the mixture to resuspend the silica gel. Pour the well-mixed mixture into a 1.5 cm i.d. × 25 cm length column in a single smooth action. Pack the silica gel under pressure. Dry the column under pressure. Flush the column with 20 ml methanol. Dry the column under pressure. Then flush the column with 50 ml CH2Cl2 to remove the methanol residue. Dry the column under pressure.
Packing an ion-exchange column with DEAE-cellulose
The DEAE-cellulose resin is prepared as follows: place 100 g dry resin into a Buchner funnel of appropriate size. Elute the resin cake with 170 ml of 1 N HCl and allow the resin cake to dry under suction. Elute the resin with 200 ml deionized water to remove HCl residue and dry the cake under suction. Elute the resin cake with 170 ml of 1N NaOH and dry the cake under suction. Elute the cake with 200 ml deionized water to remove the NaOH residue and dry the cake under suction. Repeat the same elution process (HCl, water, then NaOH and water) twice. Then wash the resin with 300 ml methanol. Dry the fluffy DEAE-cellulose in a desiccator under reduced pressure overnight. At 1 d before the purification, soak the dried DEAE-cellulose in excessive amount of glacial acetic acid (for at least 24 h). To pack a DEAE-cellulose column, swirl the bottle of resin slurry to obtain a homogeneous suspension, then pour a sufficient volume of slurry into a chromatography column (1.5 cm i.d. × 25 cm length) to obtain a settled resin bed of 20 cm length. Wash the column with deionized water until the pH is neutral (approximately three times of column volume). Then equilibrate the column with 50 mM NH4HCO3 solution (approximately 2 × column volume).  Acetic acid slurry is corrosive and an irritant. The preparation and packing must be handled with great care in a chemical fume hood. Wear appropriate gloves and safety goggles.
Acetic acid slurry is corrosive and an irritant. The preparation and packing must be handled with great care in a chemical fume hood. Wear appropriate gloves and safety goggles.
Packing a Bio-Gel P-2 gel filtration column
In all, 60-g dry Bio-Gel P-2 gel is gradually added into 320 ml deionized water in a 1,000 ml beaker. Leave the beaker at room temperature for 4–5 h. After hydration is complete, remove the fine floating particles and wash the gel twice using deionized water. The solution is transferred into a filter flask, which is attached to a water aspirator. The solution is degassed for 15 min with occasional swirling. Add 20 ml degassed deionized water into the column. Then pour the gel suspension into the column smoothly at one time.  Please also refer to the procedure of Chen et al.’s25 paper on the synthesis of sialosides for further advice on this technique if required. Avoid introducing air bubbles. After the gel bed is settled for 2 cm length, open the column outlet to allow the water to flow under gravity until the column is packed. Equilibrate the column overnight with 4 liter of degassed water before loading the sample. Scale up the column (~600 ml hydrated bed volume) for GDP–fucose purification in this protocol.
Please also refer to the procedure of Chen et al.’s25 paper on the synthesis of sialosides for further advice on this technique if required. Avoid introducing air bubbles. After the gel bed is settled for 2 cm length, open the column outlet to allow the water to flow under gravity until the column is packed. Equilibrate the column overnight with 4 liter of degassed water before loading the sample. Scale up the column (~600 ml hydrated bed volume) for GDP–fucose purification in this protocol.
PROCEDURE
Preparative-scale synthesis of N-acetylhexosamine-1-Ps  ~20 h
~20 h
-
GlcNAc/GalNAc derivatives were prepared by chemical synthesis, as described17,18 and stored at 4 °C before use (please see refs. 17,18 for experimental details of synthesizing GlcNAc/GalNAc derivatives and NMR data and spectra). Take out NahK from − 20 °C freezer and allow it to thaw on ice.
 NahK should be stored in 0.2 ml aliquots at − 20 °C. Avoid freezing and thawing repeatedly.
NahK should be stored in 0.2 ml aliquots at − 20 °C. Avoid freezing and thawing repeatedly. -
Weigh out 88.4 mg (0.4 mmol) GlcNAc, 275.5 mg (0.5 mmol, 1.25 equiv.) ATP into a 15 ml centrifuge tube.
 ATP is stored at 4 °C.
ATP is stored at 4 °C. Transfer 7 ml deionized water into the tube and dissolve the compounds completely by swirling on vortex.
Transfer 1 ml of Tris–HCl buffer solution (1 M, pH 9.0) into the tube and mix well by swirling manually.
Transfer 0.5 ml MgCl2 stock solution (200 mM) into the tube and mix well by swirling manually.
Add 0.191 ml NahK stock solution (78.5 mg ml−1). Gently mix the reaction mixture by inversion.
-
Adjust the total volume of the reaction mixture to 10 ml by adding deionized water.
 NahK stock concentration may vary for different overexpression batches. Make sure that the final concentration of the enzyme in the reaction mixture is 1.5 mg ml−1.
NahK stock concentration may vary for different overexpression batches. Make sure that the final concentration of the enzyme in the reaction mixture is 1.5 mg ml−1. -
The total volume of the reaction mixture is 10 ml, containing 40 mM GlcNAc, GalNAc or their derivatives, 50 mM ATP, 10 mM MgCl2 and 1.5 mg ml−1 enzyme in 100 mM Tris–HCl buffer (pH 9.0).
 The reaction can be proportionally scaled up or down based on the moles of the limiting substrates (GlcNAc/GalNAc or their derivatives) maintaining the final concentration of each component constant (Step 8). For example, 0.1 mmol of GalNAc will react with 0.125 mmol ATP in a total volume of 2.5 ml reaction mixture. Other sugar kinases can be used in this approach with necessary modifications. Sugar 1-kinases (e.g., NahK and FKP) are used for the synthesis of sugar-1-Ps.
The reaction can be proportionally scaled up or down based on the moles of the limiting substrates (GlcNAc/GalNAc or their derivatives) maintaining the final concentration of each component constant (Step 8). For example, 0.1 mmol of GalNAc will react with 0.125 mmol ATP in a total volume of 2.5 ml reaction mixture. Other sugar kinases can be used in this approach with necessary modifications. Sugar 1-kinases (e.g., NahK and FKP) are used for the synthesis of sugar-1-Ps. Leave the tube in a Fisher water bath at 37 °C and let the reaction proceed for 19 h.
-
Monitor the reaction using TLC analysis of the reaction mixture after 7 h and 19 h using a 2.5 cm × 4.5 cm TLC plate and n-butanol:acetic acid:water = 2:1:1 (v/v/v) as the mobile phase. Use GlcNAc (or analogues) and GlcNAc-1-P as controls. Stain with p-anisaldehyde solution. After heating (e.g., with a heat gun), there should be a new light brown spot at Rf ~0.25–0.33 on the plate showing the production of sugar-1-P. Refer to the procedure of Chen et al.’s25 paper on the synthesis of sialosides for further advice on this technique if required.
 The developing solvents are irritant and should be handled in a fume hood with care. Wear gloves and safety goggles.
The developing solvents are irritant and should be handled in a fume hood with care. Wear gloves and safety goggles. Typical Rf value for starting materials (GlcNAc/GalNAc and their analogues) ~0.53–0.8; Rf for ATP residue and the byproduct ADP ~0–0.07; starting materials usually can be consumed within 19 h (depending on differences of substrate tolerance).
Typical Rf value for starting materials (GlcNAc/GalNAc and their analogues) ~0.53–0.8; Rf for ATP residue and the byproduct ADP ~0–0.07; starting materials usually can be consumed within 19 h (depending on differences of substrate tolerance).
-
When the starting material (sugar spot) disappears or the product spot intensity stops increasing despite elongation of reaction time, stop the reaction by boiling the mixture for 2 min.
 Transfer the reaction mixture evenly to eight 1.7 ml microcentrifuge tubes. Heat the tubes in boiling water for 2 min.
Transfer the reaction mixture evenly to eight 1.7 ml microcentrifuge tubes. Heat the tubes in boiling water for 2 min. Centrifuge for 5 min at 16,200g at room temperature in an Eppendorf benchtop centrifuge. Transfer carefully and combine the clear supernatant into a 50 ml round-bottom flask.
Rinse the precipitates left in each 1.7 ml microcentrifuge tube with 200 µl deionized water. Centrifuge for 5 min at 16,200g in an Eppendorf benchtop centrifuge. Transfer carefully and combine the clear supernatant in this rinsing step into the same 50 ml round-bottom flask in Step 12.
-
Remove the water of the combined supernatant under reduced pressure using a rotary evaporator at 30 °C.
 Adding methanol (about 5 ml each time) several times to the supernatant during the rotary evaporation process can accelerate the evaporation.
Adding methanol (about 5 ml each time) several times to the supernatant during the rotary evaporation process can accelerate the evaporation. The round-bottom flask can be sealed with parafilm or a rubber stopper and stored in a 4 °C refrigerator overnight or for several days before purification by silica gel.
The round-bottom flask can be sealed with parafilm or a rubber stopper and stored in a 4 °C refrigerator overnight or for several days before purification by silica gel.Purification and characterization of N-acetylhexosamine-1-P products
 5–6 h
5–6 h -
Set up the silica gel column as described in the MATERIALS section.
 Each flushing or eluting process should be carried out under pressure. Dry the column after each flushing or eluting step and before next elution.
Each flushing or eluting process should be carried out under pressure. Dry the column after each flushing or eluting step and before next elution. Add 10 ml of methanol into the round-bottom flask to dissolve the residue from Step 14. Add about 1 g of silica gel to the resulting solution.
-
Remove the solvent of the mixture under reduced pressure using rotary evaporator at 30–35 °C to afford dry silica gel with all compounds absorbed.
 Be sure to insert a small piece of absorbent cotton into the rotary evaporator trap to contain the silica gel within the flask.
Be sure to insert a small piece of absorbent cotton into the rotary evaporator trap to contain the silica gel within the flask. Load the silica gel absorbed with sample on top of the packed silica gel bed in Step 15. Then insert a small piece of absorbent cotton into the column and cover the top of the sample.
-
Elute the column using gradient CH2Cl2/5 mM NH4HCO3 in methanol as eluant. Collect 5 ml fractions in 10 ml test tubes.
 Elute the column with 10 ml CH2Cl2 first. Then elute the column with 10 ml CH2Cl2:5 mM NH4HCO3 in methanol (7:3 v/v). Then elute the column with 10 ml CH2Cl2:5 mM NH4HCO3 in methanol (1:1 v:v). Then elute the column with 10 ml CH2Cl2:5 mM NH4HCO3 in methanol (3:7 v/v). Then elute the column with 20 ml of 5 mM NH4HCO3 in methanol. The product usually comes out in methanol or late 3:7 fractions.
Elute the column with 10 ml CH2Cl2 first. Then elute the column with 10 ml CH2Cl2:5 mM NH4HCO3 in methanol (7:3 v/v). Then elute the column with 10 ml CH2Cl2:5 mM NH4HCO3 in methanol (1:1 v:v). Then elute the column with 10 ml CH2Cl2:5 mM NH4HCO3 in methanol (3:7 v/v). Then elute the column with 20 ml of 5 mM NH4HCO3 in methanol. The product usually comes out in methanol or late 3:7 fractions. To identify carbohydrates-containing test tubes, draw 12 squares on a small TLC plate using a lead pencil and spot sequentially the samples from each tube within the squares on the TLC plate.
Dry the spots using a heat gun and stain with p-anisaldehyde as previously described (Step 10 and ref. 25). The fractions containing carbohydrates (starting material or 1-phosphate product) will appear as light brown to dark brown spots on the TLC plate.
Using n-butanol:acetic acid:water = 2:1:1 (v/v/v) as developing solvent and GlcNAc-1-P authentic sample as control, carry out TLC assay as in Step 10 and reference 25 for all the sugar-containing fractions.
Combine the fractions containing GlcNAc/GalNAc-1-P or their analogues into a 100 ml round-bottom flask (normally, 100 ml round-bottom flask should be appropriate for the reaction scale in Step 8). Remove the solvent of the combined fractions under reduced pressure using a rotary evaporator at 30–35 °C to afford the product as a colorless foam or syrup.
(Optional) Repurify the purified sugar-1-P by repeating the purification process (Steps 15–23). Carry out this step only for NMR purposes. In most cases, the purity of product from purification Steps 15–23 is about 90%. However, the NMR spectra of the sugar-1-Ps from the first silica gel purification show peaks of the same unknown impurities (please see Supplementary Data for typical NMR impurities region). In these cases, repeating the purification process (Steps 15–23) could further improve the purity to >95% and remove the impurity peaks.
-
Characterize the known sugar-1-P products using 1H NMR. Characterize all new sugar-1-P products using 1H NMR, 13C NMR, 31P NMR and high-resolution mass spectrometry (HRMS, negative mode; please see Supplementary Data for NMR and MS spectra for all new compounds).
Preparative-scale synthesis of UDP–N-acetylhexosamine analogues
 ~14 h
~14 h -
Take out GlcNAc/GalNAc-1-P or their analogues from a 4 °C refrigerator; take out UTP from a − 20 °C freezer. Allow them to warm to room temperature. Take out recombinant E. coli K-12 N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, and PPA from a − 20 °C freezer. Allow them to thaw on ice.
 Replace these reagents to − 20 °C refrigerators after use as soon as possible. Avoid long-time exposure at room temperature. GlmU and PPA should be stored in aliquots at − 20 °C. Avoid freezing and thawing repeatedly.
Replace these reagents to − 20 °C refrigerators after use as soon as possible. Avoid long-time exposure at room temperature. GlmU and PPA should be stored in aliquots at − 20 °C. Avoid freezing and thawing repeatedly. Weigh out 13.4 mg (0.04 mmol) GlcNAc-1-P and 33.0 mg (0.06 mmol, 1.5 equiv.) UTP into a 15 ml centrifuge tube.
Transfer 3 ml deionized water into the tube and dissolve the compounds completely by swirling on vortex.
Transfer 0.4 ml of Tris–HCl buffer stock solution (1 M, pH 7.5) into the tube and mix well by manually swirling.
Transfer 0.2 ml of MgCl2 stock solution (200 mM) into the tube and mix well by swirling manually.
Transfer 0.121 ml of GlmU stock solution (33 mg ml−1) and 8 µl of PPA stock solution (0.5 U µl−1). Gently mix the reaction mixture by inversion.
-
Adjust the total volume of the reaction mixture to 4 ml by adding deionized water.
 The GlmU stock concentration may vary for different overexpression batches. The final concentrations of enzymes in the reaction mixture are 1 mg ml−1 GlmU and 1 U ml−1 PPA.
The GlmU stock concentration may vary for different overexpression batches. The final concentrations of enzymes in the reaction mixture are 1 mg ml−1 GlmU and 1 U ml−1 PPA. -
The total volume of the reaction mixture is 4 ml, containing 10 mM GlcNAc/GalNAc-1-P or their analogues, 15 mM UTP, 10 mM MgCl2, 1 mg ml−1 GlmU and 1 U ml−1 PPA in 100 mM Tris–HCl buffer (pH 7.5).
 The reaction can be scaled up or down based on the moles of the limiting substrates (GlcNAc/GalNAc-1-P derivatives) maintaining the final concentration of each component constant (Step 33). For example, 0.01 mmol GalNAc-1-P will react with 0.015 mmol UTP in a total volume of 1 ml reaction mixture.
The reaction can be scaled up or down based on the moles of the limiting substrates (GlcNAc/GalNAc-1-P derivatives) maintaining the final concentration of each component constant (Step 33). For example, 0.01 mmol GalNAc-1-P will react with 0.015 mmol UTP in a total volume of 1 ml reaction mixture. Leave the tube in a Fisher water bath at 37 °C and let the reaction proceed for 12 h.
-
Using GlcNAc-1-P, GalNAc-1-P or their derivatives as controls and n-butanol:acetic acid:water = 2:1:1 (v/v/v) as developing solvent, monitor the reaction using TLC analysis as described in Step 10 and reference 25 after 6 h and 12 h. Starting materials usually can be consumed within 12 h (depending on the substrate tolerance).
 After one development, dry the plate completely with a heat gun (heat gently) and develop the plate one more time to increase the visibility of sugar nucleotide. There should be a new light brown spot at Rf ~0.18–0.20 (after two developments) on the TLC plate showing the production of UDP–sugar. Typical Rf value for starting materials (GlcNAc/GalNAc-1-P and their analogues) ~0.22–0.69; Rf value for UTP residue ~0.07.
After one development, dry the plate completely with a heat gun (heat gently) and develop the plate one more time to increase the visibility of sugar nucleotide. There should be a new light brown spot at Rf ~0.18–0.20 (after two developments) on the TLC plate showing the production of UDP–sugar. Typical Rf value for starting materials (GlcNAc/GalNAc-1-P and their analogues) ~0.22–0.69; Rf value for UTP residue ~0.07. The developing solvents are irritant and should be handled in a fume hood with care. Wear gloves and safety goggles.
The developing solvents are irritant and should be handled in a fume hood with care. Wear gloves and safety goggles. -
When the starting material (sugar-1-P spot) disappears or the product spot intensity stops increasing despite elongation of reaction time, stop the reaction by boiling the mixture for 1 min.
 Transfer the reaction mixture evenly to four 1.7 ml microcentrifuge tubes. Heat the tubes in boiling water for 1 min.
Transfer the reaction mixture evenly to four 1.7 ml microcentrifuge tubes. Heat the tubes in boiling water for 1 min. Centrifuge at 16,200g at room temperature for 5 min in an Eppendorf benchtop centrifuge. Carefully transfer and combine the clear supernatant into a 25 ml round-bottom flask.
Rinse the protein precipitates left in each 1.7 ml tube with 200 µl deionized water. Centrifuge at 16,200g for 5 min in an Eppendorf benchtop centrifuge. Carefully transfer and combine the clear supernatant in this rinsing step to the same 25 ml round-bottom flask in Step 37.
-
Lyophilize the combined supernatant solution containing all compounds (e.g., product, by-product, starting material residue and so on) using a freeze-dryer.
 The 25 ml round-bottom flask can be sealed by parafilm or a rubber stopper and stored in a − 20 °C refrigerator overnight or for several days before purification.
The 25 ml round-bottom flask can be sealed by parafilm or a rubber stopper and stored in a − 20 °C refrigerator overnight or for several days before purification.Purification and characterization of UDP–GlcNAc/GalNAc products
 14–15 h
14–15 h Open the valve of a DEAE-cellulose ion-exchange column (1.5 cm × 20 cm) to let the 50 mM NH4HCO3 solution flow down slowly to the top level of the gel bed.
Add 1 ml of 50 mM NH4HCO3 solution into the 25 ml round-bottom flask to dissolve the residue from Step 39. Carefully transfer this solution onto the DEAE-cellulose gel bed and let the sample solution flow down to the top level of the gel bed.
Rinse the 25 ml round-bottom flask with 0.5 ml 50 mM NH4HCO3 solution and carefully transfer the rinsing solution onto the gel bed. Repeat this rinsing and transferring step one more time. Make sure that all the solution flows down to the top level of the gel bed before elution.
-
Elute the column with gradient 50 ~300 mM NH4HCO3 solution. Collect 5 ml fractions in 10 ml test tubes.
 Elute the column with 10 ml 50 mM NH4HCO3 solution first. Then increase the concentration of NH4HCO3 solution by 25 mM every 10 ml until the concentration reaches 300 mM. The product usually comes out in 150–200 mM NH4HCO3 solution fractions.
Elute the column with 10 ml 50 mM NH4HCO3 solution first. Then increase the concentration of NH4HCO3 solution by 25 mM every 10 ml until the concentration reaches 300 mM. The product usually comes out in 150–200 mM NH4HCO3 solution fractions. Carry out the quick TLC screening procedure described in Step 20 and reference 25 to see which fractions contain sugar nucleotides (sugar nucleotides should be UV visible and show color spots under p-anisaldehyde stain).
Use mass spectrometry (negative mode) to confirm the fractions containing sugar nucleotides identified in Step 44 (please see Supplementary Data for MS spectra for all new compounds).
-
Combine the fractions containing UDP–GlcNAc/GalNAc or their analogues into a 100 ml round-bottom flask (normally, a 100 ml round-bottom flask should usually be appropriate for reaction scale in Step 33). Lyophilize the combined fractions containing product using a freeze-dryer to provide a usually white powder.
 The combined fractions containing product can be sealed by Parafilm or a rubber stopper in the 100 ml round-bottom flask and stored in a − 20 °C refrigerator overnight or for several days before P-2 column purification.
The combined fractions containing product can be sealed by Parafilm or a rubber stopper in the 100 ml round-bottom flask and stored in a − 20 °C refrigerator overnight or for several days before P-2 column purification. Remove the water source. Open the valve of a Bio-Gel P-2 column (1.5 cm × 90 cm). Let the water level flow down to the top of the gel bed.
Add 0.5 ml deionized water into the 100 ml round-bottom flask in Step 46 to dissolve the white powder. Carefully transfer this solution using a pipette onto the P-2 gel bed and let the solution level flow down to the top of the gel bed.
Rinse the 100 ml round-bottom flask with 0.5 ml deionized water and carefully transfer the rinsing solution onto the gel bed. Repeat this rinsing and transferring step one more time. Make sure that the sample solution flows down to the top level of the gel bed before elution.
-
Transfer gently 5 ml degassed deionized water onto the gel bed without disturbing the top of the gel bed. Connect degassed water source pipe and let about 240 ml deionized water run through via gravity from a 2-liter flask. Collect 4 ml fractions in 10 ml test tubes using a fraction collector.
 Watch the fraction collector collecting three test tubes before leaving to guarantee it works well.
Watch the fraction collector collecting three test tubes before leaving to guarantee it works well. Carry out the quick TLC screening procedure described in Step 20 and reference 25 to see which fractions contain sugar nucleotides. You will need squares for 25 spots starting from the 10th tube collected.
Use mass spectrometry (negative mode) to confirm the fractions identified in Step 51 containing sugar nucleotides.
Combine the fractions containing UDP–GlcNAc/GalNAc or their analogues product into a 100 ml round-bottom flask (normally, a 100 ml round-bottom flask should be appropriate for the reaction scale in Step 33). Lyophilize the combined solution containing pure products using a freeze-dryer to provide the usually white powder.
Add 2 ml deionized water into the 100 ml round-bottom flask to dissolve the white powder. Transfer the solution into a 20 ml vial.
Add 1 ml deionized water to rinse the round-bottom flask and transfer the rinsing solution into the same 20 ml vial. Repeat the rinsing and transferring step one more time.
-
Lyophilize the combined solution from Steps 54 and 55 containing pure products using a freeze-dryer to afford pure UDP–sugars as white powder or foam.
 Store the purified UDP–sugar (white powder or foam) in the 20 ml vial sealed by Parafilm for at least 1 year.
Store the purified UDP–sugar (white powder or foam) in the 20 ml vial sealed by Parafilm for at least 1 year. Characterize the known UDP–sugars using 1H NMR and mass spectrometry (negative mode). Characterize all new UDP–sugars by 1H NMR, 31P NMR and HRMS (negative mode; please see Supplementary Data for NMR and MS spectra for all new compounds).
![]()
Steps 1–8: 15 min
Steps 9 and 10: 19 h
Steps 11–14: 1 h
Steps 15–24: 2–3 h
Step 25: 3 h
Steps 26–33: 15 min
Step 34: 12 h
Steps 35–39: 1.5 h
Steps 40–46: 3–4 h
Steps 47–53: 8 h
Steps 54–56: 15 min
Step 57: 3 h
![]()
Troubleshooting advice can be found in Table 1.
TABLE 1.
Troubleshooting table.
| Problem | Solution |
|---|---|
|
p-Anisaldehyde sugar stain: The color of the p-anisaldehyde sugar stain changes from colorless to dark red |
It is recommended that the stain be stored at − 20 °C in a sealed bottle. If p-anisaldehyde sugar stain is left at room temperature for a long time, it may then change its color. Please refer to reference 25 for more information on preparation and storage of this stain |
|
Preparative-scale synthesis of GDP–fucose: Step 5 the enzyme does not work or precipitates completely |
|
|
Preparative-scale synthesis of GDP–fucose: Step 15 the GDP–fucose could not be isolated from the side-products and residual substrates |
|
ANTICIPATED RESULTS
UDP–GlcNAc/GalNAc
The protocol described here allows preparative-scale synthesis of GlcNAc/GalNAc-1-P and UDP–GlcNAc/GalNAc with yields ranging from 22 to 90% and from 10 to 65%, respectively. The large range of yields reflects diverse tolerance of these enzymes to the structurally modified sugar substrates (please see refs. 17–19 for detailed discussion of substrate specificity of these enzymes).
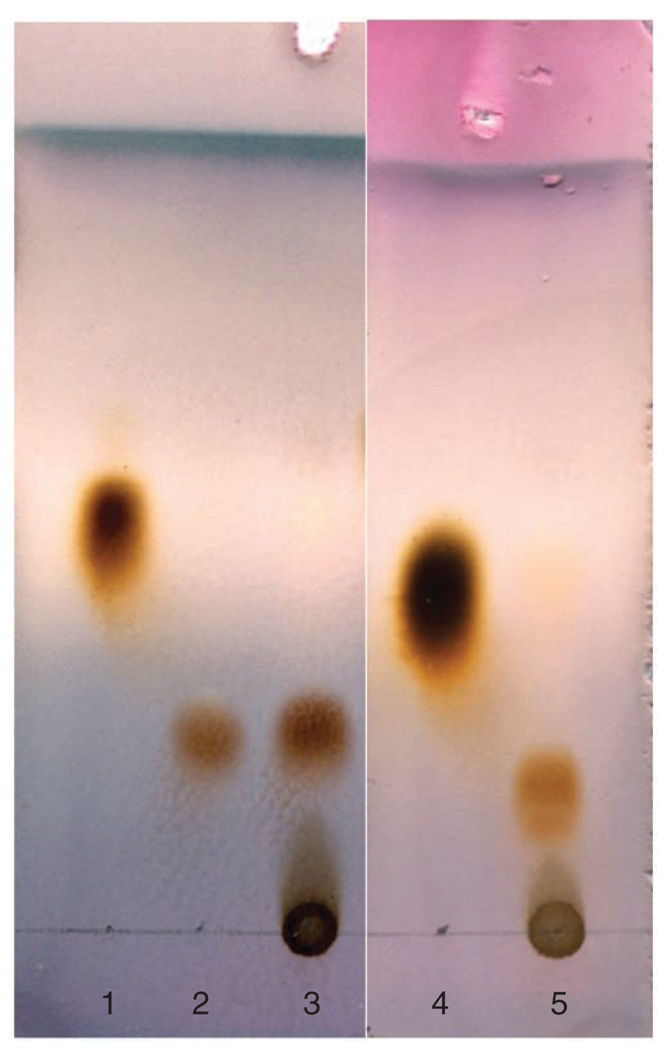
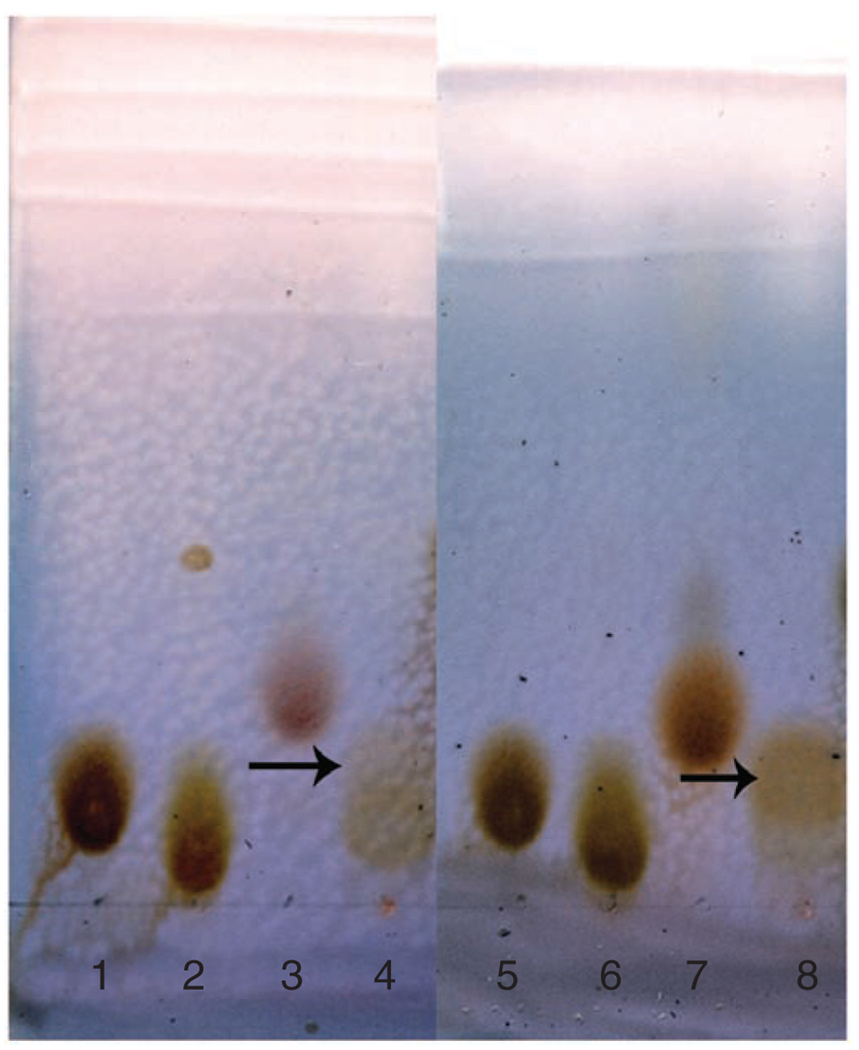
For example, the results for preparative synthesis of GlcNAc-1-P and GalNAc-1-P are shown in Figure 6. After developing in n-butanol:acetic acid:water = 2:1:1 (v/v/v) and staining with p-anisaldehyde sugar stain, the GlcNAc-1-P and GalNAc-1-P products show light brown on the TLC plates. The Rf values of GlcNAc-1-P and GalNAc-1-P are 0.26 and 0.18, respectively. The results for preparative synthesis of UDP–GlcNAc and UDP–GalNAc are shown in Figure 7. After developing in n-butanol:acetic acid:water = 2:1:1 (v/v/v) and staining with p-anisaldehyde sugar stain, the UDP–GlcNAc and UDP–GalNAc products show light brown on the TLC plates. The Rf values of UDP–GlcNAc and UDP–GalNAc are 0.19 and 0.18 (developed twice), respectively.
Figure 6.
TLC analysis of enzymatic synthesis of N-acetylglucosamine 1-phosphate (GlcNAc-1-P) and N-acetylgalactosamine 1-phosphate (GalNAc-1-P) using N-acetylhexosamine 1-kinase (NahK). The enzymatic reactions were carried out for 19 h. The developing solvent used was n-butanol:acetic acid:water = 2:1:1 (v/v/v). The plate was stained with p-anisaldehyde sugar stain. Lane: 1, N-acetylglucosamine (GlcNAc); 2, GlcNAc-1-P authentic sample; 3, reaction mixture of GlcNAc using NahK; 4, N-acetylgalactosamine (GalNAc); and 5, reaction mixture of GalNAc using NahK.
Figure 7.
TLC analysis of enzymatic synthesis of UDP-N-acetylglucosamine (UDP-GlcNAc) and UDP-N-acetylgalactosamine (UDP-GalNAc) using N-acetylglucosamine uridyltransferase (GlmU). The enzymatic reactions were carried out for 12 h. The developing solvent used was n-butanol:acetic acid: water = 2:1:1 (v/v/v). The plate was developed twice and stained with p-anisaldehyde sugar stain. Lane: 1 and 5, UDP-GlcNAc; 2 and 6, UTP; 3, N-acetylglucosamine 1-phosphate (GlcNAc-1-P); 4, reaction mixture of GlcNAc-1-P using GlmU; 7, N-acetylgalactosamine 1-phosphate (GalNAc-1-P); and 8, reaction mixture of GalNAc-1-P using GlmU. Arrows point to product spots.
Analytical data
All analytical data of GlcNAc/GalNAc-1-P and their analogues, UDP–GlcNAc/GalNAc and their analogues including NMR and MS spectra; please see Supplementary Data and references 17–19.
GDP–fucose
The protocol described here introduces preparative-scale (150–250 mg) synthesis of GDP–fucose in one-step purification.
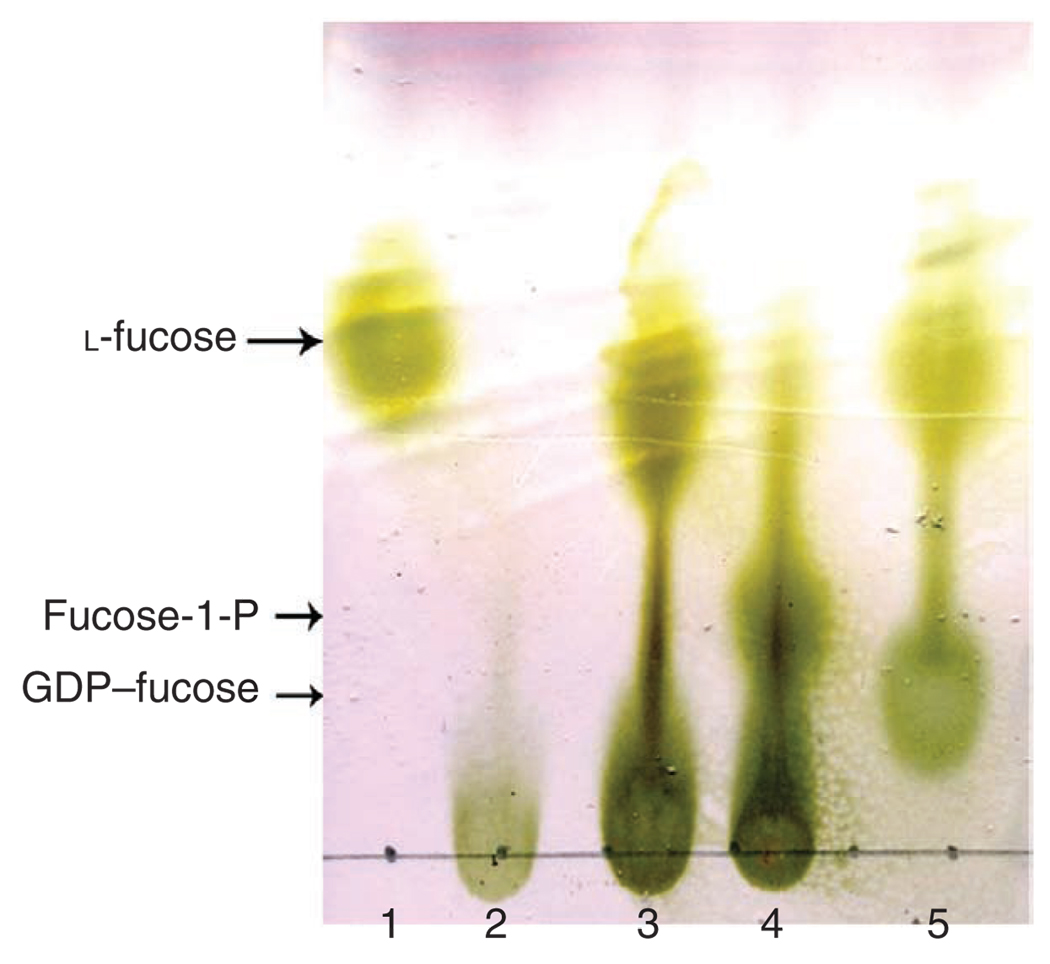
For example, the results for the synthesis of GDP–fucose in a preparative scale are shown in Figure 8. The TLC plate was developed using developing solvent: n-butanol:acetic acid: water = 2:1:1 (v/v/v). Rf for l-fucose, fucose-1-P and GDP–fucose are 0.65, 0.32 and 0.21, respectively.
Figure 8.
TLC analysis of enzymatic synthesis of GDP–fucose. The developing solvent used was n-butanol:acetic acid:water = 2:1:1 (v/v/v). The plate was stained with p-anisaldehyde sugar stain. Lane: 1, l-fucose; 2, ATP; 3, reaction mixture without l-fucose pyrophosphorylase (FKP); 4, reaction mixture without GTP; and 5, alkaline phosphatase-treated reaction mixture of GDP-fucose synthesis.
Analytical data
NMR data and spectra of GDP–fucose; please see Supplementary Data and reference 26.
Supplementary Material
ACKNOWLEDGMENTS
P.G.W. acknowledges National Cancer Institute (R01 CA118208), NSF (CHE-0616892) and NIH (R01 AI083754, R01 HD061935 and R01 GM085267) for financial support. W.G. acknowledges China Scholarship Council for financial support. We thank Robert Woodward for proofreading the manuscript.
Footnotes
Note: Supplementary information is available via the HTML version of this article.
AUTHOR CONTRIBUTIONS G.Z., W.G. and L.C. contributed equally to this work. P.G.W. supervised the project; L.C., W.G. and P.G.W. designed and carried out enzymatic synthesis of UDP–GlcNAc/GalNAc experiments and analyzed data; G.Z. and P.G.W. designed and carried out enzymatic synthesis of GDP–fucose experiments and analyzed data; L.C., W.G. and G.Z. wrote the paper; and P.G.W. revised the manuscript; all authors discussed the results and implications and commented on the manuscript at all stages.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Koeller KM, Wong CH. Complex carbohydrate synthesis tools for glycobiologists: enzyme-based approach and programmable one-pot strategies. Glycobiology. 2000;10:1157–1169. doi: 10.1093/glycob/10.11.1157. [DOI] [PubMed] [Google Scholar]
- 2.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Andreana PR, Wang PG. Carbohydrates in transplantation. Curr. Opin. Chem. Biol. 1999;3:650–658. doi: 10.1016/s1367-5931(99)00022-8. [DOI] [PubMed] [Google Scholar]
- 4.Xia C, et al. Synthesis and biological evaluation of alpha-galactosylceramide (KRN7000) and isoglobotrihexosylceramide (iGb3) Bioorg. Med. Chem. Lett. 2006;16:2195–2199. doi: 10.1016/j.bmcl.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 5.Yi W, et al. Escherichia coli O86 O-antigen biosynthetic gene cluster and stepwise enzymatic synthesis of human blood group B antigen tetrasaccharide. J. Am. Chem. Soc. 2005;127:2040–2041. doi: 10.1021/ja045021y. [DOI] [PubMed] [Google Scholar]
- 6.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 7.Drouillard S, Driguez H, Samain E. Large-scale synthesis of H-antigen oligosaccharides by expressing Helicobacter pylori alpha1,2-fucosyltransferase in metabolically engineered Escherichia coli cells. Angew. Chem. Int. Ed. Engl. 2006;45:1778–1780. doi: 10.1002/anie.200503427. [DOI] [PubMed] [Google Scholar]
- 8.Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 9.Perugino G, Trincone A, Rossi M, Moracci M. Oligosaccharide synthesis by glycosynthases. Trends Biotechnol. 2004;22:31–37. doi: 10.1016/j.tibtech.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Varki A, et al. Essentials of Glycobiology. New York, USA: Cold Spring Harbor Laboratory Press; 1999. [PubMed] [Google Scholar]
- 11.Thibodeaux CJ, Melancon CE, Liu H-W. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 12.Sim MM, Kondo H, Wong CH. Synthesis of dibenzyl glycosyl phosphites using dibenzyl N,N-diethylphosphoramidite as phosphitylating reagent: an effective route to glycosyl phosphates, nucleotides, and glycosides. J. Am. Chem. Soc. 1993;115:2260–2267. [Google Scholar]
- 13.Heidlas JE, Lees WJ, Pale P, Whitesides GM. Gram-scale synthesis of uridine 5′-diphospho-N-acetylglucosamine: comparison of enzymic and chemical routes. J. Org. Chem. 1992;57:146–151. [Google Scholar]
- 14.Wagner GK, Pesnot T, Field RA. A survey of chemical methods for sugar-nucleotide synthesis. Nat. Prod. Rep. 2009;26:1172–1194. doi: 10.1039/b909621n. [DOI] [PubMed] [Google Scholar]
- 15.Timmons SC, Jakeman DL. Stereoselective chemical synthesis of sugar nucleotides via direct displacement of acylated glycosyl bromides. Org. Lett. 2007;9:1227–1230. doi: 10.1021/ol063068d. [DOI] [PubMed] [Google Scholar]
- 16.Nishimoto M, Kitaoka M. Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl. Environ. Microbiol. 2007;73:6444–6449. doi: 10.1128/AEM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai L, et al. A chemoenzymatic route to N-acetylglucosamine-1-phosphate analogues: substrate specificity investigations of N-acetylhexosamine 1-kinase. Chem. Commun. 2009:2944–2946. doi: 10.1039/b904853g. [DOI] [PubMed] [Google Scholar]
- 18.Cai L, et al. Substrate specificity of N-acetylhexosamine kinase towards N-acetylgalactosamine derivatives. Bioorg. Med. Chem. Lett. 2009;19:5433–5435. doi: 10.1016/j.bmcl.2009.07.104. [DOI] [PubMed] [Google Scholar]
- 19.Guan W, Cai L, Fang J, Wu B, Wang PG. Enzymatic synthesis of UDP-GlcNAc/UDP-GalNAc analogs using N-acetylglucosamine 1-phosphate Uridyltransferase (GlmU) Chem. Commun. 2009:6976–6978. doi: 10.1039/b917573c. [DOI] [PubMed] [Google Scholar]
- 20.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 21.Yi W, et al. Remodeling bacterial polysaccharides by metabolic pathway engineering. Proc. Natl. Acad. Sci. USA. 2009;106:4207–4212. doi: 10.1073/pnas.0812432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, et al. Chemoenzymatic synthesis of GDP-l-fucose and the Lewis X glycan derivatives. Proc. Natl. Acad. Sci. USA. 2009;106:16096–16101. doi: 10.1073/pnas.0908248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen LR, Roderick SL. Structure of the Escherichia coli GlmU pyrophosphorylase and acetyltransferase active sites. Biochemistry. 2001;40:1913–1921. doi: 10.1021/bi002503n. [DOI] [PubMed] [Google Scholar]
- 24.Bourgeaux V, Piller F, Piller V. Two-step enzymatic synthesis of UDP-N-acetylgalactosamine. Bioorg. Med. Chem. Lett. 2005;15:5459–5462. doi: 10.1016/j.bmcl.2005.08.088. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Chokhawala HA, Huang S, Chen X. One-pot three-enzyme chemoenzymatic approach to the synthesis of sialosides containing natural and non-natural functionalities. Nat. Protoc. 2006;1:2485–2492. doi: 10.1038/nprot.2006.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu B, Zhang Y, Wang PG. Identification and characterization of GDP-D-mannose 4,6-dehydratase and GDP-L-fucose synthetase in a GDP-L-fucose biosynthetic gene cluster from Helicobacter pylori. Biochem. Biophys. Res. Commun. 2001;285:364–371. doi: 10.1006/bbrc.2001.5137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.