Abstract
Perfect direct repeats and, in particular, the prominent 13-bp repeat, are thought to cause mitochondrial DNA (mtDNA) deletions, which have been associated with the aging process. Accordingly, individuals lacking the 13-bp repeat are highly prevalent among centenarians and the number of repeats negatively correlates with mammalian longevity. However, detailed examination of the distribution of mtDNA deletions challenges the role of the 13-bp repeat and other perfect repeats in generating mtDNA deletions. Instead, deletions appear to depend on long and stable, albeit imperfect, duplexes between distant mtDNA segments. Furthermore, significant dissimilarities in breakpoint distributions suggest that multiple mechanisms are involved in creating mtDNA deletions.
Direct repeats in the mitochondrial genome and longevity
The premise that accumulation of mtDNA mutations[1] and, in particular, of large-scale deletions in mtDNA is one of the possible causes of aging has received substantial support from biochemical and longevity studies [2], [3], [4], [5]. mtDNA deletions are usually flanked by direct repeats, implying that these repeats are involved in the generation of deletions. Recombination [6], [7], slip-replication [8], and double-stranded break repair [9] have been suggested as potential alternative mechanisms involving direct repeats. In corroboration of the connection between mtDNA deletions and aging, the number of direct repeats in mtDNA of various mammal species is inversely correlated with longevity [10], [11]. Of particular interest is the so-called “common deletion”[6], the deletion most frequently detected in humans, which is flanked by a prominent 13-bp perfect direct repeat. Interestingly, the carriers of the well studied D4a mitochondrial haplogroup, who are significantly enriched among Japanese centenarians [12], lack the 13-bp direct repeat in their mtDNA, and thus presumably lack the common deletion, which seems to support the premise that deletions are involved in the aging process [13]. It should be noted, however, that although the “common” deletion is the most abundant mtDNA deletion, it typically constitutes no more than 10% of all deletions in aging tissues [5]. Therefore D4a individuals would have at most 10% fewer deletions, which perhaps is too moderate a change to affect longevity. There is another possibility, though [13]. According to an elegant hypothesis by Samuels, Schon and Chinnery, the 13-bp repeat might be responsible for the formation of nearly all mtDNA deletions, not just the common deletion [14]. Thus the absence of this repeat could have resulted in a reduction of the overall deletion burden, and, if deletions indeed are involved in the aging process, this might constitute a realistic cause of exceptional D4a longevity. The importance of this question prompted us to test the Samuels, Schon, Chinnery hypothesis.
mtDNA deletions, in general, are not related to the 13-bp repeat
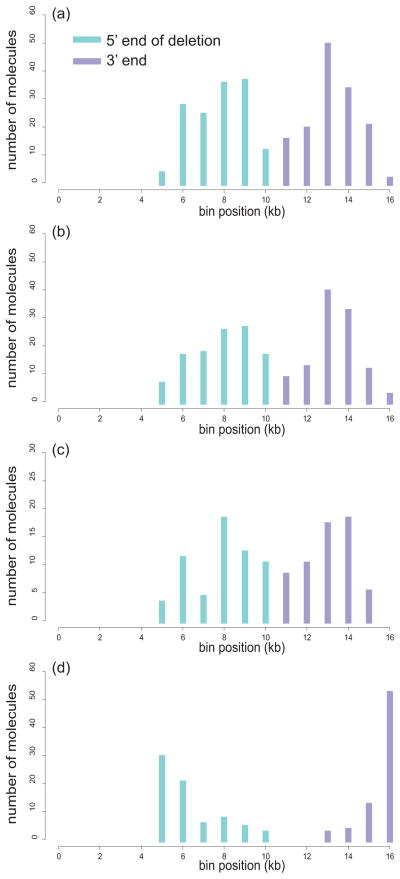
The Samuels, Schon, Chinnery hypothesis rests on the observation that the distribution of deletion breakpoints across the mitochondrial genome consists of two broad peaks centered around the 5’ and 3’ arms of the 13-bp repeat, i.e. the nucleotide positions 8469-8482 and 13447-13459 [14]. This hypothesis would therefore predict that loss of the 13-bp repeat would result in the disappearance of this characteristic pattern. To test this prediction, we compared the distribution of breakpoints in individuals with and without the 13-bp repeat. Using single molecule PCR [15] (for methods, see Supplement 2) we determined breakpoints of 242 unique mtDNA deletions from the frontal cortex tissue of four individuals. mtDNA of two of these individuals (controls) contain the common repeat, whereas the other two belong to the mitochondrial DNA haplogroup N1b in which the 5’ arm of the repeat is altered by the 8472C>T polymorphism, which is in a neighboring position to the D4a polymorphism (8473T>C) (Supplement 1). In accord with Samuels, et al. [14], the distribution of breakpoints of these deletions in control individuals is bimodal with maxima roughly coincident with the arms of the repeat (Figure 1A and Supplement 3). Unexpectedly, the distribution of deletions in individuals without the 13-bp repeats (Figure 1B) is essentially the same as in controls. Thus, lack of the 13-bp repeat has no effect on the distribution of deletion breakpoints, in contradiction with the hypothesis prediction. Note that the 8472C>T polymorphism disrupts the common repeat, i.e. significantly reduces occurrence of the common deletion. Indeed, we found only one “pseudocommon” deletion (i.e. a deletion utilizing the remaining 10-bp repeat) in individuals without the 13-bp repeat compared to 12 common deletions found in control individuals. We conclude that there is no evidence that the common 13-bp repeat causes the formation of a significant proportion of mtDNA deletions.
Figure 1. “Deletional spectra” challenge the “common” 13-bp repeat and perfect direct repeats in general as sources of mtDNA deletions.
Deletional spectra were constructed by plotting the number of breakpoints discovered in each 1-kb interval of the mitochondrial genome (“bin”) as a function of the bin position (see Supplement 3 for details). Breakpoint distributions in individuals with (a) or without (b) the “common” 13-bp repeat are very similar, a finding which contradicts any special role of the “common” repeat in generating most mtDNA deletions. (c) A subset of deletions without direct repeats at breakpoints (class II deletions [7]) appears to be distributed similarly to class I deletions (with repeats), implying that the mechanism that shapes breakpoint distribution in cortex does not depend on the presence and/or absence of perfect repeats at breakpoints. (d) An example of strikingly dissimilar spectrum of deletions from epileptic hippocampi suggests that a distinct mechanism is causing mtDNA deletions in this tissue.
Deletion breakpoints coincide with distant segments of mtDNA capable of forming stable imperfect duplexes with each other
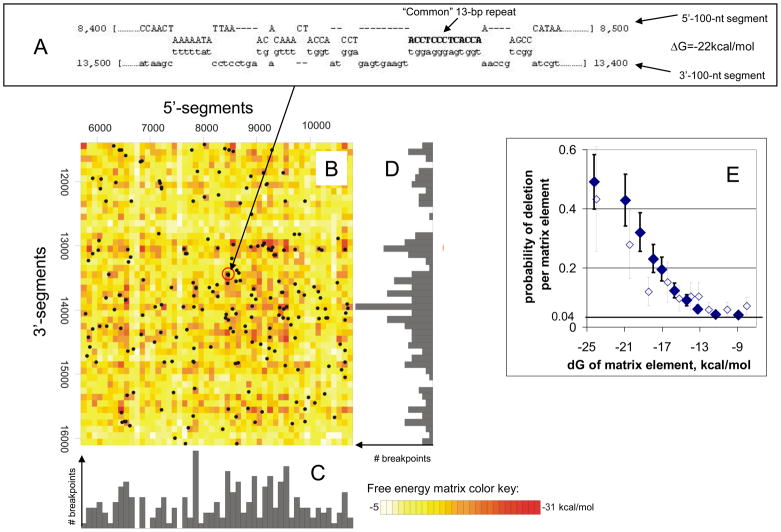
What else (if not the common repeat) could have created the characteristic distribution of breakpoints with maxima around 8–9 kb and 13–14 kb (Figure 1A–B)? BLAST analysis of our breakpoint database revealed that many deletions are flanked by long (up to 50 nt) regions of partial or interrupted homology (data not shown). Regions of imprecise homology at breakpoints have been also noticed by others [7]. The formation of long imperfect duplexes between these regions of homology might have brought together distant segments of the mitochondrial genome, which is necessary for formation of deletions by any of the proposed mechanisms [6], [7], [8]. We therefore explored in silico the distribution of possible secondary structures between 100-bp long segments of mtDNA from the region of 5’ breakpoints (base pairs 5700-10737) and the region of the 3’ breakpoints (base pairs 11400-16100) (Supplement 4). The resulting best fit duplexes typically include several short double stranded regions separated by loops of various sizes (Figure 2A and Supplement 5). Stability of all such duplexes can be conveniently depicted by a “matrix of free energies” (Figure 2B). Analysis of this matrix reveals a strong correlation between the stability of inter-segment duplexes and the distribution of mtDNA deletions (Figure 2C–E).
Figure 2. mtDNA deletions coincide with long and stable imperfect duplexes between distant segments of the mitochondrial genome.
(a) An example of a long imperfect duplex between two distant 100-bp long segments of mtDNA discovered by an in silico search (see also Supplement 4) is shown. For demonstration purposes we selected the duplex between the segments containing the arms of the common 13-bp repeat. The 5’- and 3’-100-nt segments of mtDNA that participate in the formation of this duplex and the corresponding free energy ΔG are indicated. (b) The distribution and stabilities of all possible intersegment duplexes is depicted by the matrix of free energies (see also Supplement 4). The color of each matrix element represents the free energy (ΔG) of the best possible DNA duplex between the corresponding 100-bp 5’-segment and the 100-bp 3’-segment (the lower the ΔG, the higher the stability, the more intense the color – see color key). Black dots depict positions of the real cortical deletions which apparently correlate with dark-colored matrix elements: deletions “prefer” to form between segments that have the potential to form a stable duplex with each other. Similarly, maxima or minima of the distributions of 5’-(c) and 3’-(d) deletion breakpoints appear to correlate with dark or pale “stripes” that run vertically or horizontally across the matrix, correspondingly. In other words, the probability that a deletion falls within a certain matrix element increases rapidly and significantly (p~2 × 10−16) with increasing duplex stability (decreasing ΔG), as quantitatively shown (e, filled diamonds; see also Supplement 6). This finding strongly implicates long imperfect duplexes in mtDNA deletion formation. Interestingly, deletions without repeats at breakpoints are also much more likely to fall within stable matrix elements (e, empty diamonds), implying that short perfect repeats per se do not significantly affect the generation of deletions.
Perfect repeats or stable imperfect duplexes?
The stability of long imperfect duplexes between distant segments of the mitochondrial genome closely predicts the distribution of mtDNA deletions (Figure 2). Because perfect repeats are widely considered to be the culprit of mtDNA deletions, we also performed regression analysis of the distribution of breakpoints versus the distribution of perfect direct repeats >=5bp (Supplement 7) and found a significant correlation between the probability of a deletion to occur in a matrix element and the number of direct repeats in the same element. So what causes deletions: perfect repeats or long stable albeit imperfect duplexes? Precise sequence homology or duplex stability? Distinguishing between the two is difficult because long duplexes actually consist of appropriately arranged perfect repeats, although not every arrangement of short perfect repeats results in a long stable duplex. To sort out these possibilities, we used multiple logistic regression analysis (Supplement 7) of the probability of observing at least one deletion within an element of the matrix of free energies (Supplement 4) as a function of two variables: the ΔG (the free energy of the best duplex formation) and the number of perfect repeats within the same element. It appears that the contribution of the ΔG of the best imperfect duplex remains as highly significant as in the single-variable analysis (p values ~10−16), whereas the contribution of the number of perfect repeats completely loses its significance (p=0.2) in the presence of ΔG as a variable. We therefore conclude that mtDNA deletion formation depends primarily on availability of stable imperfect duplexes rather than on the presence of perfect repeats.
The above conclusion, however, seems to contradict an observation that almost 75% of deletions in the dataset, i.e. much more than expected by chance, contain perfect repeats exactly at breakpoints . To reconcile these observations, we note that there is no shortage of perfect repeats in mtDNA: an average 100 × 100 matrix element contains about 70 direct repeats >=5 bp (Supplement 8). Therefore it is tempting to speculate that although deletions might form preferably at perfect repeats, the vast abundance of such repeats renders this requirement non-limiting. Instead deletions likely depend on the formation of long and stable, albeit imperfect, duplexes, possibly needed to hold distant mtDNA segments together. Once such a duplex has formed, a deletion most likely will be created precisely at one of the many perfect repeats available nearby. This explains both the presence of perfect repeats at breakpoints and the weakness of correlation between the distributions of breakpoints and perfect repeats. In support of this view, deletions without perfect repeats at breakpoints are distributed similarly to deletions with repeats (Figure 1A and B versus C, see also [14]), and just like deletions with repeats at breakpoints, deletions without repeats are much more likely to form in elements with stable intersegment duplexes than elsewhere in the genome (Figure 2E).
Other factors affecting formation of mtDNA deletions: multiple mechanisms?
It is worth noting however, that the proposed relationship between the stability of intersegment duplexes and the distribution of mtDNA deletion breakpoints in mtDNA deletions from normal cortex is by no means an all-encompassing one. Other tissues or individuals might present with highly dissimilar deletional spectra. A remarkable example is given by the distribution of mtDNA deletion breakpoints in the hippocampi of patients with temporal lobe epilepsy and Ammon’s horn sclerosis (Figure 1D). These hippocampi contain over 10-fold more mtDNA deletions than normal hippocampus, which might be related to increased oxidative stress [16]. The significant differences in the distribution of deletion breakpoints to the normal frontal cortex pattern are particularly evident at the 3’-end as there is a huge excess of breakpoints at the long-known breakpoint hotspot at position 16,070 [17]. This deletion hotspot is suggested to be generated by double-strand breaks [18] that can be formed by attacks of reactive oxygen species [19]. It is tempting to speculate that differences in the distribution of breakpoints indicate that different mechanisms can direct the generation of mtDNA deletions.
Concluding remarks
Formation of deletions in the mitochondrial genome appears to be strongly dependent on stable secondary structures that might potentially form between distant segments of the genome, rather than by relatively short perfect direct repeats as is widely assumed. This observation will certainly affect thinking related to mechanisms of deletion formation. Furthermore, previously discovered associations between the presence or absence of the common repeat or other perfect direct repeats in mtDNA and longevity [10], [11], [13] might need to be revisited. Methodologically, this work demonstrates the power of analysis of deletion breakpoint distributions, i.e. “deletional spectra”. Spectral dissimilarities between different tissues imply the existence of multiple factors that can cause mtDNA deletions and/or shape their distribution. This opens up promising research avenues where one could explore mechanisms of deletion formation by comparing deletional spectra created by various potential sources such as oxidative stress, double-stranded breaks and other types of DNA damage, or defective mtDNA maintenance proteins to delineate the various factors responsible for the accumulation of mtDNA deletions in aging and disease.
Supplementary Material
Acknowledgments
KK has been supported in part by NIH grants AG019787 and NS058988, XG by a visiting scholar fellowship from the China Scholarship Council Program, WSK by Deutsche Forschungsgemeinschaft SFB TR3 A11, DMT by Newcastle University Centre for Brain Ageing and Vitality supported by the BBSRC, EPSRC, ESRC, and MRC as part of the cross-council Lifelong Health and Wellbeing Initiative, and KP by RAS Molecular and Cellular Biology Program, RFBR Grants 08-04-01651- and 10-04-00276-, RMSF (P2442), Russian Science Support Foundation and Grant of the President of the RF (MK-5423.2008.4).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Linnane AW, et al. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- 2.Wanagat J, et al. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. Faseb J. 2001;15:322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
- 3.Herbst A, et al. Accumulation of mitochondrial DNA deletion mutations in aged muscle fibers: evidence for a causal role in muscle fiber loss. J Gerontol A Biol Sci Med Sci. 2007;62:235–245. doi: 10.1093/gerona/62.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender A, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 5.Kraytsberg Y, et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 6.Schon EA, et al. A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science. 1989;244:346–349. doi: 10.1126/science.2711184. [DOI] [PubMed] [Google Scholar]
- 7.Mita S, et al. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 1990;18:561–567. doi: 10.1093/nar/18.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoffner JM, et al. Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proc Natl Acad Sci U S A. 1989;86:7952–7956. doi: 10.1073/pnas.86.20.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnan KJ, et al. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 10.Samuels DC. Mitochondrial DNA repeats constrain the life span of mammals. Trends Genet. 2004;20:226–229. doi: 10.1016/j.tig.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Khaidakov M, et al. Direct repeats in mitochondrial DNA and mammalian lifespan. Mech Ageing Dev. 2006;127:808–812. doi: 10.1016/j.mad.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Bilal E, et al. Mitochondrial DNA haplogroup D4a is a marker for extreme longevity in Japan. PLoS ONE. 2008;3:e2421. doi: 10.1371/journal.pone.0002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popadin K, Bazykin G. Nucleotide Repeats in Mitochondrial Genome Determine Human Lifespan. Nature preceedings. 2009 http://precedings.nature.com/documents/2399/version/2391.
- 14.Samuels DC, et al. Two direct repeats cause most human mtDNA deletions. Trends Genet. 2004;20:393–398. doi: 10.1016/j.tig.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Kraytsberg Y, Khrapko K. Single-molecule PCR: an artifact-free PCR approach for the analysis of somatic mutations. Expert Rev Mol Diagn. 2005;5:809–815. doi: 10.1586/14737159.5.5.809. [DOI] [PubMed] [Google Scholar]
- 16.Kudin AP, et al. Mitochondrial involvement in temporal lobe epilepsy. Exp Neurol. 2009;218:326–332. doi: 10.1016/j.expneurol.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 17.Zeviani M, et al. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava S, Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum Mol Genet. 2005;14:893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.