Summary
A-to-I editing is an adenosine-to-inosine modification of mRNA particularly widespread in the human brain, where it affects thousands of genes. A growing body of evidence suggests that A-to-I RNA editing is necessary for normal development and maintenance in mammals and that its deficiencies contribute to a number of pathological states. In this study, we examined whether mRNA editing levels of two mRNA species, CYFIP2 and GABRA3, change with aging. CYFIP2 has been implicated in synaptic maintenance, while GABRA3 is a GABA receptor subunit, a part of the major inhibitory neurotransmitter system in the CNS. The levels of mRNA editing were assessed in cortex samples of 20 subjects 22 to 102 years old. The data show an age-dependent statistically significant decrease in editing in CYFIP2. GABRA3 editing remained much more stable with age, implying that age-related decline of RNA editing is gene-specific. This is the first report of age-dependent decline in A-to-I editing. Further examination of these and other vulnerable genes may reveal specific RNA editing mechanisms that contribute to the aging phenotype.
Keywords: Aging, RNA editing, human brain
Adenosine-to-inosine (A-to-I) RNA editing is a post-transcriptional processing pathway particularly widespread in the human brain wherein adenosine in pre-mRNA is modified to yield inosine, which is equivalent of guanosine for the splicing and translational machineries. This process is catalyzed by the members of the double-stranded RNA (dsRNA)- specific ADAR (adenosine deaminase acting on RNA) family (Bass, 2002). In the past few years, bioinformatic and experimental studies have revealed tens of thousands of editing sites affecting over 1600 different genes (Athanasiadis et al., 2004; Blow et al., 2004; Kim et al., 2004; Levanon et al., 2004; Morse and Bass, 1999). Deregulation (or dysregulation) of RNA editing has been linked to a few diseases of the central nervous system (Maas et al., 2006), such as depression (Gurevich et al., 2002; Niswender et al., 2001), epilepsy (Brusa et al., 1995), Glioblastoma (Cenci et al., 2008; Maas et al., 2001; Paz et al., 2007) and amyotrophic lateral sclerosis (Kawahara et al., 2007). It is thus conceivable that if RNA editing becomes deregulated with age, then it may in part explain the decline in some brain functions attributed to aging.
Changes in other types of mRNA editing have been previously discovered. These include developmental and age-related changes for a much more rare type of RNA editing such as (C-to-U) of the apoB in mice which are believed to represent a regulated process of lipoprotein biogenesis (Higuchi et al., 1992) and other possible editing type e.g. (van Leeuwen et al., 1998). However, although A-to-I editing is much more abundant, there is almost no knowledge about its regulation during aging.
In the present study, we tested whether levels of RNA editing change with age by examining two carefully selected transcripts, GABRA3 and CYFIP2. CYFIP2 is a p53-inducible protein (Jackson et al., 2007), which has been implicated in synaptic maintenance (Schenck et al., 2001), and its editing levels are regulated in murine development (Wahlstedt et al., 2009). GABRA3 is a variant of the alpha subunit of a GABA-A receptor, a part of the major inhibitory neurotransmitter system in the CNS (Akabas and Ronald, 2004). GABRA3 RNA editing is also tightly regulated (increased A to I) during early development, and the edited and the non-edited form may have different functional profiles in the mouse (Rula et al., 2008), (Wahlstedt et al., 2009). CYFIP2 and GABRA3 transcripts were selected because editing in both is regulated during development, both function in the brain, and both are located in extremely conserved genomic regions suggesting an important function. In addition, both transcripts have shown high levels of RNA editing in young adult individuals and each contained a single edited site thus allowing for straightforward quantitative analysis.
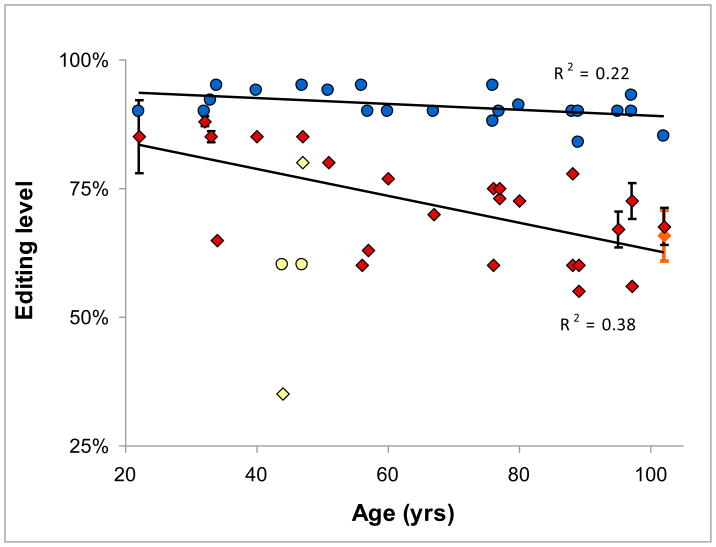
Levels of RNA editing were measured in 25 frontal cortex samples from individuals aged 22 to 102 years old (Table 1) by quantitative analysis of the sequencing chromatograms of the RT-PCR products (see materials and methods on-line for detailed description of the samples and the procedures). The level of editing as a function of age is graphically presented in figure 1. The graph suggests that there is a decrease in the level of editing with age in CYPFIP2, but no change in GABRA3. Indeed the decrease of CYPFIP2 with age is statistically significant using both Pearson’s (p=0.048) and Spearman’s (p=0.006) tests, while this is not true for GABRA (p=0.28, Spearman test; Pearson not applicable since the data was not normally distributed).
Table 1.
List of samples.
| Editing level | ||||
|---|---|---|---|---|
| Age | cause of death | gender | GABRA | CYFIP2 |
| 22 | suicide | m | 0.9 | 0.85 |
| 32 | suicide | f | 0.9 | 0.88 |
| 33 | suicide | m | 0.92 | 0.85 |
| 34 | cystic fibrosis | m | 0.95 | 0.65 |
| 40 | suicide | m | 0.94 | 0.85 |
| 44 | skin cancer/ventilator | f | 0.6 | 0.35 |
| 47 | heart failure | ? | 0.95 | 0.85 |
| 47 | hypoxia ischemia | m | 0.6 | 0.8 |
| 51 | heart failure | m | 0.94 | 0.8 |
| 56 | bone cancer | m | 0.95 | 0.6 |
| 57 | heart failure | f | 0.9 | 0.63 |
| 60 | heart failure | f | 0.9 | 0.77 |
| 67 | heart failure | m | 0.9 | 0.7 |
| 76 | heart failure | m | 0.88 | 0.75 |
| 76 | unknown | m | 0.95 | 0.6 |
| 77 | heart failure | m | 0.9 | 0.73 |
| 77 | heart failure | m | 0.9 | 0.75 |
| 80 | heart failure | ? | 0.91 | 0.725 |
| 80 | heart failure | f | 0.95 | 0.7 |
| 88 | heart failure | f | 0.9 | 0.78 |
| 88 | heart failure | f | 0.9 | 0.6 |
| 89 | heart failure | ? | 0.84 | 0.6 |
| 89 | leukemia | f | 0.9 | 0.55 |
| 95 | heart failure | ? | 0.9 | 0.67 |
| 97 | heart failure | f | 0.9 | 0.725 |
| 97 | heart failure | f | 0.93 | 0.56 |
| 102 | heart failure | m | 0.85 | 0.675 |
Samples considered outliers highlighted yellow.
Figure 1.
Levels of editing of the CYFIP2 (diamonds) and GABRA3 (circles) mRNAs as a function of age. While the decrease of CYFPI level with age is statistically significant, there is no significant decrease in the editing levels of GABRA. GABRA decrease become significant upon removal of two outlier samples (yellow symbols). These two samples were considered outliers as they lie further than 3 standard deviations away from average. Regression lines constructed in Excel for data with outliers excluded. Error bars show formal standard deviations for samples where two or more measurements were taken. The orange data point represents an average of 3 different areas, each with duplicate measurements, of the 102 yo individual (this data point was not included in regression analysis).
Visual inspection of Figure 1 reveals that two samples are clear outliers (marked yellow, these same samples are also highlighted in table 1). In both samples, at least one value (CYPFIP2 or GABRA3 editing level) is more than 3 standard deviations away from the regression line (constructed for all samples). Interestingly, both outlier samples originate from patients who likely suffered unusually prolonged or disruptive treatment/pathology affecting the brain: one patient spent a full month on a ventilator until her death, and in the other one brain tissue was subject to ischemia. These observations imply that aging per se is probably not the only factor affecting RNA editing. We therefore decided to re-evaluate the data upon removal of the outliers. As expected, removal of outliers improved the significance of the CYFIP2 trend according to both tests, Pearson’s (p=0.001) and Spearman’s (p=0.002). Furthermore, now we were able to detect a statistically significant decrease in GABRA3 editing (p=0.035 and 0.043, correspondingly), though the GABRA3 slope is 4 times as moderate as that of CYPFIP2, implying that age-related changes in the levels of editing are gene-specific.
We further inquired whether the trends we see could have been confounded by the fact that most of our younger samples originate from suicide victims. Perhaps suicidal behavior, not their young age could have been related to the levels of editing in their brain. Analysis shows, however, that the trend for CYPFIP2 remains statistically significant even if suicide victims are removed from the dataset, (p = 0.007 using Pearson and 0.016 using Spearman’s test), though the correlation between GABRA and age loses significance.
Previous work by others showed constant levels of A-to-I editing with age. In a mouse model, aging failed to result in a change in AMPA glutamate receptors editing (Carlson et al., 2000). In contrast, our data imply that A-to I editing declines with aging in a gene-specific manner (CYFIP2 affected much more profoundly than GABRA3). Of note, editing of CYFIP2 is performed by the enzyme ADAR2 (Nishimoto et al., 2008; Riedmann et al., 2008), while GARBA3 is a substrate for editing by both ADAR1 and ADAR2 (Ohlson et al., 2007), thus the differences we observe might reflect different changes in the activities or abundance of these enzymes. The fact that levels of RNA editing level of GABRA3 is much lower than that of CYFIP2, suggests that an overall non-specific age-related decline alone cannot account for the changes.
This work is merely the first step in exploring still one more dimension of the human aging process, i.e. changes in the RNA editing levels. Recent technological progress in the RNA editing field opens exciting opportunities of exhaustive, whole-genome studies of RNA editing changes (Li et al., 2009). This approach can hopefully be used to monitor editing levels in all editing sites across the whole human genome simultaneously in multiple samples. The results presented in our report make a case for using these novel approaches to study aged-related changes in RNA editing.
Supplementary Material
Acknowledgments
The authors thank Massachusetts Alzheimer’s Tissue and Resource Center for providing brain samples. This work has been supported in part by the NIH grant AG19787 to KK and supplement to AN. E.Y.L. was supported by the Machiah foundation. JPM is supported by the BBSRC and a Marie Curie International Reintegration Grant within EC-FP7.
Footnotes
Note: as this manuscript was under review, a newly published linkage analysis study (Sebastiani et al., 2009) reported strong evidence for association of polymorphisms in ADARB1 and ADARB2 with extreme old age. This finding implies that the age related changes that were observed may potentially be a part of whole genome changes of RNA editing, which do not merely accompany, but actually cause certain processes that limit human longevity.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akabas MH, Ronald JB. International Review of Neurobiology. Academic Press; 2004. GABAA Receptor Structure-Function Studies: A Reexamination in Light of New Acetylcholine Receptor Structures; pp. 1–43. [DOI] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA Editing of Alu-Containing mRNAs in the Human Transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- Carlson NG, Howard J, Gahring LC, Rogers SW. RNA editing (Q/R site) and flop/flip splicing of AMPA receptor transcripts in young and old brains. Neurobiol Aging. 2000;21:599–606. doi: 10.1016/s0197-4580(00)00127-5. [DOI] [PubMed] [Google Scholar]
- Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, Di Rocco C, O’Connell MA, Gallo A. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem. 2008 doi: 10.1074/jbc.M708316200. [DOI] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- Higuchi K, Kitagawa K, Kogishi K, Takeda T. Developmental and age-related changes in apolipoprotein B mRNA editing in mice. J Lipid Res. 1992;33:1753–1764. [PubMed] [Google Scholar]
- Jackson RS, 2nd, Cho YJ, Stein S, Liang P. CYFIP2, a direct p53 target, is leptomycin-B sensitive. Cell Cycle. 2007;6:95–103. doi: 10.4161/cc.6.1.3665. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of Silencing Targets by Adenosine-to-Inosine Editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Bass BL. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc Natl Acad Sci U S A. 1999;96:6048–6053. doi: 10.1073/pnas.96.11.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto Y, Yamashita T, Hideyama T, Tsuji S, Suzuki N, Kwak S. Determination of editors at the novel A-to-I editing positions. Neurosci Res. 2008;61:201–206. doi: 10.1016/j.neures.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Niswender CM, Herrick-Davis K, Dilley GE, Meltzer HY, Overholser JC, Stockmeier CA, Emeson RB, Sanders-Bush E. RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology. 2001;24:478–491. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- Ohlson J, Pedersen JS, Haussler D, Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. Rna. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedmann EM, Schopoff S, Hartner JC, Jantsch MF. Specificity of ADAR-mediated RNA editing in newly identified targets. Rna. 2008;14:1110–1118. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rula EY, Lagrange AH, Jacobs MM, Hu N, Macdonald RL, Emeson RB. Developmental modulation of GABA(A) receptor function by RNA editing. J Neurosci. 2008;28:6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Montano M, Puca A, Solovieff N, Kojima T, Wang MC, Melista E, Meltzer M, Fischer SEJ, Andersen S, et al. RNA Editing Genes Associated with Extreme Old Age in Humans and with Lifespan in C. elegans. PLoS ONE. 2009;4:e8210. doi: 10.1371/journal.pone.0008210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen FW, de Kleijn DP, nbsp V, van den Hurk HH, Neubauer A, Sonnemans MA, nbsp F, Sluijs JA, et al. Frameshift Mutants of {beta}?Amyloid Precursor Protein and Ubiquitin-B in Alzheimer’s and Down Patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- Wahlstedt H, Daniel C, Enstero M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009 doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.