Abstract
The Myc–Max–Mad network of proteins activates or represses gene transcription depending on whether the dimerization partner of Max is c-Myc or Mad. To elucidate the physical properties of these protein–protein interactions, fluorescence anisotropy of TRITC-labeled Max was used. The binding affinities and thermodynamics of dimerization of the Max–Max homodimer and c-Myc–Max and Mad–Max heterodimers were determined. Our results indicate that c-Myc and Max form the most stable heterodimer. Previous work [Kohler, J. J., Metallo, S. J., Schneider, T. L., and Schepartz, A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11735–9] has shown that instead of dimerizing first and then binding to DNA, these proteins use a monomer pathway in which a monomer binds to DNA followed by dimerization on the surface of the DNA. The DNA E-box affects the dimerization, but nonspecific effects may also play a role. The influence of polyions, poly-l-lysine and poly-l-glutamic acid, were investigated to determine the effects of charged polymers other than DNA on homodimerization and heterodimerization. While the positively charged poly-l-lysine, PLL, did not show any significant effect, negatively charged poly-l-glutamic acid, PLG, stabilized both heterodimers and homodimers by 2–3 kJ/mol. These data suggest that in the cell nucleus the presence of negatively charged DNA or RNA could nonspecifically aid in association of these proteins. Calculations of ΔH° and ΔS° from the temperature dependence of Kd indicated that although the thermodynamic parameters for the dimer are different, the reactions for all three dimers are driven by negative (favorable) enthalpic and negative (unfavorable) entropic contributions. In the presence of PLG, entropy became more negative with the effect being largest for c-Myc–Max heterodimers. This suggests that van der Waals and H-bonding interactions are predominant in dimerization of these proteins.
Initiation of transcription requires the ordered assembly of transcription factors, architectural proteins, and template DNA. Many of the transcription factors bind DNA as dimers, and it appears that dimerization through switching between homo- and heterodimeric structures provides a means for diverse target site recognition and function with the smallest investment of the genome (1). Myc, Max, and Mad are three such transcription modulators that bind DNA as dimers. These proteins belong to a subset of proteins having a C-terminal basic helix–loop–helix/leucine zipper (b/HLH/z) 1 motif. Interest in the Max network grew out of studies on the Myc oncogene family. Overexpression of all members of this family, including the cellular homologue c-Myc, is correlated with cell proliferation (2). The presence of an N-terminal trans-activation domain (TAD) (3) and a C-terminal b/HLH/z domain (4) in c-Myc led to the assumption that this protein would bind DNA as a dimer. However, its poor homodimerization and DNA binding ability at physiological concentrations initiated a search for a heterodimerization partner, eventually leading to the discovery of Max (5). Max is a b/HLH/z protein similar to Myc, however lacking the transactivation domain. Max was found to bind specifically with Myc family proteins, forming heterodimers that recognize the promoter hexameric palindromic sequence CACGTG (E-box) (5). Subsequently, it was found that binding of the Myc–Max heterodimer to the E-Box activates transcription of a series of genes related to growth, cell cycle activation, and apoptosis (6). Transcription activation is mediated at least in part by histone acetyl transferase (HAT) recruitment via the TAD of Myc (7).
The fact that Max is expressed even in the absence of Myc prompted researchers to search for new Max interacting proteins which led to the discovery of a Mad family of proteins consisting of Mad1, Mxi1, Mad3, and Mad4 (8–10). All of these proteins are similar to Myc in the sense that they homodimerize and bind DNA poorly; however, DNA E-box promoter binding is greatly enhanced when they heterodimerize with Max. From a functional point of view, the Mad proteins are strikingly different from Myc. Expression of Mad proteins is related to terminal differentiation where the mechanism of the Mad–Max heterodimer is to repress gene transcription by associating with the mSin3 corepressor complex via histone deacetylation (8, 11–13).
Previous work in our laboratory (14) and elsewhere (1) supports assembly of b/HLH/z proteins through a monomer binding pathway where one monomer binds DNA followed by dimerization with a second monomer on the DNA. However, even in the absence of DNA, Max and Max, Myc and Max, and Mad and Max form dimers. Both the homodimer and the heterodimers form an asymmetric parallel left-handed four-helix bundle consisting of two pairs of right-handed α-helices H1 and H2 and a C-terminal extension of the α-helices, the left-handed coiled coil leucine zipper (17). Crystal structures of the Max homodimer (15) and Myc–Max and Mad–Max heterodimers (16) along with solution NMR data of Myc–Max synthetic leucine zipper dimer (17) confirmed the putative role of LZ in dimerization specificity. LZ is thought to be specifically responsible for molecular recognition (17, 18).
Crystallographic and NMR (15–17) studies have revealed important data about specific amino acids involved in Max–Max, Myc–Max, and Mad–Max dimerization. Our previous studies (14) showed the importance of protein–protein interactions in the assembly of the transcription initiation complex. Moreover, the HLH/z domain of the Myc–Max heterodimer is a potential target for inhibitors (19, 20). To further understand these interactions, binding constants and thermodynamic data have been obtained. In this study, Max was labeled with TRITC and titrated with unlabeled Max, Myc, and Mad at various temperatures to determine the equilibrium dissociation constants and ΔH°, ΔS°, and ΔG° values of Max2, Myc–Max, and Mad–Max dimers. Our results indicate that the Myc and Max form the most stable heterodimer followed by Mad and Max and Max and Max by factors of 2.2 and 3.7, respectively, in terms of Kd.
Studies in our laboratory by Hu et al. (14) showed that binding of the Max monomer to E-box DNA significantly reduced the level of binding of the second monomer, whether the monomer was Max, Myc, or Mad. Other studies (20) have shown that dimerization rates of bz and b/HLH/z proteins increase in the presence of negatively charged polymers, including specific and nonspecific DNA, presumably through charge neutralization. To further understand the effects of DNA on the dimerization and the protein–protein interactions, the effect of polyions on dimer formation was examined. There were no significant changes in protein dimerization in the presence of the positively charged polyion, poly-l-lysine. However, the negatively charged polyion, poly-l-glutamic acid, stabilized all three dimers by 2–3 kJ/mol. This would indicate that the presence of a negatively charged environment of DNA and RNA in vivo could nonspecifically enhance the protein–protein interactions of these dimers.
MATERIALS AND METHODS
Protein Purification
Max (amino acids 22–113) and c-Myc (amino acids 347–439) in vectors pET2a and pGex2T, respectively, were obtained from S. K. Burley (Rockefeller University, New York, NY). Mad1 (henceforth Mad) was cloned in pET30a from Mad1 cDNA purchased from Open Biosystems (Huntsville, AL). All three proteins were expressed in BL21-pLys Escherichia coli cells and purified according to the protocol described elsewhere (14). Briefly, Max was purified by HiTrap SP ion exchange chromatography (15). c-Myc was purified using a GST affinity column purchased from Amersham Biosciences according to the manufacturer’s protocol using 50 mM reduced glutathione and 200 mM Tris-HCl (pH 8.5) as the elution buffer. Mad was purified by Ni–His affinity column chromatography (Novagen) according to the manufacturer’s protocol using 1 mM imidazole and 20 mM Tris-HCl (pH 7.9) as the elution buffer. All three purified proteins were analyzed by 12–15% SDS–PAGE and dialyzed extensively in HEPES buffer (pH 7.6). Protein concentrations were determined by the Bradford assay.
Fluorescent Labeling of the Max Protein
The N-terminus of the Max protein was labeled with tetramethylrhodamine 5-isothiocyanate (5-TRITC). To 1–5 mg/mL Max protein in 50 mM HEPES buffer (pH 7.6) was added 1–1.5 mg/mL TRITC dissolved in anhydrous DMSO. The solution was incubated overnight in the dark at 4 °C with continuous gentle stirring. Labeled protein was separated from free dye by Sephadex G-15 gel filtration with HEPES buffer. The collected fractions were concentrated with Centricon YM filters (Amicon Corp.). The final protein concentration and labeling ratio were calculated using the formula Aprotein = (A280 – A540)CF, where CF = (A280freedye)/(A540freedye) = 0.30 for TRITC and [protein] = Aprotein/1.4 mg/mL, and the F/P was calculated using an ε of 9300 for TRITC. Labeled Max is termed Max-TRITC.
Fluorescence Titrations
Protein–protein interactions of Max–Max homodimers and Max–c-Myc and Max–Mad heterodimers were assessed by observing the change in anisotropy (r) of 50 nM Max-TRITC by titrating with increasing concentrations of unlabeled Max, c-Myc, or Mad. The excitation wavelength was 540 nm, and emission was monitored at 570 nm. All fluorescence measurements were performed in a Spex Fluorolog τ2 spectrofluorimeter equipped with excitation and emission polarizers. The excitation and emission slits were set at 3 and 5 nm, respectively. Anisotropy was measured using an L format detection configuration. All titration measurements were performed in 20 mM HEPES-KOH, 50 mM KCl, 0.5 mM MgCl2, and 1 mM DTT buffer (pH 7.6) (titration buffer). For studying the temperature dependence of the protein–protein interaction, the samples were thermostated at different temperatures as described in Results. The temperature was measured using a thermocouple inside the cuvette.
For studying the effect of polyions on the dimerization equilibrium of Max2, Max–c-Myc, and Max–Mad dimers, titration experiments were performed under similar conditions as described above in the presence of 150 µM poly-l-lysine or 150 µM poly-l-glutamic acid, both purchased from Sigma.
Data Analysis
Data were fitted to the following equation (21, 22)
| (1) |
where b = KD + [A] + [B] and KD is the dissociation constant of [A][B], where [A] = [Max-TRITC] and [B] = [Max], [Myc], or [Mad].
Thermodynamic parameters for binding of Max-TRITC to unlabeled Max, Myc, or Mad were analyzed according to the van’t Hoff isobaric equation assuming that ΔH° and ΔS° remain unchanged over the temperature range that was studied:
| (2) |
Data were fit using Kaleidagraph (version 2.1.3, Synergy Software).
RESULTS AND DISCUSSION
Myc, Max, and Mad are relatively unstructured as monomers. Their α-helices are properly folded only when their dimerization interfaces are properly aligned whether they are bound to DNA. Schepartz et al. (1) have shown by CD measurements that at low concentrations (25–100 µM range) Max is largely unstructured and therefore presumed to be in a monomeric state. Similar experiments (data not shown) in our laboratory confirm this observation. Thus, to determine monomer–dimer dissociation constants, TRITC-labeled Max at 50 nM was titrated with unlabeled Max, Myc, or Mad. Thermodynamic parameters were obtained from the temperature dependence.
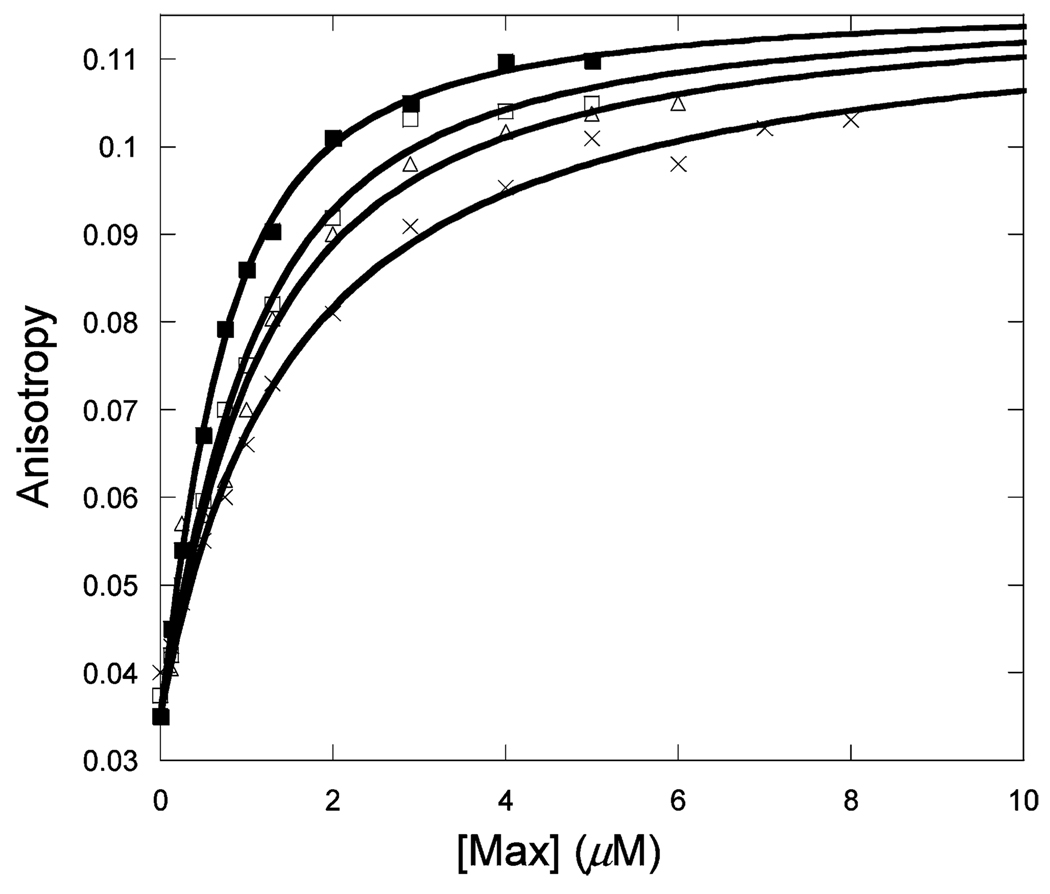
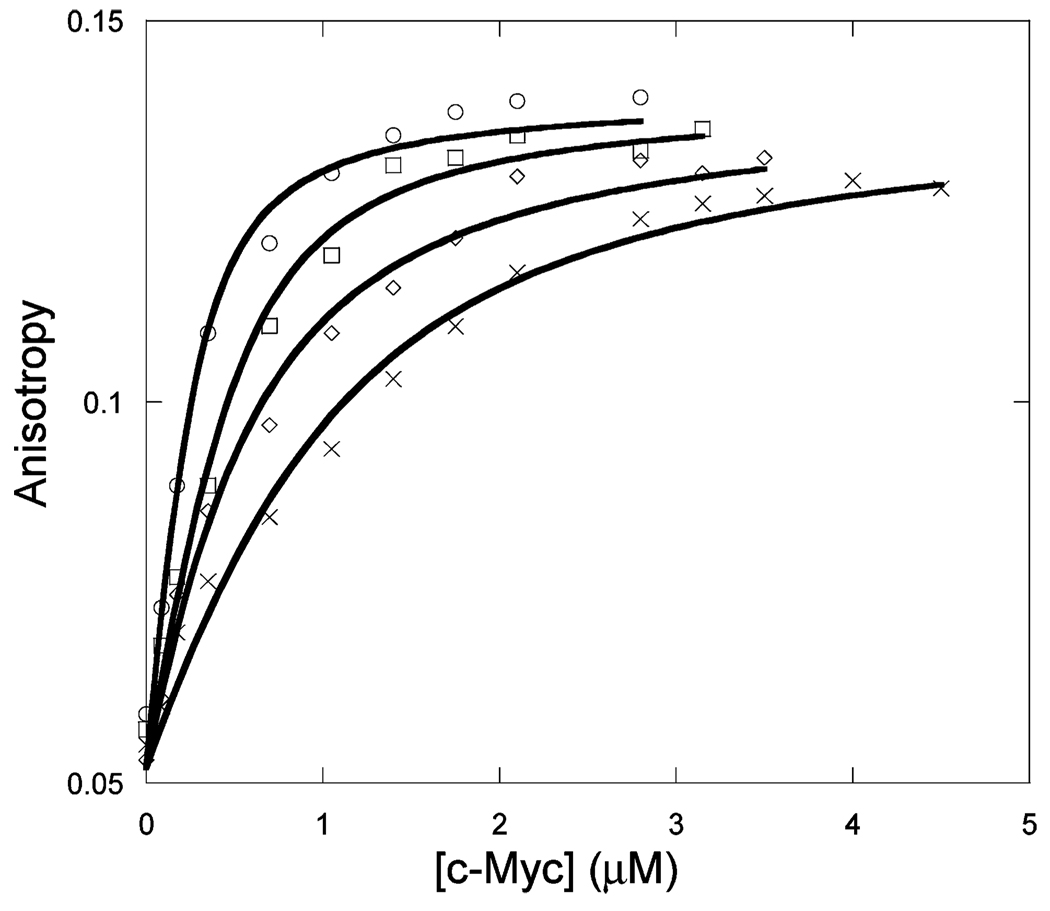
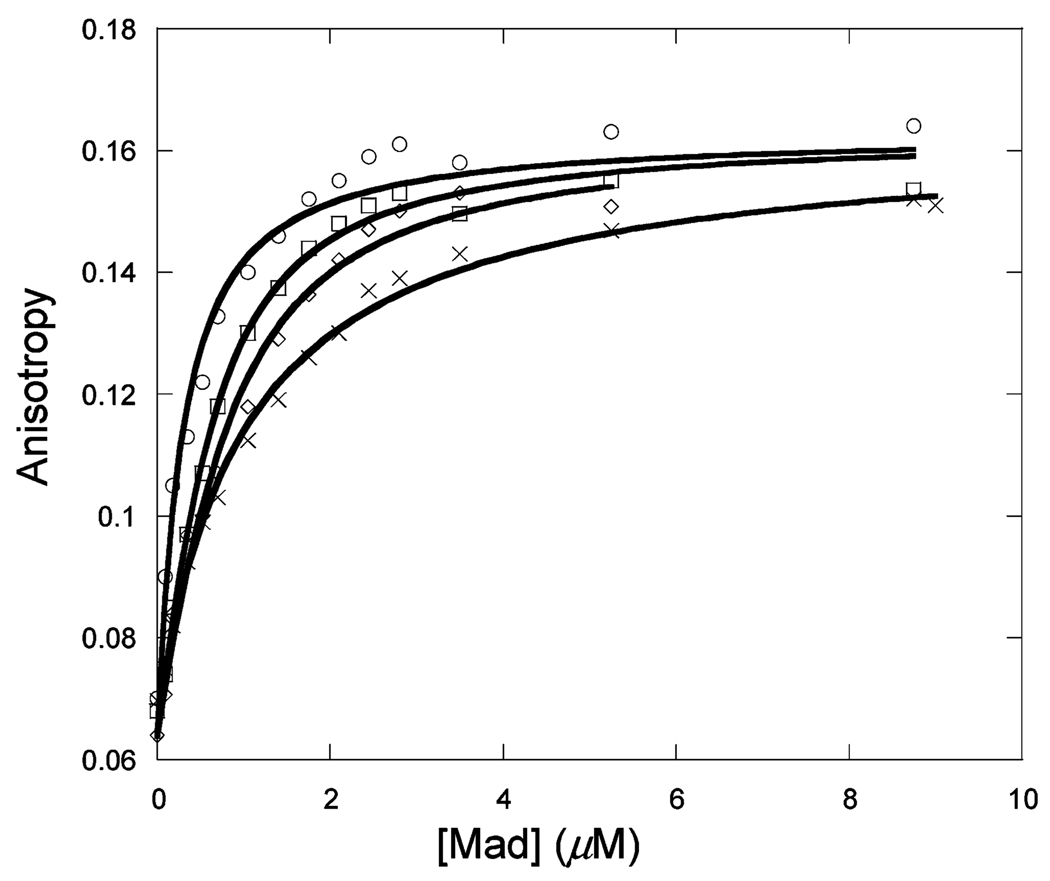
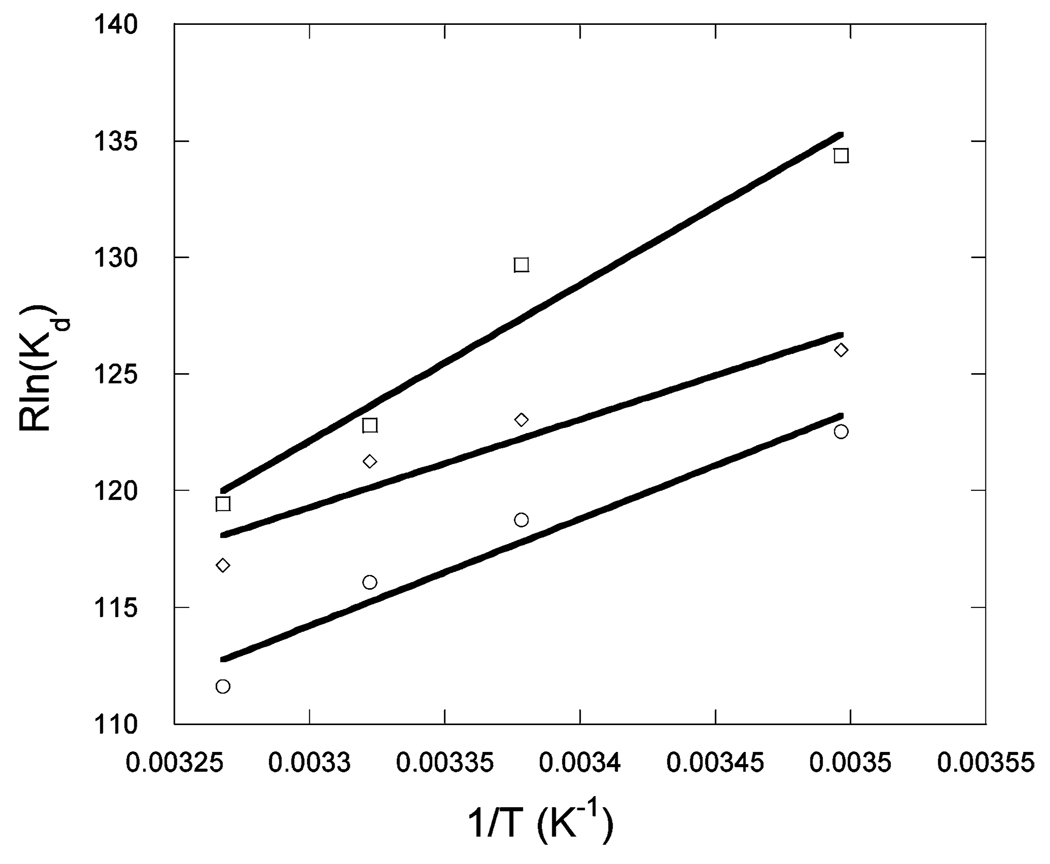
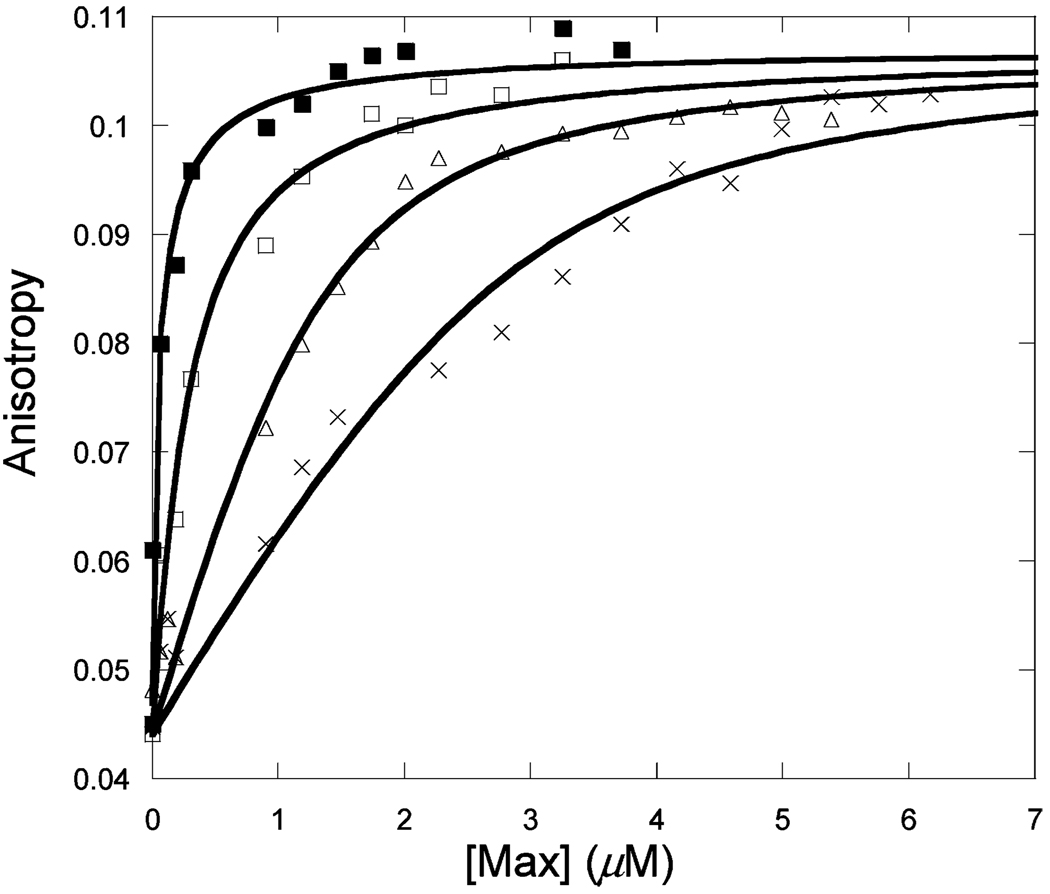
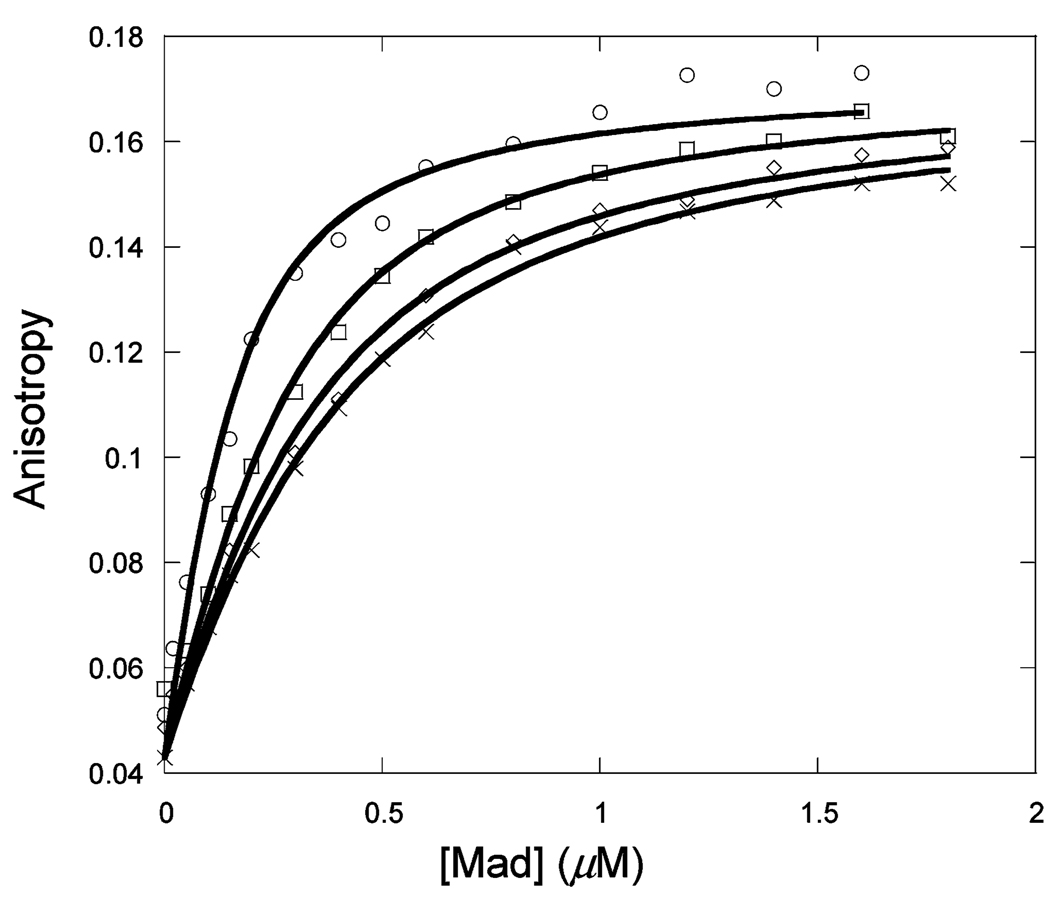
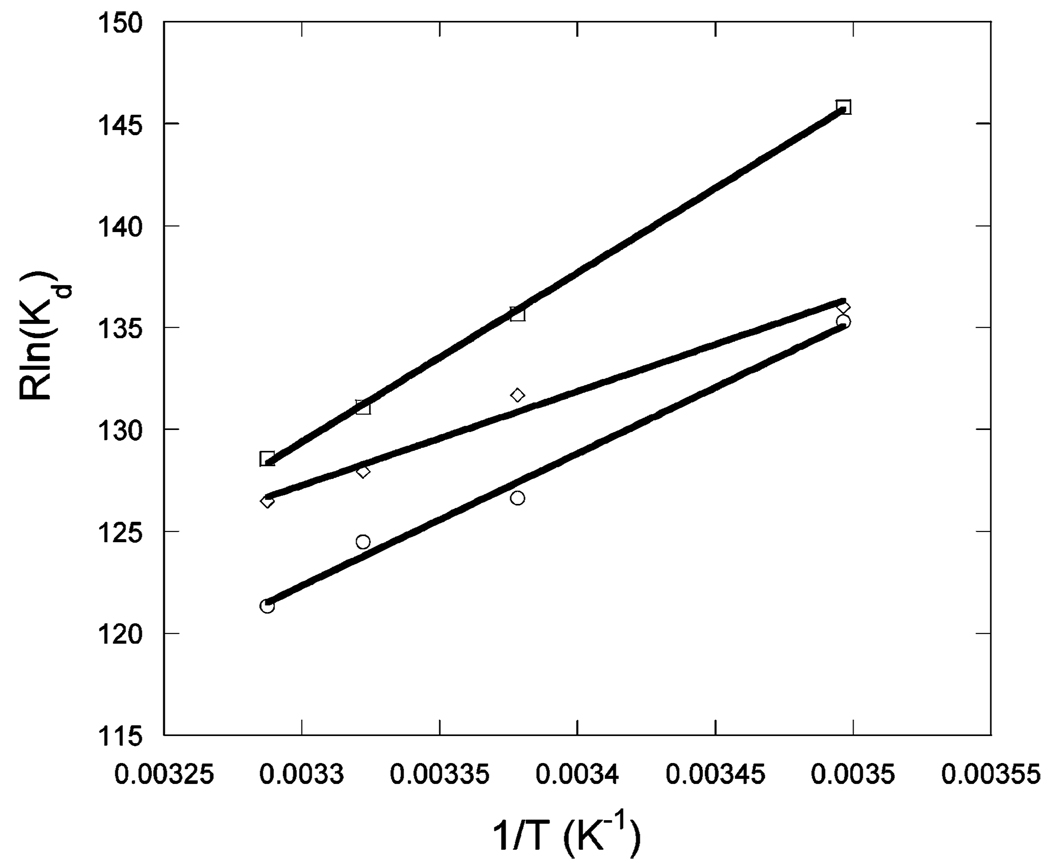
Figure 1 shows the binding of unlabeled Max to Max-TRITC as a function of temperature. The observed anisotropy change robs of Max-TRITC was plotted versus the concentration of unlabeled Max. The data were fitted to eq 1, and Kd values were determined. The lines indicate the fitted curves. The fitted anisotropy when extrapolated to 100% binding was the same within experimental error for all temperatures. Similar sets of data were obtained for Myc (Figure 2) and Mad (Figure 3) titrations with labeled Max-TRITC. The temperature dependences of the reactions were plotted (Figure 4) and used to determine ΔG°, ΔH°, and ΔS° according to eq 2. Table 1 shows the Kd values and the enthalpy and entropy of Max2, Myc–Max, and Mad–Max dimerization. Comparison of the individual dissociation constants indicates that at 33 °C, which is close to the physiological temperature, the binding of Myc to Max exhibits an affinity that is ~2.5 and ~1.4 times greater than the binding affinity of Max to Max and of Mad to Max, respectively. The association of all three dimers becomes even stronger at lower temperatures. However, the difference in association between Max2 and Mad–Max dimers becomes smaller. These trends are reflected in the thermodynamic parameters. All three dimerizations are enthalpically favored and entropically unfavorable. Table 2 shows the calculated ΔG° values. Interestingly, the ΔG° values at 37 °C are the same within experimental error for Myc and Mad associating with Max and only slightly less favorable for Max homodimer formation. The unfavorable entropy is compensated by the favorable enthalpy. Mad has a significantly lower entropic contribution than Myc or Max.
FIGURE 1.
Titration of 50 nM Max-TRITC with unlabeled Max at 13 (■), 23 (□), 28 (Δ), and 33 °C (×) in pH 7.6 titration buffer.
FIGURE 2.
Titration of 50 nM Max-TRITC with unlabeled c-Myc at 13 (◊), 23 (○), 28 (×), and 33 °C (□) in pH 7.6 titration buffer.
FIGURE 3.
Titration of 50 nM Max-TRITC with unlabeled Mad at 13 (○), 23 (□), 28 (◊), and 33 °C (×) in pH 7.6 titration buffer.
FIGURE 4.
vant Hoff’s plot for titration of 50 nM Max-TRITC with c-Myc (□), Max (○), and Mad (◊) in pH 7.6 titration buffer.
Table 1.
Thermodynamic Parameters for Association of Max, c-Myc, and Mad with Max-TRITC
| Kd (µM) | ||||||
|---|---|---|---|---|---|---|
| 13 °C | 23 °C | 28 °C | 33 °C | ΔH° (kJ/mol) | ΔS° (J/mol) | |
| Max | 0.392 ± 0.02 | 0.622 ± 0.06 | 0.857 ± 0.16 | 1.47 ± 0.14 | −45.7 ± 7.6 | −36.8 ± 15.5 |
| c-Myc | 0.095 ± 0.02 | 0.167 ± 0.03 | 0.382 ± 0.05 | 0.573 ± 0.09 | −66.8 ± 1.1 | −98.6 ± 17.4 |
| Mad | 0.259 ± 0.04 | 0.371 ± 0.03 | 0.460 ± 0.07 | 0.786 ± 0.09 | −37.6 ± 8.3 | −4.9 ± 10 |
Table 2.
ΔG° Values Calculated from Table 1
| ΔG° (kJ/mol) | |||
|---|---|---|---|
| 10 °C | 20 °C | 37 °C | |
| Max | −35.28 | −34.9 | −34.29 |
| c-Myc | −38.89 | −37.90 | −36.23 |
| Mad | −36.2 | −36.16 | −36.08 |
Formation of more stable heterodimers could be explained in terms of specific amino acid association as observed in the Myc–Max, Mad–Max, and Max–Max crystal structures (16). The three-dimensional structures of the Max homodimer and Max–Myc and Max–Mad heterodimers reveal hydrophobic interactions along the extensive protein–protein interface (buried solvent accessible area) in addition to hydrogen bonding in the periphery of the leucine zipper that is responsible for the stability and specificity of dimerization (15, 16).
In the Max homodimer, a tetrad association occurs between Gln91-Asn92-Gln91-Asn92 with Gln91 of one LZ at position g and Asn92 of the other LZ at position a. This creates a packing defect in the homodimer interface. However, in the Myc–Max heterodimer, Gln91 and Asn92 in Max can form stronger H bonds with positively charged Arg423 and Arg424 in Myc, resulting in closer protein–protein interactions. A similar situation arises in the case of the Mad–Max heterodimer in which Glu125 hydrogen bonds with Asn92.
In a NMR study with vMyc, Bister et al. (23) demonstrated that the b/HLH/z domain of vMyc was prestructured in solution with two α-helical subdomains. The presence of such a well-ordered structure may also occur in c-Myc and might serve as the nucleus for specific association with Max.
Negative entropy changes are generally associated with the loss of rotational and translational degrees of freedom (25). Bister et al. (23) have argued that in the case of vMyc, the presence of preformed secondary structure might restrict the conformational mobility of the protein. In our study, the highest negative unfavorable entropy was found for the Myc–Max dimer (Table 1). This suggests that the preformed structure of Myc can influence significantly the association with other proteins. Further, the data reveal that among the three protein dimers, the Mad–Max dimer probably has the most conformational flexibility which could account for its relatively less negative (−4.9 kJ/mol) entropy of dimerization. It will be interesting to see if NMR or other solution studies can confirm this prediction.
Our earlier studies (14) showed the importance of E-box DNA in protein–protein binding affinity. Looked at from the perspective of the Max protein, DNA binding reduced the binding affinity of the second monomer regardless of whether the monomer was Myc, Mad, or Max. To further investigate whether DNA itself was responsible for the weaker binding of the second monomer or if charge neutralization was sufficient, the effects of polyions were investigated. Schepartz et al. (20) have shown that proteins that use the monomeric pathway for DNA binding and dimerization are able to form dimers at a greater rate in the presence of negatively charged polymers. These polymers essentially substitute for DNA (20). Negatively charged polymers can neutralize the charge on the basic contact regions of the proteins and bring the dimers together more easily. Rentzeperis et al. (24) have shown that in the case of the Arc repressor, refolding of the protein into a functional dimer is accelerated in the presence of polyanions.
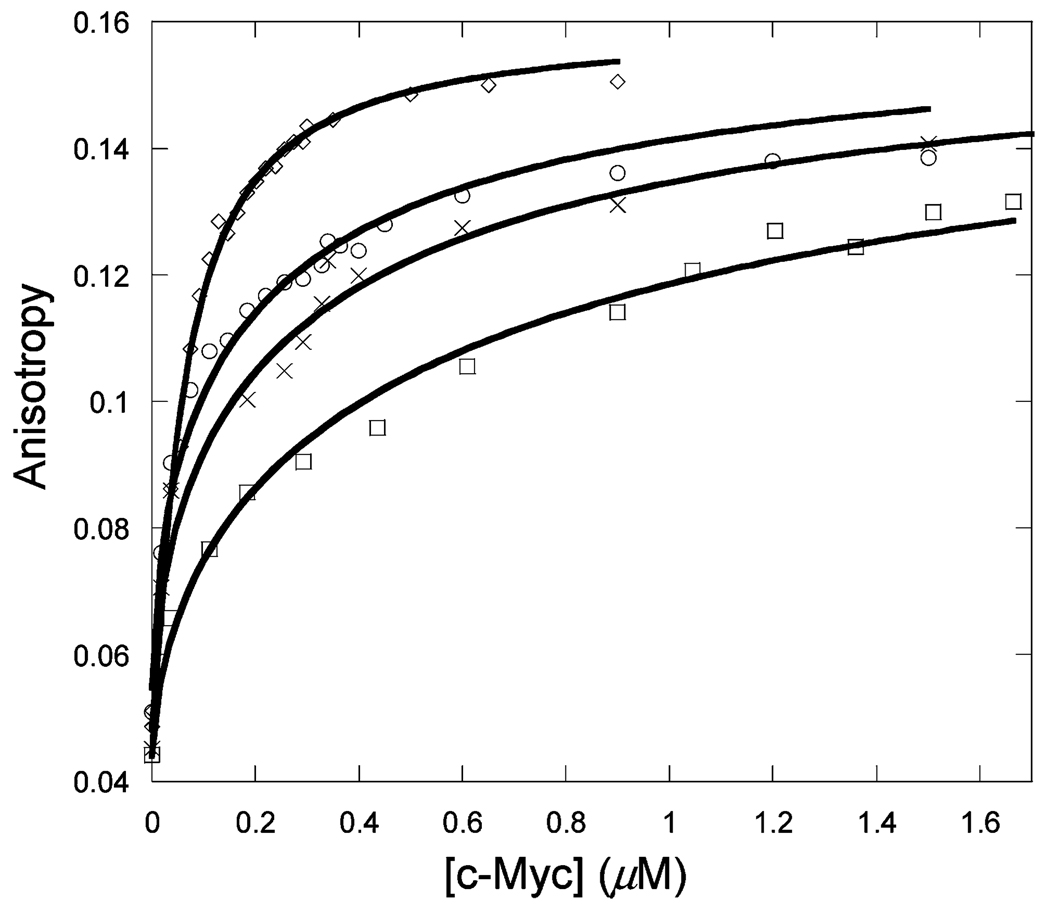
The effects of both positive and negative polyions on the equilibrium binding of Max, Myc, and Mad to TRITC-labeled Max were investigated. Positively charged poly-l-lysine showed little effect on the Kd of c-Max, Mad–Max, and Max2 dimers (data not shown). However, poly-l-glutamic acid, PLG, significantly reduced the Kd values for all three protein dimers. There were marked differences in the thermodynamic parameters for all three protein complexes. Figures 5–7 show the effects of PLG on Max homodimer and Myc–Max and Mad–Max heterodimer formation, respectively. Table 3 lists the Kd values calculated from the plots and the thermodynamic parameters calculated from the temperature dependence (Figure 8). One important observation is that although the protein–protein interactions in the presence of PLG become tighter for all three dimers, the difference in binding affinity among the Myc–Max, Mad–Max, and Max–Max dimers remains in the same order as in the absence of PLG (the Myc–Max Kd is 2.4 times the Max–Max Kd and 1.28 times the Max–Mad Kd). Table 3 shows that the enthalpy and entropy for each dimer are more negative in the presence of PLG. PLG enhances the stability of all three dimers by 2–3 kJ/mol (Table 4). A similar enhancement of binding was found when calf thymus DNA was used (data not shown). However, low-temperature data and hence thermodynamic parameters were not reliably attained, probably because of aggregation of the DNA at lower temperatures.
FIGURE 5.
Titration of 50 nM Max-TRITC with unlabeled Max in the presence of 150 µM poly-l-glutamic acid at 13 (■), 23 (□), 28 (Δ), and 33 °C (×) in pH 7.6 titration buffer.
FIGURE 7.
Titration of 50 nM Max-TRITC with unlabeled Mad in the presence of 150 µM poly-l-glutamic acid at 13 (○), 23 (□), 28 (◊), and 33 °C (×) in pH 7.6 titration buffer.
Table 3.
Thermodynamic Parameters for Association of Max, c-Myc, and Mad with Max-TRITC in the Presence of 150 µM Poly-l-glutamic Acid
| Kd (µM) | ||||||
|---|---|---|---|---|---|---|
| 13 °C | 23 °C | 28 °C | 33 °C | ΔH° (kJ/mol) | ΔS° (J/mol) | |
| Max | 0.085 ± 0.01 | 0.241 ± 0.05 | 0.312 ± 0.06 | 0.455 ± 0.15 | −64.9 ± 4.8 | −91.8 ± 16.4 |
| c-Myc | 0.024 ± 0.09 | 0.081 ± 0.05 | 0.141 ± 0.02 | 0.191 ± 0.05 | −83.0 ± 1.4 | −144.7 ± 5.0 |
| Mad | 0.078 ± 0.01 | 0.131 ± 0.02 | 0.206 ± 0.02 | 0.245 ± 0.01 | −46.1 ± 4.3 | −24.8 ± 14.5 |
FIGURE 8.
van’t Hoff plot for titration of 50 nM Max-TRITC in the presence of 150 µM poly-l-glutamic acid with c-Myc (□), Max (○), and Mad (◊) in pH 7.6 titration buffer.
Table 4.
ΔG° Values Calculated from Table 3
| ΔG° (kJ/mol) | |||
|---|---|---|---|
| 10 °C | 20 °C | 37 °C | |
| Max | −38.90 | −37.98 | −36.42 |
| c-Myc | −42.10 | −40.65 | −38.19 |
| Mad | −39.05 | −38.81 | −38.38 |
CONCLUSIONS
The low stability of the Max homodimer permits reassortment with monomers of its interacting partners, and this destabilization of the homodimer can serve as a prominent driving force for efficient heterodimerization (17). Negative enthalpy and entropy are characteristic of the involvement of van der Waals forces and hydrogen bonding in protein–protein interactions (25). Crystal structures and NMR studies have revealed the importance of hydrogen bonding between charged and uncharged polar residues in these protein dimers. Ross et al. also hypothesized (25) that hydrogen bond formation in a low-dielectric environment is an important source of negative enthalpy and entropy. Mobile polyanions such as PLG can create a low-dielectric environment by shielding positive charges on the protein–protein interacting interfaces which is a possible reason for the more negative ΔH° and ΔS° for Myc–Max, Mad–Max, and Max–Max association in the presence of PLG. As monomers, the individual protein subunits exist in a fullly or partially unfolded form. Such unstructured or partially structured monomers can nonspecifically collide on the surface of the polyanions more rapidly through electrostatic interactions. These collisions appear to be more productive for proper folding and stronger association of the monomers. Dimerization on the DNA surface occurs through a similar mechanism. From our results, it can be concluded that the presence of negatively charged DNA and RNA in the cell nucleus at a high concentration or changes in the environment such as histone deactylation, protein phosphorylation, or other events can nonspecifically help in dimerization of Myc–Max, Mad–Max, and Max2 dimers. Further, the significant differences in entropy for formation of Myc–Max and Mad–Max heterodimers suggest that small molecules that restrict the conformational flexibility could selectively affect heterodimer formation and possibly biological function.
FIGURE 6.
Titration of 50 nM Max-TRITC with unlabeled cMyc in the presence of 150 µM poly-l-glutamic acid at 13 (○), 23 (□), 28 (◊), and 33 °C (×) in pH 7.6 titration buffer.
Footnotes
This work was supported in part by National Science Foundation Grant MCB 0413982 (to D.J.G.). Research Center in Minority Institutions Award RR-03037 from the National Center for Research Resources of the National Institutes of Health supports infrastructure at Hunter College.
Abbreviations: b/HLH/z, basic helix–loop–helix/leucine zipper; LZ, leucine zipper; DNA E-box, 6 bp double-stranded DNA sequence CACGTG known as the enhancer box; TRITC, tetramethylrhodamine isothiocyanate; HEPES, 4-(2-hydroxyethyl)piperazineethanesulfonic acid; EDTA, ethylenediaminetetraacetic acid; DTT, dithiothreitol; PBS, phosphate-buffered saline; PLG, poly-l-glutamic acid; DMSO, dimethyl sulfoxide.
REFERENCES
- 1.Kohler JJ, Metallo SJ, Schneider TL, Schepartz A. DNA specificity enhanced by sequential binding of protein monomers. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11735–11739. doi: 10.1073/pnas.96.21.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/max/Mad network and the transcriptional control of cell behavior. Annu. ReV. Cell DeV. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 3.Kato GJ, Barret J, Villa-Garcia M, Dang CV. An amino terminal c-Myc domain required for neoplastic transformation activates transcription. Mol. Cell. Biol. 1990;10:5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murre C, McCam PS, Baltimore D. A New DNA Binding and Dimerization Motif in Immunoglobulin Enhancer Binding, Daughterless, MyoD and Myc Proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 5.Blackwood EM, Eisenman RN. Max: A helix loop helix zipper protein that forms sequence specific DNA binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 6.Amati B, Littlewood TD, Evan GI, Land H. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J. 1993;12:5083–5087. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMahon SB, Wood MA, Cole MD. The Essential Cofactor TRRAP Recruits the Histone Acetyltransferase HGCN5 to cMyc. Mol. Cell. Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayer DE, Eisenman RN. A switch from Myc: Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes DeV. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- 9.Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 10.Hurlin PJ, Queva C, Koskinen PJ, Steingrimsson E, Ayer DE, et al. Mad3 and Mad4, novel Max interacting transcription repressor, that suppress c-Myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McArthur GA, Laherty CD, Queva C, Hurlin PJ, Loo L, et al. The Mad protein family links transcriptional repression to cell differentiation. Cold Spring Harbor Symp. Quant. Biol. 1998;63:423–433. doi: 10.1101/sqb.1998.63.423. [DOI] [PubMed] [Google Scholar]
- 12.Larsson L-G, Pettersson M, Oberg F, Nilsson K, Luscher B. Expression of mad, mxi1, max and c-myc during induced differentiation of haemopoetic cells: Opposite regulation of mad and c-myc. Oncogene. 1994;9:1247–1252. [PubMed] [Google Scholar]
- 13.Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes DeV. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu J, Banerjee A, Goss DJ. Assembly of b/HLH/z Proteins c-Myc, Max, and Mad1 with Cognate DNA: Importance of Protein-Protein and Protein-DNA Interaction. Biochemistry. 2005;44:11855–11863. doi: 10.1021/bi050206i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferre-D’Amare AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 16.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 17.Lavigne P, Crump M, Hodges RS, Kay CM, Sykes BD. Insights into the mechanism of heterodimerization from 1H NMR solution structure of the c-Myc-Max heterodimeric leucine zipper. J. Mol. Biol. 1998;281:165–181. doi: 10.1006/jmbi.1998.1914. [DOI] [PubMed] [Google Scholar]
- 18.Berg T, Boger DL, Vogt P. Small-molecule antagonist of Myc/Max dimerization inhibit Myc induced transformation of chicken fibroblast. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boger DL, Lee JK, Goldberg J, Jin Q. Two Comparisons of the Performance of Positional Scanning and Deletion Synthesis for the Identification of Active Constituents in Mixture Combinatorial Libraries. J. Org. Chem. 2000;65:1467–1474. doi: 10.1021/jo9916481. [DOI] [PubMed] [Google Scholar]
- 20.Kohler JJ, Schepartz A. Effects of Nucleic Acids and Polyanions on Dimer Formation and DNA Binding by bZip and bHLHZip Transcription Factors. Bioorg. Med. Chem. 2001;9:2435–5. doi: 10.1016/s0968-0896(01)00221-8. [DOI] [PubMed] [Google Scholar]
- 21.Sha M, Wang Y, Xiang T, van Heerden A, Browning K, Goss DJ. Interaction of wheat germ protein synthesis initiation factor eIF-(iso)4F and its subunits p28 and p86 with m7GTP and mRNA analogs. J. Biol. Chem. 1995;270:29904–29909. doi: 10.1074/jbc.270.50.29904. [DOI] [PubMed] [Google Scholar]
- 22.Kohler J, Schepartz A. Kinetic Studies of Fos-Jun- DNA Complex Formation: DNA Binding Prior to Dimerization. Biochemistry. 2001;40:130–142. doi: 10.1021/bi001881p. [DOI] [PubMed] [Google Scholar]
- 23.Fieber W, Schneider ML, Matt T, Krautler B, Konrat R, Bister K. Structure, Function, and Dynamics of the Dimerization and DNA binding Domain of Oncogenic Transcription Factor v-Myc. J. Mol. Biol. 2001:1308–1310. doi: 10.1006/jmbi.2001.4537. [DOI] [PubMed] [Google Scholar]
- 24.Rentzeperis D, Jonsson T, Sauer RT. Acceleration of the refolding of Arc repressor by nucleic acids and other polyanions. Nat. Struct. Biol. 1999;6:560–573. doi: 10.1038/9353. [DOI] [PubMed] [Google Scholar]
- 25.Subramanin S, Ross PD. Thermodynamics of Protein Association Reactions: Forces Contributing to Stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]