Abstract
The growing number of high-resolution crystal structures of large RNA molecules provides much information for understanding the principles of structural organization of these complex molecules. Several in-depth analyses of nucleobase-centered RNA structural motifs and backbone conformations have been published based on this information, including a systematic classification of base pairs by Leontis and Westhof. However, hydrogen bonds involving sugar–phosphate backbone atoms of RNA have not been analyzed systematically until recently, although such hydrogen bonds appear to be common both in local and tertiary interactions. Here we review some backbone structural motifs discussed in the literature and analyze a set of eight high-resolution multi-domain RNA structures. The analyzed RNAs are highly structured: among 5372 nucleotides in this set, 89% are involved in at least one “long-range” RNA–RNA hydrogen bond, i.e., hydrogen bonds between atoms in the same residue or sequential residues are ignored. These long-range hydrogen bonds frequently use backbone atoms as hydrogen bond acceptors, i.e., OP1, OP2, O2′, O3′, O4′, or O5′, or as a donor (2′OH). A surprisingly large number of such hydrogen bonds are found, considering that neither single-stranded nor double-stranded regions will contain such hydrogen bonds unless additional interactions with other residues exist. Among 8327 long-range hydrogen bonds found in this set of structures, 2811, or about one-third, are hydrogen bonds entailing RNA backbone atoms; they involve 39% of all nucleotides in the structures. The majority of them (2111) are hydrogen bonds entailing ribose hydroxyl groups, which can be used either as a donor or an acceptor; they constitute 25% of all hydrogen bonds and involve 31% of all nucleotides. The phosphate oxygens OP1 or OP2 are used as hydrogen bond acceptors in 12% of all nucleotides, and the ribose ring oxygen O4′ and phosphodiester oxygens O3′ and O5′ are used in 4%, 4%, and 1% of all nucleotides, respectively. Distributions of geometric parameters and some examples of such hydrogen bonds are presented in this report. A novel motif involving backbone hydrogen bonds, the ribose–phosphate zipper, is also identified.
Introduction
Advances in molecular biology and crystallographic methods have led to a dramatic increase in the number of high-resolution crystal structures of large RNA molecules solved in the past decade.1,2 These structures provide a wealth of information for understanding principles of organization of complex RNA structures and have motivated detailed analysis of nucleobase-centered RNA structural motifs and backbone conformations; for example, see ref. 3–12. This information can be used for knowledge-based prediction of three-dimensional (3D) RNA structures.13–15 In many instances, such methods can currently predict native structures of relatively small one-domain RNA within an atomic root mean square deviation (RMSD) of 2 Å,14 although the RMSD increases to 8–16 Å for larger multi-domain RNA.15 It is likely that the sugar–phosphate backbone plays important roles in inter-domain tertiary interactions of RNA, and systematic classification of such motifs may improve predictive methods. Indeed, many backbone motifs, especially those involving the ribose hydroxyl groups, have been identified.16–25 Yet, until recently, there were no attempts to systematically analyze hydrogen bonds (H-bonds) involving the backbone atoms; a recent paper reports compiled distributions of phosphate oxygens around individual RNA bases.26 Here, we review some of the previously identified structural motifs emphasizing the usage of the backbone H-bonds (BH-bonds) and also analyze a set of high-resolution multi-domain RNA structures. Consideration of BH-bonds greatly increases the number of structural motifs in RNA. Indeed, the analyzed set of RNA structures has 2444 base pairs stabilized by at least one H-bond when allowing donor and acceptor atoms only on nucleobases. The number of pairs of residues (as opposed to base pairs) is approximately doubled to 4294 when BH-bonds are included (Table 1). We do not attempt here to present a full systematic classification of structural motifs with BH-bonds. Instead, we present some typical examples, including some discussed previously in the literature. The purpose of this short report is to increase awareness of the roles of the sugar–phosphate backbone atoms in hydrogen bonding interactions, which contribute to structure, in RNA and to stimulate efforts to create a systematic classification of such interactions.
Table 1.
Selection of multi-domain RNA structures used for H-bond analysis
| PDB | Resolution/Å | Description | nt | Pairs | H-bonds | BH-bonds | Reference |
|---|---|---|---|---|---|---|---|
| 3DIL | 1.90 | Lysine riboswitch | 173 | 118 | 257 | 46 | 39 |
| 1J1U | 1.95 | tRNATyr | 74 | 51 | 101 | 20 | 40 |
| 2R8S | 1.95 | Group I intron P4–P6 domain | 159 | 102 | 205 | 57 | 41 |
| 3D2V | 2.00 | TPP riboswitch | 154 | 122 | 235 | 84 | 42 |
| 1M5O | 2.20 | Hairpin ribozyme complex with U1A protein | 224 | 114 | 241 | 23 | 43 |
| 1VQ8 | 2.20 | Haloarcula marismortui large ribosomal subunit | 2874 | 2426 | 4661 | 1724 | 44 |
| 2VQE | 2.50 | Escherichia coli small ribosomal subunit | 1522 | 1252 | 2396 | 814 | 33 |
| 2V3C | 2.50 | 7S RNA of SRP | 192 | 109 | 231 | 43 | 45 |
| Total | 5372 | 4294 | 8327 | 2811 |
Results and discussion
Table 1 lists the high-resolution multi-domain RNA crystal structures selected for the analysis as described in the Methods section. The structures include rRNA from small and large ribosomal subunits, 7S RNA from the signal recognition particle, hairpin ribozyme, P4–P6 domain from the group I intron, lysine and thiamine pyrophosphate riboswitches, and tRNA.
Hydrogen bond statistics
The analyzed RNAs are highly structured, which is perhaps not surprising since they were able to crystallize. In 5372 nucleotides (nt) analyzed, 8327 H-bonds have been found. Residues connected by at least one H-bond form 4294 hydrogen-bonded pairs, which involve 4796 nt and leave less than 11% of the residues unpaired. A surprisingly large number of these H-bonds were BH-bonds, which are defined as having an acceptor OP1 (pro-Sp), OP2 (pro-Rp), O3′, O5′, or O2′ or a donor 2′OH. It has been shown that base protons H2, H5, H6, and H8 can also form stabilizing interactions with phosphate oxygens OP1 and OP2,26 but these interactions were not included in the present analysis. BH-bonds constitute 34% of the total number of RNA–RNA H-bonds and involve 39% of all RNA residues. These statistics are, of course, strongly biased towards large multi-domain rRNA molecules such as the small and large ribosomal subunits (Table 1), but even the average percent among the eight structures analyzed (25% of all H-bonds and 31% of all residues) is greater than we intuitively expected prior to this analysis. The lowest number of BH-bonds, 10% of all H-bonds, is observed in hairpin ribozyme, which has few inter-domain contacts. The absolute majority, 2111 BH-bonds, use the ribose 2′OH hydroxyl group, either as donor (1766 times), acceptor (704 times), or both (359 times; see Table 2); 31% of all ribose hydroxyl groups participate in hydrogen bonding. Nevertheless, other backbone acceptor atoms are also used quite frequently. The frequencies of the phosphate oxygens OP1 and OP2, ribose ring oxygen O4′, and phosphodiester oxygen O3′ normalized to the total number of residues are comparable to those of guanine N3 and N7 normalized to the total number of guanines (Table 2). Together, BH-bonds with OP1 and OP2 acceptors account for 10% of all H-bonds and involve 12% of all nucleotides. The phosphodiester oxygen O5′ is the least used acceptor in either any BH-bonds or in 2′OH–O5′ BH-bonds (Table 2); the reasons for this are currently not entirely clear. Purine amino groups are the most common donors in BH-bonds, although the cytosine amino group and guanine and uracil imino protons are also observed (Table 2).
Table 2.
Total numbers and frequencies of acceptor and donor atoms in long-range RNA–RNA H-bondsa
| All H-bonds |
BH-bondsb |
|||
|---|---|---|---|---|
| Atom | Number | Frequency (%) | Number | Frequency (%) |
| N7A | 407 | 32.0 | 85 | 6.7 |
| N7G | 93 | 5.3 | 69 | 3.9 |
| N6A | 1088 | 85.6 | 301 | 23.7 |
| O6G | 1333 | 75.9 | 65 | 3.7 |
| N4C | 1247 | 88.3 | 103 | 7.3 |
| O4U | 430 | 46.1 | 33 | 3.5 |
| N1A | 675 | 53.1 | 195 | 15.3 |
| N1G | 1509 | 85.9 | 180 | 10.3 |
| N3C | 1207 | 85.5 | 26 | 1.8 |
| N3U | 699 | 75.0 | 61 | 6.5 |
| N2G | 2018 | 114.9c | 400 | 22.8 |
| O2C | 1313 | 93.0 | 98 | 6.9 |
| O2U | 390 | 41.8 | 34 | 3.6 |
| N3A | 280 | 22.0 | 144 | 11.3 |
| N3G | 171 | 9.7 | 34 | 1.9 |
| O2′ | 704 | 13.1 | 359 | 6.7 |
| O2′ | 1766 | 32.9 | 983d | 18.3d |
| O4′ | 234 | 4.4 | 122 | 2.3 |
| O3′ | 205 | 3.8 | 148 | 2.8 |
| O5′ | 52 | 1.0 | 14 | 0.3 |
| OP1 | 369 | 6.9 | 216 | 4.0 |
| OP2 | 464 | 8.6 | 124 | 2.3 |
Non-intraresidue, non-sequential RNA–RNA H-bonds have been identified as described in the Methods section for the eight multi-domain RNA structures listed in Table 1. Table rows with the acceptor atoms are aligned left, and rows with donor groups are aligned right. The frequencies are calculated as percent of the total number of relevant residues, i.e., 1271 A, 1412 C, 1756 G, and 932 U residues for specific nucleobase acceptors and donors and 5372 nt for the acceptors and donors in the sugar-phosphate backbone.
For the acceptor atoms, total numbers and frequencies of the H-bonds with the sugar–phosphate donors (i.e., 2′OH) are shown. For the donor groups, the data are shown for the H-bonds with acceptors on the sugar-phosphate backbone (i.e., O2′, O4′, O3′, O5′, OP1, and OP2).
Frequency is greater than 100% here, because each guanine amino group participates in more than one H-bond on average.
O2′–O2′ H-bonds.
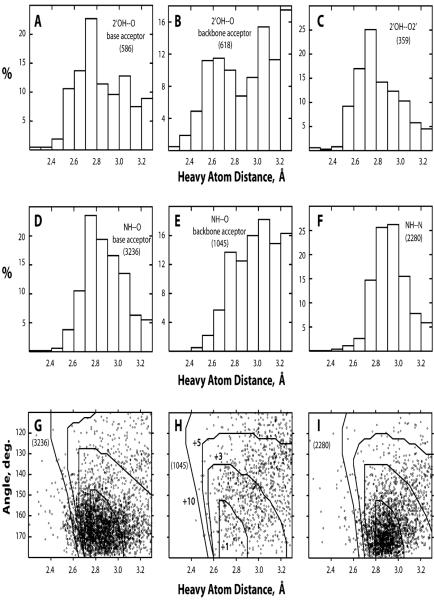
The distributions of geometries of observed H-bonds are summarized in Fig. 1. A noticeable number of observed H-bonds appear not to have optimal geometries due to short heavy atom distances but also due to small H-bond angle; some of them should probably be classified as steric clashes rather than H-bonds. The reasons for the nonoptimal geometries are not clear; a possible explanation has to do with refinement deficiencies due to the moderate resolution of these structures (Table 1). In any case, BH-bonds (Fig. 1H) do not appear to have this problem more severely than nucleobase–nucleobase H-bonds (Fig. 1G and I). A specific peculiarity of BH-bonds is a more frequently observed long heavy atom distance. This feature is pronounced in both OH–O and NH–O BH-bonds (Fig. 1B and E), but, interestingly, not in the 2′OH–O2′ BH-bonds specifically (Fig. 1C).
Fig. 1.
Histograms of H-bond heavy atom distances (A–F) and scatter plots of H-bond angle versus heavy atom distance (G–I). The histograms are shown for BH-bonds 2′OH–O with nucleobase acceptors (A) and backbone acceptors excluding O2′ (B), BH-bonds 2′OH–O2′ (C), H-bonds NH–O with nucleobase acceptors (D), BH-bonds NH–O with backbone acceptors (E), and H-bonds NH–N (F). The scatter plots are shown for H-bonds NH–O with nucleobase acceptors (G), BH-bonds NH–O with backbone acceptors (H), and H-bonds NH–N (I). Superimposed on the scatter plots are the lines of equal energy calculated for isolated H-bonds. The energy increments relative to the H-bond with the optimal geometry are +1, +3, +5, and +10 kcal mol−1 for the four lines, respectively; the energy labels are shown only in H for clarity. In each case, the total number of relevant H-bonds is shown in parentheses.
Below, we will show some examples of BH-bonds, including reviewing some motifs identified previously. In discussing various types of base pairing, we will follow the classification of Leontis and Westhof,3 according to which base pairs are divided into 12 groups based on their interacting edges (Watson–Crick, Hoogsteen, or sugar) and orientation of glycosidic bonds (cis or trans).
Ribose atoms in hydrogen bonds
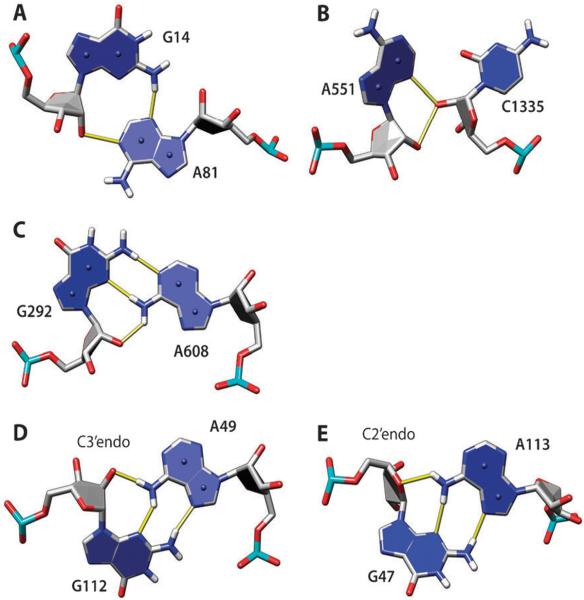
Ribose hydroxyl groups are frequently used in BH-bonds, either as donor or acceptor (Table 2). Indeed, several structural motifs with 2′OH group have been identified in the past. A-minor refers to a series of structural motifs where the ribose and minor groove edge of an adenosine interact with the sugar edge of a Watson–Crick pair, most commonly GC, within an RNA stem.18,27 In effect, the adenine forms a minor-groove triple in this motif. In the analyzed set of structures, there are 226 minor-groove triples (not necessarily all within the context of an RNA stem); in 184 of them, there are BH-bonds present. The majority of these triples, 108, have adenine as the third residue, and 89 of them are GC–A triples. In the A-minor I motif, the adenine interacts with both residues of the Watson–Crick GC pair of the stem making H-bonds AN1–GO2′ and AN3–GN2 and bifurcated H-bonds AO2′–CO2 and AO2′–CO2′. Similar interactions, but without the AN3–GN2 H-bond, are observed when an adenine forms a minor-groove triple with a Watson–Crick AU pair within an RNA stem,27 but they are less common. The AG interaction within the A-minor I motif can be also classified as Leontis–Westhof group 12 AG pair (trans-sugar edge/sugar edge). Such pairs are also found outside of the context of RNA stems and even outside of base triples. In the data set analyzed here, there are 69 occurrences of such pairs (an example is shown in Fig. 2A); they account for about one-third of all AN1–O2′ H-bonds (Table 2).
Fig. 2.
Examples of adenine H-bonds with ribose atoms: N1–O2′ (A), N3–O2′ (B), N6–O2′ (C and D), and N6–O4′ (E). The hydroxyl oxygen O2′ serves as an H-bond donor (A, B) or acceptor (C, D). In (A), adenine and guanine make a group 12 AG pair (trans-sugar edge/sugar edge), which is part of the A-minor I motif with G14 Watson–Crick-base paired to C78 and hydroxyl group of C78 making additional H-bonds with the A81 ribose (not shown); G14–C78 is part of a GC-rich helix in the lysine riboswitch (PDB 3DIL). In (B), the unclassified AC pair is part of the A-minor II motif, with A551 interacting with the minor groove of 8-base pair helix (not shown) in 23S rRNA (PDB 1VQ8). In (C), A and G form a group 6 AG pair (trans-Watson–Crick/sugar edge) in 16S rRNA (PDB 2VQE). In (D and E), A and G form group 10 AG pair (trans-Hoogsteen/sugar edge); both are from 23S rRNA (PDB 1VQ8). In (D), the AN1–O2′ H-bond is formed, and in (E), the AN1–O4′ H-bond is formed instead, which is correlated with C2′–endo sugar pucker of G47. Here, and in other molecular graphics, the numbering scheme from the PDB files is used.
In the A-minor II motif, the adenine interacts only with the ribose of the cytosine of the GC base pair, with the adenine N3 and O2′ atoms making bifurcated H-bonds with the 2′OH of the cytosine.18 Such adenine–ribose interactions can be also frequently found outside of the context of RNA stems or triples; they are not cytosine-specific, although cytosines are the most common. In the set of structures analyzed here, 106 such pairs are present (Fig. 2B) (16 A, 47 C, 18 G, and 25 U). These interactions account for the majority of the 144 AN3–O2′ observed H-bonds; an example is shown in Fig. 2B.
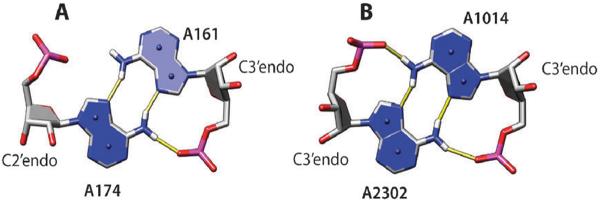
The adenine amino group is one of the most common donors in BH-bonds, involving more than 20% adenines (Table 2); in the majority of cases (59%), ribose hydroxyl groups serve as acceptors of such H-bonds. Fig. 2C shows one example of such an interaction; the AG is a group 6 pair (trans-Watson–Crick/sugar edge) in this case. Such pairs are relatively infrequent; out of 13 group 6 AG pairs found, eight have the AN6–O2′ H-bond. Fig. 2D shows a much more frequent group 10 AG pair (trans-Hoogsteen/sugar edge). Out of 95 such pairs in the data set analyzed, 67 pairs have the AN6–O2′ H-bond. It appears that this H-bond requires a C3′–endo sugar conformation in the G residue. If the sugar is C2′–endo, the AN6–O4′ H-bond is formed instead (Fig. 2E; observed in 13 out of 95 group 10 AG pairs).
The G-ribo motif defines a side-by-side arrangement of two RNA stems stabilized by the sugar edge interaction of a guanine in a GC pair of one stem with ribose atoms of another stem,24 such motifs are present in pseudoknot structures in rRNA.28 The G-ribo interaction is characterized by two BH-bonds, GN2–O4′ and GO2′–O2′ (Fig. 3A), and it can also be found outside of the context of RNA stem packing. Somewhat different variants of guanine–ribose interactions are also possible, one with the reverse order of hydrogen bonding, GN2–O2′ and GO2′–O4′ (Fig. 3B), and another with three BH-bonds: GN2–O3′, GN3–O2′, and GO2′–O2′ (Fig. 3C). These motifs are relatively infrequent; 11, 4 and 4 examples of the three variants, respectively, were found in the analyzed set of structures. Interestingly, they are present only in rRNA, 16S and 23S.
Fig. 3.
Examples of guanine–ribose interactions: the original G-ribo interaction with GN2–O4′ and GO2′–O2′ H-bonds (A); “reverse G-ribo” interaction with GN2–O2′ and GO2′–O4′ H-bonds (B); a variant G-ribo with three H-bonds, GN2–O3′, GN3–O2′, and GO2′–O2′ (C). All examples are from 23S rRNA, PDB 1VQ8.
Along-groove packing motifs (AGPM), or P-interactions, entail backbone interactions between two RNA helices packed against each other. Such motifs entail two base pairs interacting via their sugar edges, either two Watson–Crick GC pairs or a GC pair and a wobble GU pair.19,22 A number of specific arrangements have been identified for such pairs of base pairs.19,22,23,25 Two recently identified specific arrangements of a pair of GC pairs were termed ribo-base (see Fig. 8 in ref. 25). In ribo-base type 1, only guanines interact directly forming H-bonds GN2–GO2′ and GO2′–GN2. In ribo-base type 2, one GC pair is flipped 180° around the dyad axis and shifted such that G from the first pair interacts with both G and C in the second GC pair forming H-bonds GO2′–GN2 and GN2–CO2′. Two more arrangements have been described for a pair of GC pairs and two more for a combination of GC and GU pairs (see Fig. 2 in ref. 22). Various arrangements can be used in tandem (four base pairs altogether) to facilitate packing of helices; such motifs appear to be associated with a perpendicular arrangement of helices.25 To avoid confusion with terminology, we suggest reserving the terms AGPM or P-interactions to describe backbone-mediated packing of RNA helices and to use ribo-base to describe specific arrangements of two base pairs, especially because the latter can be potentially found outside of the helix packing context.
Fig. 8.
Ribose–phosphate zipper motifs with side-by-side O2′–OP1 (A), O2′–OP2 (B) and mixed O2′–OP1/OP2 interactions (C). The examples are from tRNATyr, PDB 1J1U (A), and 23S rRNA, PDB 1VQ8 (B and C).
A special role for wobble GU pairs in P-interactions has been highlighted by Mokdad et al.23 who showed that participation in helix packing leads to a stronger conservation of the GU pairs in rRNA sequences. In addition, two local structural motifs involving BH-bonds with the shallow minor groove side of wobble GU pairs have been identified in this work (see Fig. 1 in ref. 23). One is the O2′-in-pocket interaction and another is the phosphate-in-pocket interaction (see also the next section on phosphate groups in H-bonds). Although relatively infrequent, these local motifs can be observed both within and outside of the helix packing context. We located 32 similar motifs (all of them in rRNA) among 147 wobble GU pairs present in the set of structures from Table 1: 16 are O2′-in-pocket motifs, 8 are phosphate-in-pocket motifs, and 8 more have both ribose and phosphate atoms forming BH-bonds with base atoms on the minor groove side of the GU pair (not shown). This count is somewhat different from that published previously,23 because of somewhat different criteria used: only base donor and acceptor atoms of GU pairs were considered, to exclude purely ribose–ribose interactions. Formation of BH-bonds with the major (non-glycosidic) groove side of the GU pairs is extremely rare: only two such cases are observed with the O2′ forming H-bonds with O4, O6 or N7 atoms in the analyzed structures, one in the 23S rRNA and another in the group I intron structure (not shown). It has been observed that sometimes protein side chains and Mg2+ ions form H-bonds with this side of GU pairs.23
Ribose zipper is a frequent structural motif defined as hydrogen bonding between ribose 2′-hydroxyl groups of at least two consecutive residues to the 2′-hydroxyl groups of at least two other residues antiparallel to the first two.16,20,29 There are 61 ribose zipper motifs in the set of structures analyzed here, mostly in rRNA of large and small ribosomal subunits, and also in the P4–P6 domain of group I intron and in the hairpin ribozyme. These interactions account for about one-third of all O2′–O2′ H-bonds in these structures.
Phosphate groups in hydrogen bonds
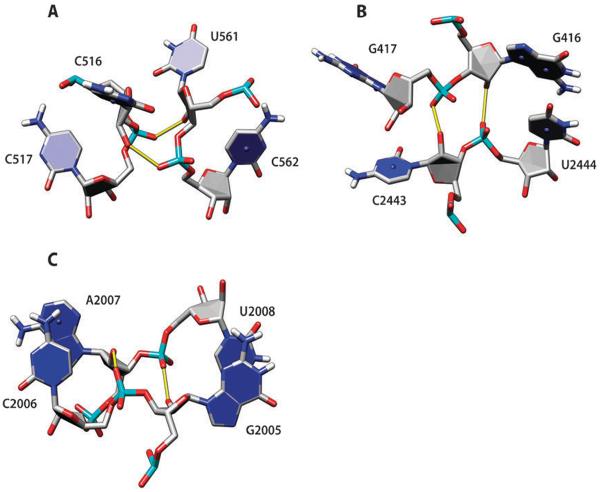
Although not as frequently as 2′OH hydroxyl groups, oxygens on phosphoryl groups often participate in hydrogen bonding (Table 2). In this section, we present some examples of such BH-bonds. Group 8 symmetric AA pairs (trans-Hoogsteen/Hoogsteen), in addition to N7-amino interactions, can be additionally stabilized by amino-to-OP1/OP2 H-bonds (Fig. 4); see also ref. 3. There are twenty group 8 AA pairs in the analyzed structures; one AN6–AOP1/OP2 H-bond (Fig. 4A) is observed in 13 of them, and two such H-bonds (Fig. 4B) are observed in three cases. It appears that a C3′–endo sugar pucker is required for such hydrogen bonding. Most often, OP2 oxygen is used as acceptor (in 17 out of 19 H-bonds).
Fig. 4.
Examples of group 8 AA base pairs (trans-Hoogsteen/Hoogsteen) from 23S rRNA in the large ribosomal subunit, PDB 1VQ8. In addition to the Hoogsteen edge/sugar edge H-bonds, these pairs are stabilized by one (A) or two (B) H-bonds between the adenine amino proton and phosphate OP2 oxygen.
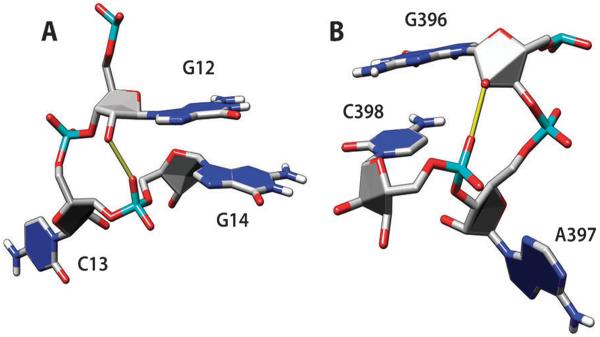
Group 10 AG base pairs (trans-Hoogsteen/sugar edge) were discussed above relative to the amino-to-hydroxyl hydrogen bonding. In many instances (in 43 cases out of 67 AG pairs; shown in Fig. 2D), the phosphate oxygen OP2 of the residue immediately upstream of A is making additional bifurcated H-bonds with imino and amino protons of G (Fig. 5). This XA–G motif can be a part of a more complex structure called the sarcin–ricin loop motif,17,21,30 but the XA–G block is much more frequent than the complete sarcin–ricin loop.
Fig. 5.
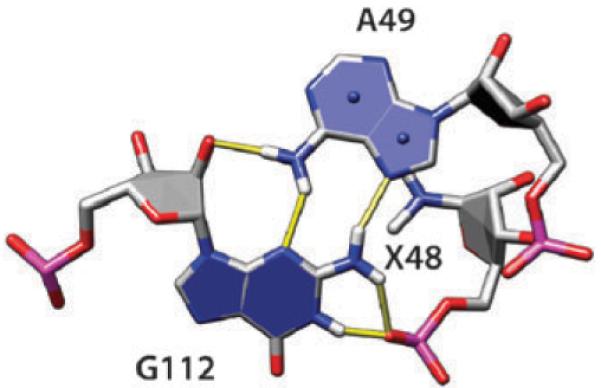
Group 10 AG base pair (the same as shown in Fig. 2D). The phosphate oxygen OP2 of the residue immediately upstream of A is making bifurcated H-bonds with imino and amino protons of G. The nucleobase atoms of the upstream residue, labeled X48, are omitted for clarity.
Major-groove triples are less common than minor-groove triples in the analyzed RNA structures; 96 such triples were found with the third residue interacting with the purine Hoogsteen edge of a Watson–Crick pair. In 39 of them, there are additional BH-bonds stabilizing the triples, including 13 instances with phosphate oxygens, usually OP2, serving as acceptors (Fig. 6). Modeling has shown that such triples with additional O2′–OP2 H-bonds can form extended triple helices,31 although such H-bonds have not been found experimentally in RNA triple helices.
Fig. 6.
Examples of major-groove triples AU–U (A) and GC–U (B) with additional O2′–OP2 H-bonds stabilizing the triples. Both examples are from 23S rRNA, PDB 1VQ8.
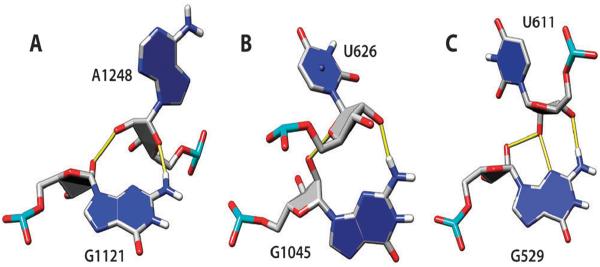
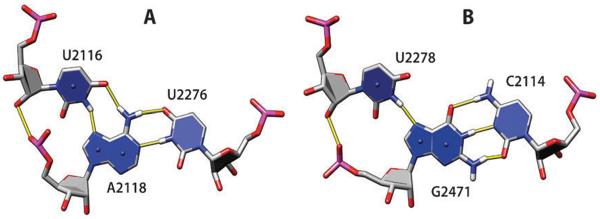
Ribose hydroxyl–phosphate oxygen interactions can frequently stabilize bulged-out conformations. We identified 34 stacked n and n + 2 bases with O2′–OP1/OP2 H-bonds; OP1 and OP2 acceptors are used with similar frequency (Fig. 7). Finally, by analogy to the ribose zipper, we identified a ribose–phosphate zipper. In this motif, consecutive residues from two strands form O2′–OP1/OP2 H-bonds. Fifteen such motifs are present in the analyzed set of structures, seven O2′–OP1 zippers (Fig. 8A), three O2′–OP2 zippers (Fig. 8B), and five mixed O2′–OP1/OP2 zippers (Fig. 8C). The orientation of strands in ribose–phosphate zippers can be either parallel (Fig. 8A) or antiparallel (Fig. 8B and C). Flexibility in the choice of OP1 or OP2 acceptor atom increases the number of feasible variants when packing together two RNA segments.
Fig. 7.
Stabilization of bulged-out bases with O2′–OP2 (A) or O2′–OP1 H-bond. The examples are from the lysine riboswitch, PDB 3DIL (A), and 16S rRNA, PDB 2VQE (B).
Conclusions
Hydrogen bonding entailing sugar–phosphate backbone donor and acceptor atoms is very common in multi-domain RNA structures. Such interactions create a multitude of structural motifs, some of which are reviewed here. Systematic classification of such motifs, which is still lacking, will help elucidate principles of organization of complex RNA conformations and improve methods predicting RNA structures.
Methods
A set of high-resolution multi-domain RNA crystal structures was selected from the Protein Data Bank (PDB)32 for the analysis. A resolution cutoff of 2.5 Å was chosen such as to include the highest-resolution structure of the small ribosomal subunit.33 To avoid redundancy only one structure of each type of RNA was selected (Table 1).
The atomic coordinates of the RNA structures were downloaded from the PDB database32 and analyzed with tools written in the Python programming language (mf3d, Motif Finder in 3D structures). These tools represent a collection of functions searching for various structural features in 3D RNA structures, such as H-bonds, base pairs or triples, stems, apical and internal loops, etc. The flexibility of Python allows for quick development of additional functions tailored to specific research questions.
Imino and amino protons were added to RNA residues from purely geometric considerations, i.e., using the N–H distance of 1.01 Å, the H–N–H angle of 120° in amino groups, and assuming a planar geometry for amino groups. H-bonds involving imino and amino donor protons were defined as having the distance between the heavy atoms below 3.3 Å and the H-bond angle above 110°. No protons were added to the ribose 2′-hydroxyl groups, because their placement must depend on the energy and cannot be based on purely geometric considerations. H-bonds with the 2′OH donor were accepted if the distance between the heavy atoms was below 3.3 Å. Such H-bond definitions are rather arbitrary, because calculations with empirical force fields show that the H-bond configurations remain weakly stabilizing even with the heavy-atom distance beyond 3.3 Å, when the H-bond angle is close to 180° (data not shown). H-bonds formed within a residue or between sequential residues were excluded from the analysis; also, no attempts were made to analyze H-bonds between the RNA and proteins or other ligands present in the structures. The backbone hydrogen bonding statistical analysis, therefore, counts only H-bonds between residue n and residue n + 2 or greater.
Hydrogen-bonded pairs of nucleobases were classified following Leontis and Westhof3 by specifying the interacting edge for each base as “Watson–Crick edge”, “Hoogsteen edge”, or “sugar edge”. In addition to the interacting edges, base pairs were classified as cis or trans according to the orientation of the glycosidic bonds relative to the direction of H-bonds. We have not included in this analysis base protons H2, H5, H6 and H8 that have weak positive partial charges, although their interaction with oxygen and nitrogen atoms with negative partial charges can be considered as weak H-bonds (see, e.g., ref. 26, 34 and 35) and are also likely to contribute to stabilization of RNA conformations. Pseudo-rotation parameters of sugars were calculated with the Fitparam program.36 UCSF Chimera37,38 was used to prepare molecular graphics.
Acknowledgements
This work was supported in parts by the National Institutes of Health grant AI46967 and Binational Science Foundation grant 2001065.
Footnotes
This paper is dedicated to Professor Wojciech J. Stec on his 70th birthday. This article is part of a themed issue on Biophosphates.
References
- 1.Holbrook SR. Curr. Opin. Struct. Biol. 2005;15:302–308. doi: 10.1016/j.sbi.2005.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holbrook SR. Annu. Rev. Biophys. Biomol. Struct. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- 3.Leontis NB, Westhof E. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray LJ, Arendall WB, 3rd, Richardson DC, Richardson JS. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13904–13909. doi: 10.1073/pnas.1835769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sims GE, Kim SH. Nucleic Acids Res. 2003;31:5607–5616. doi: 10.1093/nar/gkg750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider B, Moravek Z, Berman HM. Nucleic Acids Res. 2004;32:1666–1677. doi: 10.1093/nar/gkh333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrix DK, Brenner SE, Holbrook SR. Q. Rev. Biophys. 2005;38:221–243. doi: 10.1017/S0033583506004215. [DOI] [PubMed] [Google Scholar]
- 8.Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, Keating KS, Pyle AM, Micallef D, Westbrook J, Berman HM. RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xin Y, Laing C, Leontis NB, Schlick T. RNA. 2008;14:2465–2477. doi: 10.1261/rna.1249208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olson WK, Esguerra M, Xin Y, Lu XJ. Methods. 2009;47:177–186. doi: 10.1016/j.ymeth.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stombaugh J, Zirbel CL, Westhof E, Leontis NB. Nucleic Acids Res. 2009;37:2294–2312. doi: 10.1093/nar/gkp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xin Y, Olson WK. Nucleic Acids Res. 2009;37:D83–D88. doi: 10.1093/nar/gkn676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das R, Baker D. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parisien M, Major F. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 15.Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB. RNA. 2009;15:189–199. doi: 10.1261/rna.1270809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 17.Correll CC, Munishkin A, Chan YL, Ren Z, Wool IG, Steitz TA. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gagnon MG, Steinberg SV. RNA. 2002;8:873–877. doi: 10.1017/s135583820202602x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura M, Holbrook SR. J. Mol. Biol. 2002;320:455–474. doi: 10.1016/s0022-2836(02)00515-6. [DOI] [PubMed] [Google Scholar]
- 21.Correll CC, Beneken J, Plantinga MJ, Lubbers M, Chan YL. Nucleic Acids Res. 2003;31:6806–6818. doi: 10.1093/nar/gkg908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagnon MG, Mukhopadhyay A, Steinberg SV. J. Biol. Chem. 2006;281:39349–39357. doi: 10.1074/jbc.M607725200. [DOI] [PubMed] [Google Scholar]
- 23.Mokdad A, Krasovska MV, Sponer J, Leontis NB. Nucleic Acids Res. 2006;34:1326–1341. doi: 10.1093/nar/gkl025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinberg SV, Boutorine YI. RNA. 2007;13:549–554. doi: 10.1261/rna.387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laing C, Jung S, Iqbal A, Schlick T. J. Mol. Biol. 2009;393:67–82. doi: 10.1016/j.jmb.2009.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zirbel CL, Sponer JE, Sponer J, Stombaugh J, Leontis NB. Nucleic Acids Res. 2009;37:4898–4918. doi: 10.1093/nar/gkp468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doherty EA, Batey RT, Masquida B, Doudna JA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 28.Steinberg SV, Boutorine YI. RNA. 2007;13:1036–1042. doi: 10.1261/rna.495207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pley HW, Flaherty KM, McKay DB. Nature. 1994;372:111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]
- 30.Leontis NB, Westhof E. J. Mol. Biol. 1998;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- 31.Shefer K, Brown Y, Gorkovoy V, Nussbaum T, Ulyanov NB, Tzfati Y. Mol. Cell. Biol. 2007;27:2130–2143. doi: 10.1128/MCB.01826-06. Epub 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T. J. Biol. Chem. 2008;283:18801–18811. doi: 10.1074/jbc.M800233200. [DOI] [PubMed] [Google Scholar]
- 34.Wahl MC, Sundaralingam M. Trends Biochem. Sci. 1997;22:97–102. doi: 10.1016/s0968-0004(97)01004-9. [DOI] [PubMed] [Google Scholar]
- 35.Mandel-Gutfreund Y, Margalit H, Jernigan RL, Zhurkin VB. J. Mol. Biol. 1998;277:1129–1140. doi: 10.1006/jmbi.1998.1660. [DOI] [PubMed] [Google Scholar]
- 36.Ulyanov NB, James TL. Methods Enzymol. 1995;261:90–120. doi: 10.1016/s0076-6879(95)61006-5. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Couch GS, Hendrix DK, Ferrin TE. Nucleic Acids Res. 2006;34:e29. doi: 10.1093/nar/gnj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serganov A, Huang L, Patel DJ. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi T, Nureki O, Ishitani R, Yaremchuk A, Tukalo M, Cusack S, Sakamoto K, Yokoyama S. Nat. Struct. Biol. 2003;10:425–432. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- 41.Ye JD, Tereshko V, Frederiksen JK, Koide A, Fellouse FA, Sidhu SS, Koide S, Kossiakoff AA, Piccirilli JA. Proc. Natl. Acad. Sci. U. S. A. 2008;105:82–87. doi: 10.1073/pnas.0709082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thore S, Frick C, Ban N. J. Am. Chem. Soc. 2008;130:8116–8117. doi: 10.1021/ja801708e. [DOI] [PubMed] [Google Scholar]
- 43.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D'Amare AR. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 44.Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. Mol. Cell. 2005;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Hainzl T, Huang S, Sauer-Eriksson AE. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14911–14916. doi: 10.1073/pnas.0702467104. [DOI] [PMC free article] [PubMed] [Google Scholar]