Abstract
Diffusion tensor imaging (DTI) is gaining increasing importance for anatomical imaging of the developing mouse brain. However, the application of DTI to mouse brain imaging at microscopic levels is hindered by the limitation on achievable spatial resolution. In this study, fast diffusion tensor microimaging (DTMI) of the mouse brain based on a diffusion-weighted gradient and spin echo (DW-GRASE) technique with twin-navigator echo phase correction is presented. Compared to echo planar and spin echo acquisition, the DW-GRASE acquisition resulted in significant reduction in scan time and had minimal image distortion, thereby allowing acquisition at higher spatial resolution. In this study, three dimensional DTMI of the mouse brains at spatial resolutions of 50 – 60 µm revealed unprecedented anatomical details. Thin fiber bundles in the adult striatum and white matter tracts in the embryonic day 12 mouse brains were visualized for the first time. The study demonstrated that data acquired using the DTMI technique allow three dimensional mapping of gene expression data and can serve as a platform to study gene expression patterns in the context of neuroanatomy in the developing mouse brain.
Keywords: diffusion tensor imaging, mouse, brain, gene expression mapping
Introduction
The laboratory mouse plays an essential role in the ongoing efforts to understand the genetic control of the organization, development, and disease processes of the mammalian central nervous system (CNS). With the availability of mouse genome sequences and high throughput gene expression techniques, such as in situ hybridization, researchers have identified many genes that control CNS development, and one of the current challenges is to study the precise functions of these genes in the context of neuroanatomy (1). Imaging techniques that can provide rich anatomical information in the mouse brain are of critical importance for such endeavor.
Several imaging techniques are currently being used for acquiring anatomical information from the mouse brain, including the conventional 2D histological methods (2,3), optical projection tomography (OPT) (4,5), and magnetic resonance microimaging (6,7). Both histology and OPT can provide high resolution images of the mouse brain, but also have their respective limitations. Conventional 2D histology based methods are limited by the number and orientation of available sections, sectioning related tissue damage and deformation, and may not accurately capture the rapidly changing anatomy during development even with complex 3D reconstruction methods. OPT provides high anatomical fidelity, but has limited tissue contrast for differentiation of substructures within the brain. In recent years, magnetic resonance microimaging, and especially diffusion tensor imaging (DTI) (8,9), is gaining increasing importance as a tool for studying the anatomy of the mouse brain (10). There have been several adult mouse brain atlases based on T1, T2, and diffusion MRI and reports on integrating gene expression data into MRI based atlases of the adult mouse brain (11–13). Diffusion tensor microimaging has been shown to provide superior contrasts and reveal more fine anatomical details in the embryonic mouse brain (14), as well as in the postnatal and adult mouse brains (15,16). There have been previous studies demonstrating the efficacy of DTI for imaging the adult mouse brain (17–19), and a few studies based on application of DTI to the embryonic and neonatal mouse brain (14,20).
Although the anatomy of the developing mouse brain has been a subject of decades of studies, it is surprising to know that a significant portion of embryonic brain tissues have not been well characterized. In fact, there are only limited sources of available brain atlases (2,21), which often have discrepancies in anatomical definitions. In early developmental stages (embryonic day 10 – 16), when the simple neural tube starts to differentiate, the rate of development is especially rapid and it contains many emerging and transient structures. DTI, which can provide 3D views with superb tissue contrasts, is therefore expected to decipher the intricate sequences of the tissue differentiation processes. High resolution 3D DTI can also serve as an anatomical framework to map gene expression data with spatially restricted patterns (22–24), for assignment of emerging structures and delineation of developmental boundaries. Here again, the rich anatomical information from the DTI contrast is crucial to accurately map gene expression information from multiple histological sections.
Currently, the greatest challenge to apply DTI for anatomical characterization of the mouse brain is the limitation in spatial resolution. DTI is an inherently low signal-to-noise ratio (SNR) technique. Although it is possible to achieve 20 to 60 µm resolution using conventional T1 and T2 MRI (6,7), there are only a few DTI studies with spatial resolutions of up to 80 µm (14,18), which may still be too coarse to study miniature structures in the developing mouse brain. The implementation of DTI at finer resolutions, especially for imaging embryonic and neonatal mouse brains, is faced with significant technical challenges. In order to achieve a level of spatial resolution that can provide anatomically useful information, considerably long scan times are required. For instance, to acquire images at 60 µm resolution with SNR comparable to images acquired at 80 µm will require a 4 fold increase in scan time. With increasing scan durations, image degradation due to field drifts and gradient instability becomes significant and cannot be ignored. Currently used imaging protocols for DTI of mouse brain are based on echo planar imaging (EPI) or spin echo acquisitions. Diffusion-weighted EPI is sensitive to susceptibility-induced magnetic field inhomogeneities that become particularly problematic at high field strengths, resulting in geometric distortions in the reconstructed images. Multiple RF echo based techniques, such as fast spin echo (FSE), provide superior image quality compared to EPI, but with reduced scanning efficiency. When combined with heavy diffusion weighting, which often causes phase shift due to eddy currents, the Carr-Purcell-Meilboom-Gill (CPMG) condition of FSE tends to break down (25), prohibiting a series of long echo trains, and therefore limiting potential reduction in scan time.
The aim of the present study was to develop a high resolution DTI technique for anatomical characterization of the mouse brain at the microscopic level. To this effect, we developed a 3D diffusion-weighted gradient and spin echo (DW-GRASE) sequence with twin-navigator echo based phase correction for high field diffusion tensor microimaging (DTMI) applications. The GRASE sequence is a combination of EPI and FSE. It interleaves a train of RF refocusing pulses with readout gradient lobes of alternating polarity, and can lead to shorter acquisition time (26). The implementation of GRASE, however, particularly in diffusion-weighting sequences, needs a careful design of coherence pathway selection, and phase correction scheme for phase errors due to gradient instability, off-resonance effects and eddy currents induced by diffusion gradients. In this paper, we present DTMI for anatomical characterization of the mouse brain based on the 3D DW-GRASE sequence. We also demonstrate the three dimensional mapping of gene expression patterns obtained using in situ hybridization to anatomical DTMI data in the early embryonic mouse brain. The long term goal of this study is to establish DTMI based high resolution three-dimensional atlases of developing mouse brains, which will serve as platforms to analyze spatial and temporal varying gene expression patterns. We believe that DTMI can play a vital role to enhance our knowledge in the mechanisms of brain development.
Materials and Methods
Animals and sample preparation
All experimental procedures were approved by the Animal Research Committee at the Johns Hopkins University School of Medicine. For in vivo experiments, adult C57BL/6 mice (2 month old, female, n=8, The Jackson Laboratory, Bar Harbor, ME, USA) were used. Ex vivo tissue specimens from mice at different developmental stages: C57BL/6 adult brain samples (n=3) and postnatal day 12 (P12) cerebellums (n=3), and CD-1 whole embryo samples at embryonic day 12 (E12) (n=3, Charles River Laboratories, Inc., Wilmington, MA, USA) were used in this study. The adult and postnatal mice were perfusion fixed with 4% paraformaldehyde (PFA), followed by immersion fixation of the excised tissue samples for four weeks. The mouse embryos were immersion fixed and kept in 4% PFA before imaging. Prior to imaging, the specimens were placed in PBS with 1mM Gd-DTPA (27) for more than 24 hours to enhance the MR signal and then transferred to 10 mm diameter glass tubes for MR imaging. The tubes were filled with fomblin (Fomblin Profludropolyether, Ausimont, Thorofare, NJ, USA) to prevent dehydration.
Pulse sequence details and phase correction
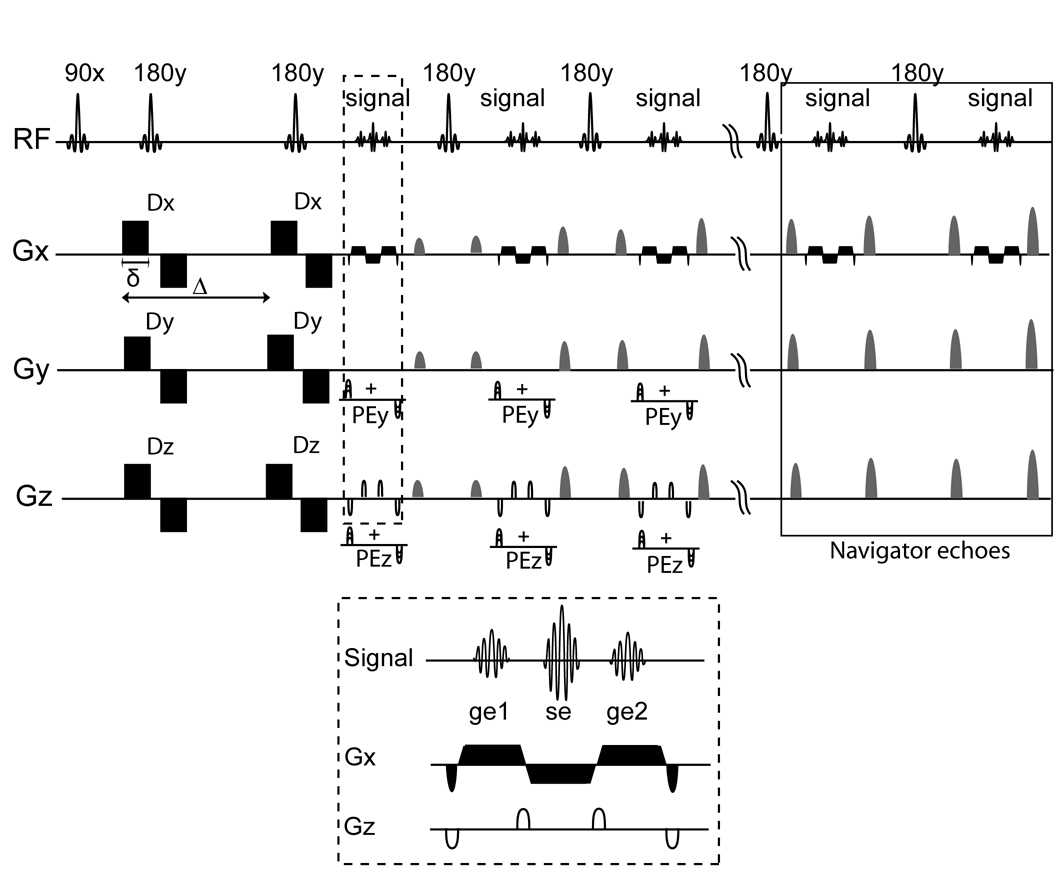
A 3D DW-GRASE sequence with twin navigator echo phase correction was developed for high-field DTI of the mouse brain. A schematic diagram of the pulse sequence is shown in Fig. 1. The echo train consisted of Nrf refocusing pulses, each followed by acquisition of one RF-refocused spin echo, which is flanked by two gradient echoes. k-space data acquired using GRASE suffers from phase and amplitude modulation that has been described previously (28). For 3D acquisition, we used the so-called SORT phase encoding strategy to separate the T2-dependent amplitude modulation and the phase modulation due to off-resonance spins along different Fourier encoding axes as described in (29). In order to minimize the effect of diffusion gradient induced eddy currents on GRASE acquisition, a double refocusing diffusion-weighting scheme shown in Fig. 1 was used (30). The strength of the crusher gradient pairs around each refocusing pulse was varied along the echo train for rejection of the stimulated echo component. Two consecutive refocusing pulses at the end of each echo train were used for navigator echo acquisition in the absence of the phase encoding gradients (25).
Figure 1.
Pulse sequence diagram of the 3D diffusion-weighted GRASE sequence with twin-navigator echo phase correction. Gradients Gx, Gy and Gz are used for the readout, primary and secondary phase encoding respectively. Details of the readout scheme used for GRASE acquisition (inside the dashed box, Top) are enlarged and shown at the bottom (dashed box, Bottom). Blip gradients along Gz are applied as shown for acquisition of an RF refocused spin echo (se) and two gradient echoes (ge1, ge2) after each refocusing pulse at different locations along the secondary phase encoding direction. The last two refocusing pulses are used for navigator echo acquisition in the absence of phase encoding gradients (solid box). Crusher gradient pairs of varying gradient strength centered about the RF refocusing pulses are used for rejection of the stimulated echo component. Dx, Dy, Dz are bipolar diffusion-weighting gradients, PEy and PEz denote the primary and secondary phase encoding gradients respectively.
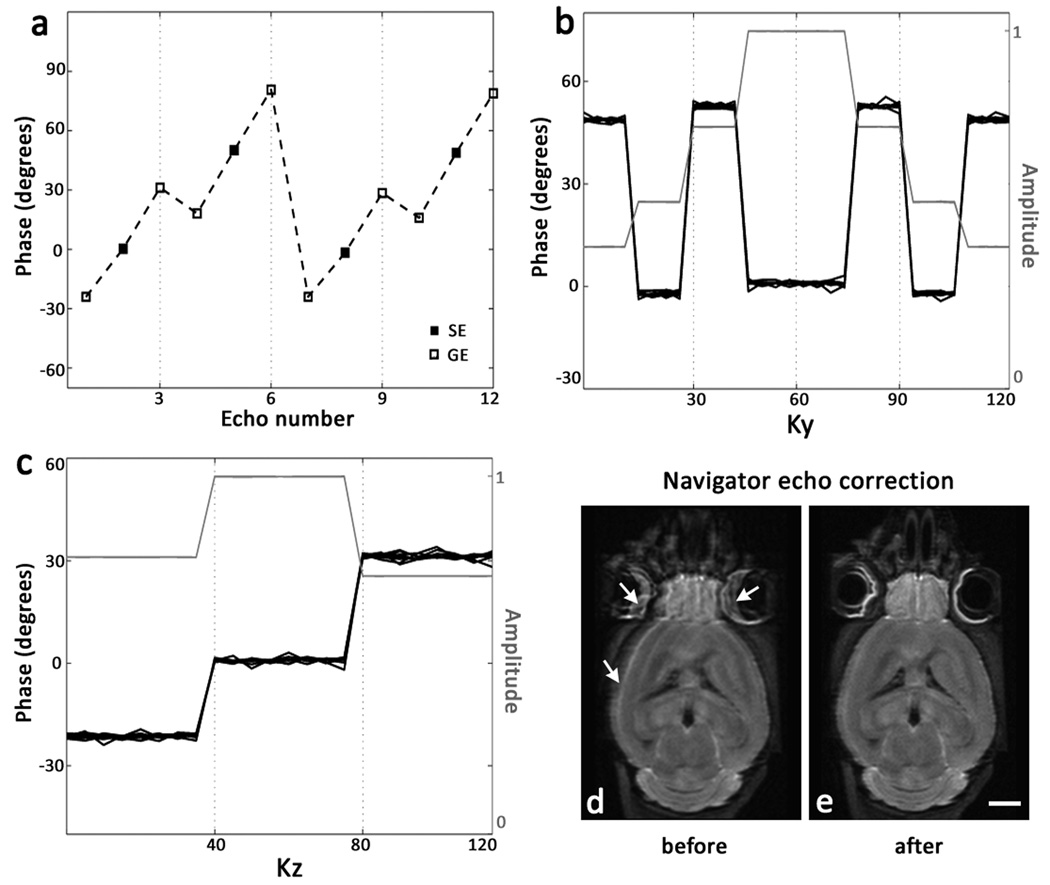
The k-space data acquired with the GRASE sequence are susceptible to phase modulation from off-resonance spins, similar to EPI, which results in phase differences between gradient and spin echoes. If uncorrected for, these phase discontinuities appear as a 'stair-step' pattern across the k-space and can cause severe ghosting artifacts in the reconstructed images (29). Additionally, for DW imaging, breakdown of the CPMG condition introduces an initial phase which results in phase oscillations between echoes from even-numbered and odd-numbered refocusing pulses. In Fig. 2, an example of phase evolution along the echo train and phase modulation of the resulting k-space data acquired from an ex vivo adult mouse brain specimen using the 3D DW-GRASE sequence is shown. A phase correction scheme based on twin-navigator echo acquisition similar to (25) was applied to the k-space data. Echoes from an even- and an odd- numbered refocusing pulse in the absence of phase encoding were acquired at the end of each echo train. The navigator echoes were Fourier transformed along the readout direction to map the phase evolution along the echo train, and the resulting phase corrections were applied to the k-space data for each echo train before Fourier transform along the phase encoding dimensions. Fig. 2d shows reconstructed DW images of the mouse brain before and after navigator echo based phase correction.
Figure 2.
Examples of phase modulation across the k-space and navigator-echo based phase correction in the DW-GRASE sequence. The phase data shown are acquired using an ex vivo mouse brain specimen, with a "center-out" k-space acquisition order chosen to maximize the echo amplitude at the center of the k-space along both the primary (Ky) and secondary (Kz) phase encoding axes. Echo phases shown (in (a), (b) and (c)) are obtained from an arbitrary position along the readout direction after the first Fourier transform. For reference, the phase of the spin echo after the first refocusing pulse is set to zero. In (b) and (c), the amplitude is normalized with respect to the amplitude of the first spin echo. a) Evolution of phase along the echo train with Nrf =4; showing phase oscillations between echoes from odd and even refocusing pulses, and phase differences between spin echoes (SE) and gradient echoes (GE) due to off-resonance spins. b) Phase and T2-dependent amplitude modulation along Ky, for echoes at the center along Kz. c) Phase and T2*-dependent amplitude modulation along Kz, for echoes at the center along Ky. d) and e) Representative DW image from an adult mouse brain before and after navigator echo phase correction. Phase discontinuities result in ghost artifacts and blurred tissue edges in the uncorrected image (white arrows in (d)), which are corrected by navigator echo phase correction.
DTI acquisition
In vivo mouse brain DTI was performed on a 9.4 T NMR spectrometer (Bruker Biospin, Billerica, MA, USA) equipped with a Micro2.5 gradient system (120 G/cm maximum gradient strength), a manufacturer-provided animal imaging probe, and a physiological monitoring system (EKG, respiration, and body temperature). During imaging, mice were anesthetized with isoflurane (1%) together with mixed oxygen and air at a 1:3 ratio via a vaporizer. Images were acquired using a 20 mm diameter birdcage coil as the radiofrequency transmitter and receiver. Imaging parameters for DTI were as follows: echo time (TE) of 32 ms, repetition time (TR) of 800 ms, Nrf of 4, bandwidth of 100 kHz, and 2 signal averages with phase cycling were used. The field of view and matrix size were 15 mm × 15 mm × 15 mm and 128 × 120 × 40, and the native resolution was 117 × 125 × 375 µm3. Two non diffusion-weighted and 6 diffusion directions (b-value 1000 s/mm2) were acquired with δ = 3 ms and Δ = 15 ms. The total imaging time was approximately 2.2 hours with respiratory gating. T2-weighted images were acquired using the RARE sequence with TE of 40 ms, TR of 2000 ms, 4 signal averages, echo train length of 4, native resolution of 100 × 100 × 300 µm3, and total imaging time of 20 minutes. The T2-weighted images were co-registered to the DTI dataset by tri-linear interpolation.
All studies on postmortem specimens were performed on an 11.7 T NMR spectrometer (Bruker BioSpin, Billerica, MA, USA) equipped with a Micro5 gradient system (300 G/cm maximum gradient strength), using a 10 mm diameter saddle coil as the radiofrequency transmitter and receiver. Imaging parameters for DTMI were as follows: TE of 32 ms, TR of 800 ms, Nrf of 4, bandwidth of 100 kHz, and 4 signal averages with phase cycling were used. For the adult mouse brain samples, the imaging field of view and matrix size were 7 mm × 9.2 mm × 14.5 mm and 128 × 168 × 264 respectively, and the native resolution was 55 × 55 × 55 µm3. The spectral data were apodized by a symmetric trapezoidal function with 10% ramp widths and zero-filled before Fourier transformation. For DTI, 12 diffusion directions (b-value 1700 s/mm2), and two non diffusion-weighted images were acquired with δ = 3 ms and Δ = 15 ms. The total imaging time was 45.5 hours (3.25 hours for each image). From the same spectral data, we reconstructed a 3D dataset at lower spatial resolution (80 × 80 × 80 µm3 native) by cropping the original k-space data to a 88 × 116 × 182 matrix before apodization by a trapezoidal function with 10% ramp widths and zero-filling prior to Fourier transformation. For comparison between GRASE and spin echo based acquisitions, relatively low resolution (125 × 125 × 125 µm3) data were acquired using the DW-GRASE sequence and diffusion weighted multiple spin echo sequence (MSE) with the same field of view as the DTMI experiments in the present study. For both GRASE and MSE acquisitions, a TE of 32 ms, TR of 800 ms, Nrf of 4 were used, and 6 diffusion directions (b-value 1700 s/mm2) plus two non diffusion-weighted images were acquired. The number of signal averages were 4 for GRASE and 2 for MSE. The total imaging time for the GRASE and MSE acquisitions were 5 and 15 hours respectively. For the embryonic samples, the field of view and matrix size were 7.6 mm × 8.0 mm × 10.9 mm, and 128 × 136 × 186 respectively, and the native resolution was 60 × 60 × 60 µm3. 16 DW imaging directions (b-value ~1200 s/mm2) (31), and two non diffusion-weighted images were acquired with δ = 3 ms and Δ = 15 ms. The total imaging time was 33 hours (1.83 hours for each image).
The diffusion tensor was calculated by a multivariate linear fitting method using the DTIStudio software (www.mristudio.org) (32), and diagonalized to obtain three pairs of eigenvalues and corresponding eigenvectors. Direction-encoded color (DEC) map images were computed from the primary eigenvector and fractional anisotropy (FA) images. For each voxel, the ratio between the red, green and blue components was defined by the ratio of absolute values of x, y and z components of the primary eigenvector, and the intensity was proportional to FA. Red was assigned to the medial-lateral orientation, green to the rostral-caudal, and blue to the dorsal-ventral orientation. To reconstruct axonal tracts from tensor data, the FACT algorithm (33) was used to trace fibers passing through manually defined regions of interest.
Coregistration of gene expression data
DTMI data of one E12 mouse embryo were used as the anatomical reference for mapping the regional expression of homeobox transcription factors Isl1, Pitx2 and Lhx8, and the Shh gene, which are known to be involved in regulation of embryonic brain development in the mouse. Serial in situ hybridization (ISH) sections from E12 brains examined for the expression of these genes were used (1 µm × 1 µm in plane resolution and 40 µm between adjacent sections) (34). The 2D ISH sections were first reconstructed into a 3D volume using the rigid alignment function provided by Automated Image Registration (AIR) software (35). The reconstructed ISH volume was then aligned to the MRI data using 3D affine transformation. The ISH volume was nonlinearly deformed to closely match the MRI volume using nonlinear landmark-based image mapping technique based on 300 landmarks manually placed throughout the entire brain (36).
Results
Comparison of GRASE and spin echo DTI, and in vivo mouse brain DTI
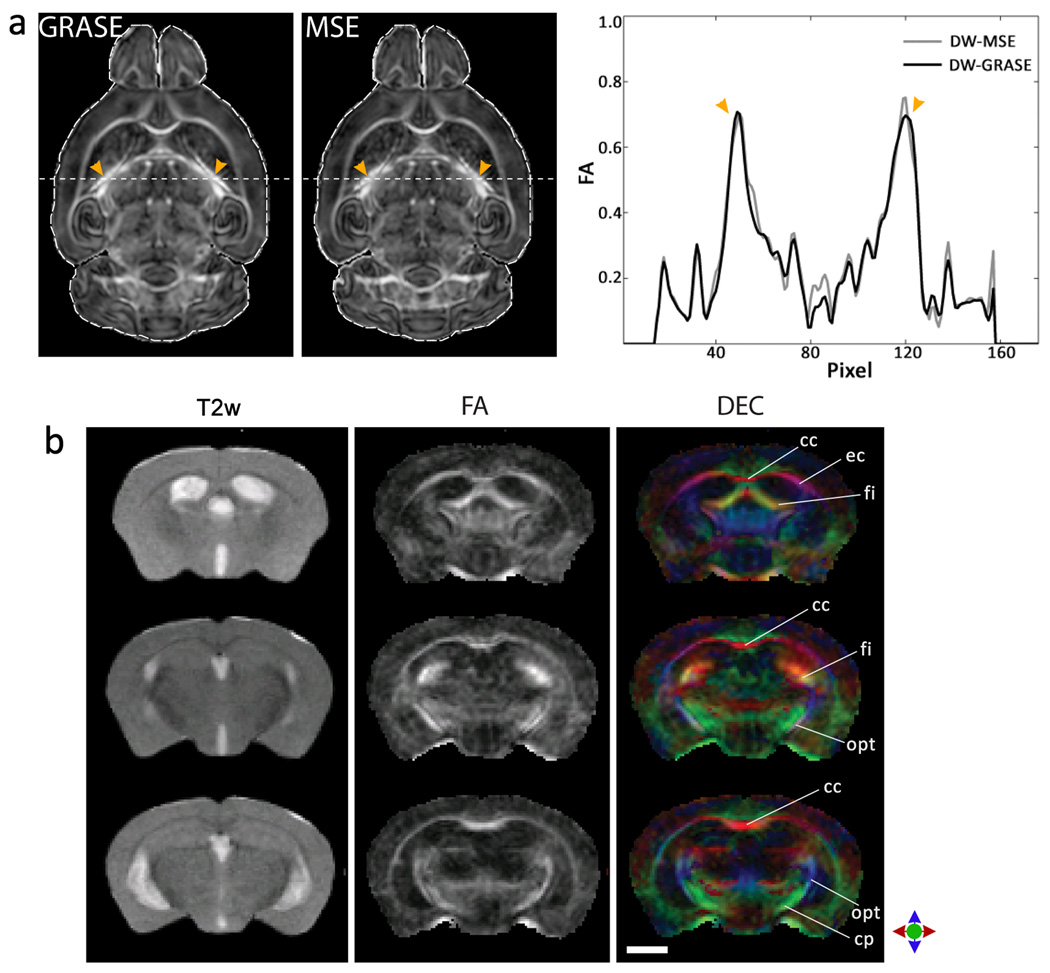
In Fig. 3a, DTI images of an adult mouse brain acquired with the DW-GRASE sequence are compared to those acquired using the diffusion-weighted multiple spin echo (DW-MSE) sequence (37), that were used as the ‘gold standard’. For comparison, the GRASE and MSE datasets were acquired with the same field of view, spatial resolution (125 × 125 × 125 µm3), and diffusion-weighting directions. The total imaging time for the GRASE acquisition was 5 hours (SNR~76 for the b0 image), while the total imaging time for the MSE acquisition was 15 hours (SNR~80 for the b0 image). Compared to MSE images, we found no discernible geometric distortions in the GRASE images (Fig 3, a and b). A voxel-wise comparison of fractional anisotropy (FA) values from the two datasets revealed no apparent differences between MSE- and GRASE- based DTI mapping within the brain volume (6.2 ± 2.1% difference in FA in voxels with FA > 0.2). For illustration, the FA values along an arbitrary profile through the two images are plotted in Fig. 3c. These results demonstrate that using the GRASE acquisition scheme, it was possible to achieve a threefold reduction in scan time as compared to the MSE based acquisition, without any major difference in DTI-derived anisotropy contrasts.
Figure 3.
a) Comparison of fractional anisotropy (FA) images from MSE- and GRASE- based ex vivo DTI. Corresponding axial sections from FA maps of an adult mouse brain for DW-GRASE and DW-MSE acquisitions are shown. Overlaying the brain contour from the MSE image onto the GRASE image reveals no discernible geometric distortions. Pixel-wise comparison of FA profiles along the dashed lines are shown on the right. Arrowheads indicate the brain structures with high diffusion anisotropy corresponding to the two peaks in the FA profiles. For DTI mapping, GRASE images are acquired at a spatial resolution of 0.125 × 0.125 × 0.125 mm3, with 6 diffusion directions and 4 signal averages. MSE images are acquired with the same spatial resolution and diffusion directions, and 2 signal averages. Total scan time is 15 hours for MSE-based DTI and 5 hours for GRASE-based DTI. b) GRASE-based in vivo DTI showing coronal sections from FA and DEC maps of an adult mouse brain. Comparison with in vivo T2-weighted images (left) acquired using the RARE sequence reveals no discernible geometric distortions. In the DEC maps, red, blue and green represent diffusion anisotropy along the medial-lateral, dorsal-ventral and rostral-caudal axes respectively. Structural abbreviations for labeled white matter tracts are: cc: corpus callosum, cp: cerebral peduncle, ec: external capsule, fi: fimbria, opt: optic tract. Scale bar = 2 mm.
Fig. 3b shows in vivo DTI images of a selected adult mouse brain acquired using the DW-GRASE sequence. Compared to T2-weighted images acquired using the RARE sequence, no discernible geometric distortion was observed. Average FA and diffusivity measurements of major white matter tracts obtained via manually placed regions of interest and listed in Table 1 were comparable to those reported in previous studies of in vivo mouse brain DTI (38,39).
Table 1.
Means and standard deviations of FA and diffusivity measurements of major white matter tracts in the in vivo adult mouse brain (two month old, female, n = 8). The two diffusivity measurements, λ║ and λ⊥, are the primary eigenvalue and the average of the secondary and tertiary eigenvalues respectively. Structural abbreviations are: gcc: genu of the corpus callosum; bcc: body of the corpus callosum; scc: splenium of the corpus callosum; ec: external capsule; cp: cerebral peduncle; opt: optic tract; fi: fimbria.
| White matter tract | FA | λ║(10−5 cm2/s) | λ⊥ (10−5 cm2/s) |
|---|---|---|---|
| gcc | 0.62 ± 0.11 | 1.3 ± 0.16 | 0.42 ± 0.17 |
| bcc | 0.51 ± 0.09 | 1.1 ± 0.15 | 0.48 ± 0.16 |
| scc | 0.59 ± 0.10 | 1.1 ± 0.15 | 0.41 ± 0.16 |
| ec | 0.51 ± 0.09 | 1.1 ± 0.16 | 0.46 ± 0.16 |
| opt | 0.57 ± 0.10 | 1.1 ± 0.18 | 0.42 ± 0.16 |
| cp | 0.54 ± 0.10 | 1.0 ± 0.15 | 0.42 ± 0.16 |
| fi | 0.62 ± 0.09 | 1.4 ± 0.19 | 0.45 ± 0.18 |
DT-microimaging of the adult mouse brain
Fig. 4 illustrates the application of DTMI for whole brain anatomical characterization in the adult mouse brain. For comparison, DTI-derived DEC maps of orthogonal sections through the brain are shown alongside isotropically diffusion-weighted (iDW) images that are shown in mirrored semi-sections on the left (Fig. 4a). High resolution anisotropy mapping revealed the underlying three dimensional microstructural organization within the brain. In the DEC maps, white matter tracts such as the cingulum, fasciculus retroflexus and the fimbria could be distinctly delineated based on differences in their structural orientation, but were not discernible in conventional T2-w or iDW images. To evaluate the efficacy of DTI at higher spatial resolution for detailed anatomical characterization of the brain, we compared the DTMI results from the present study with the same DTI k-space data reconstructed to lower resolution (80 × 80 × 80 µm3) and with images from our current DTI-based mouse brain atlas (125 × 125 × 125 µm3) (15). The comparisons show that DTMI revealed finer anatomical details within the brain (Fig. 4b). The most striking difference at higher resolutions was seen in the striatum. The striatal volume is traversed by numerous fascicles that fan out from the internal capsule toward the cerebral cortex (white arrows, Fig. 4b). This mesh of very fine fibers traversing through the striatum cannot be resolved in images acquired at lower resolutions.
Figure 4.
DTMI of an adult mouse brain acquired at 55 µm resolution. a) DTI based contrasts in orthogonal sections through the brain. Isotropically diffusion weighted (iDW) images and direction encoded color (DEC) maps are shown in the left and right semi-sections of the brain respectively. In the DEC maps, red, blue and green represent diffusion anisotropy along the medial-lateral, dorsal-ventral and rostral-caudal axes respectively. b) Comparison of anatomical details revealed by DTI at 55 µm (left panel), 80 µm (middle panel), and 125 µm (right panel) resolutions. Sagittal sections at the level of the striatum are shown; high resolution DTI revealed fine microstructural organization of white matter fibers within the striatum (white arrows, right panel), which could not be resolved at lower resolutions (middle, left panels). Abbreviations used are Cx: cortex, cc: corpus callosum, SC: superior colliculus. Scale bars = 1 mm.
DTMI of the postnatal mouse cerebellum
At high resolutions, as the voxel size is further reduced, the number of phase encoding steps required to cover the entire brain volume often becomes a limiting factor for reduction of scan time. Therefore, imaging subsections of the brain may become a useful approach for localized anatomical characterization. To explore this possibility, we imaged an excised cerebellum from a P12 mouse brain, with a resolution of 50 µm. The cytoarchitecture of the mammalian cerebellum consists of a well-defined network of densely interwoven axonal and dendritic processes. In the mouse, neuronal connectivity in the cerebellum is established primarily during the first three weeks of postnatal development. At P12, DTMI of the ex vivo mouse cerebellum revealed a characteristic layered architecture (Fig. 5). In a coronal cross-section through the cerebellum (Fig 5b), the distinctly layered organization of the cerebellar cortex can be observed. Fig. 5c shows a comparison of T2-w, FA and DEC contrasts in a sagittal section through the vermis of the cerebellar cortex. In the DEC map, three distinct layers in the cerebellar lobules are distinguishable based on characteristic patterns of orientation of their underlying neuronal fibers. The red layer corresponds to the orientation of very thin (0.2 – 0.3 µm in diameter) and densely packed parallel fibers that course parallel to the long axes of the folia. At the center of each cerebellar folium, the deep cerebellar white matter consisting of myelinated axonal fibers can be delineated as a layer of high anisotropy, and between these two layers is a low anisotropy region containing primarily the dendritic ramifications of the Purkinje cells that fan out at right angles to the axes of the folia. These results illustrate that DTMI can distinctly reveal the complex cytoarchitectural organization within the developing mouse cerebellum, and may be used to study the temporal evolution of cerebellar cortical organization in the early postnatal mouse brain.
Figure 5.
DTMI of an isolated mouse cerebellum at postnatal day 12 (P12) acquired at 50 µm resolution. a) 3D volume rendering of the cerebellum. b) DEC map of a coronal section through the cerebellum. Red, green and blue represent diffusion anisotropy along the medial-lateral, rostral-caudal and dorsal-ventral axes respectively. C) Comparison of mid-sagittal T2-weighted image, fractional anisotropy image and DEC map through the cerebellar vermis. Scale bar = 1 mm.
DTMI for anatomical characterization of the embryonic mouse brain
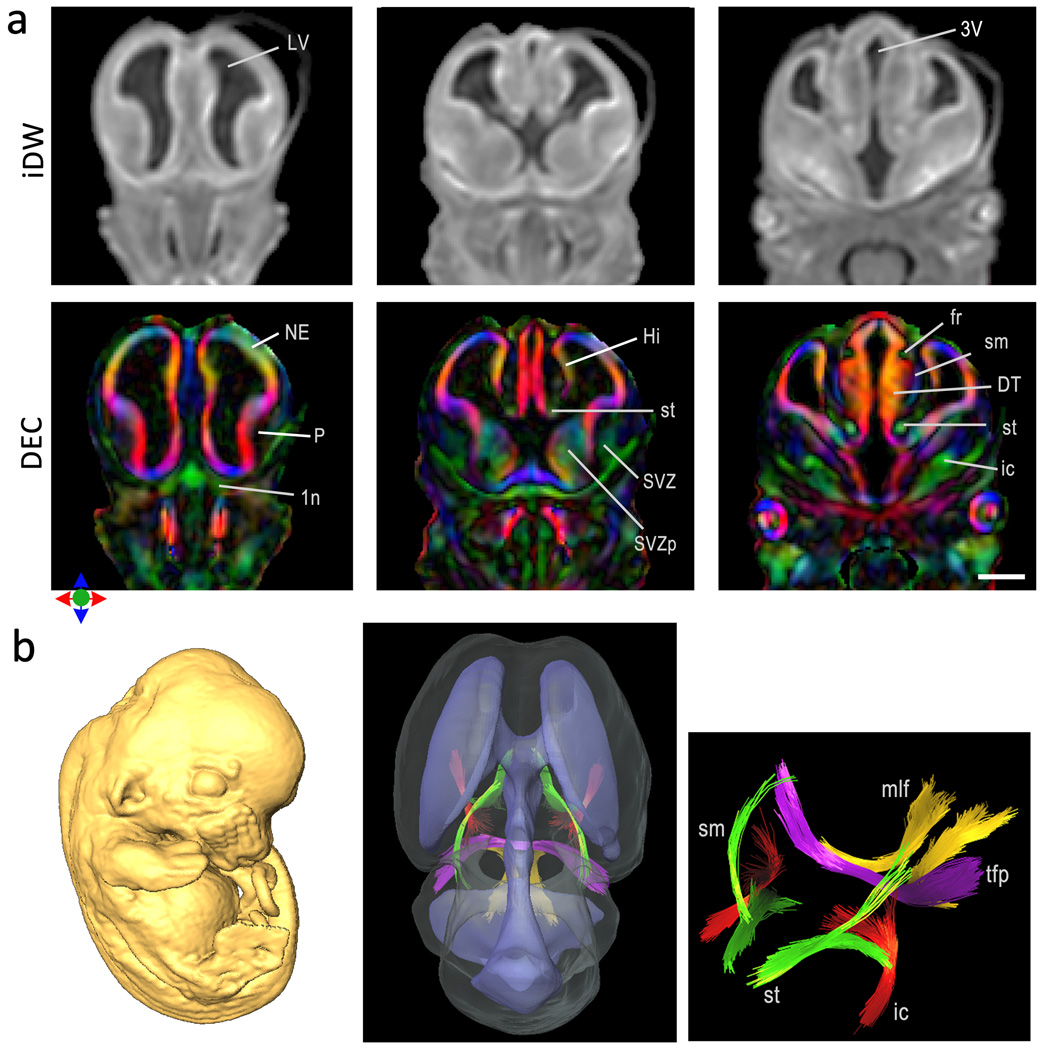
At early stages of embryonic development, well before the onset of myelination, conventional T2-w and iDW images provide limited anatomical contrasts within the brain. As shown in Fig. 6a, in iDW images of an E12 mouse embryo, the overall brain morphology and ventricular volume can be appreciated, but there is limited contrast for differentiation of gray or white matter structures within the CNS. In comparison, orientation contrasts derived from DTMI revealed rich anatomical information for characterization of gray and white matter organization at the microscopic level. The neuroepithelium, which is the zone of active neuronal proliferation can be clearly differentiated as a region marked by high anisotropy that is radially oriented perpendicular to the ventricular surface (Fig. 6a). Early developing white matter tracts such as the stria medullaris, internal capsule, fasciculus retroflexus and stria terminalis can also be delineated in the DEC maps. Using fiber tracking in high resolution tensor data, it was possible to trace the 3D trajectories of early developing axonal tracts in the E12 mouse brain, which have not been reported before. Fig. 6c shows the spatial organization of the reconstructed white matter tracts and the ventricles within the E12 mouse brain. These results indicate that high resolution DTI can be used to trace the emergence and early growth of white matter tracts during embryonic development in the mouse CNS.
Figure 6.
DTMI of the embryonic mouse brain at E12 acquired at 60 µm resolution. a) Coronal sections through the E12 brain showing isotropically diffusion-weighted (iDW) images and direction encoded color (DEC) maps. In the DEC maps, red, green and blue represent diffusion anisotropy along the medial-lateral, rostral-caudal and dorsal-ventral axes respectively. Brain structures that can be delineated from diffusion orientation based contrasts are labeled. Scale bar = 0.5 mm. Structural abbreviations are: LV: lateral ventricle, 3V: third ventricle, NE: neuroepithelium, P: Pallidum, 1n: olfactory nerve, Hi: Hippocampus, st: stria terminalis, SVZ: striatal subventricular zone, SVZp: pallidal subventricular zone, fr: fasciculus retroflexus, DT: dorsal thalamus, ic: internal capsule, sm: stria medularis. b) 3D reconstruction of structures inside the E12 mouse brain. Surface rendering of the brain and ventricular volumes shows the 3D spatial trajectories of early developing white matter tracts delineated by fiber tracking. Magnified view of the delineated axonal tracts shows the st (green), sm (green), ic (red), medial longitudinal fasciculus (mlf, yellow) and transverse pontine fibers (tfp, purple).
Gene expression mapping
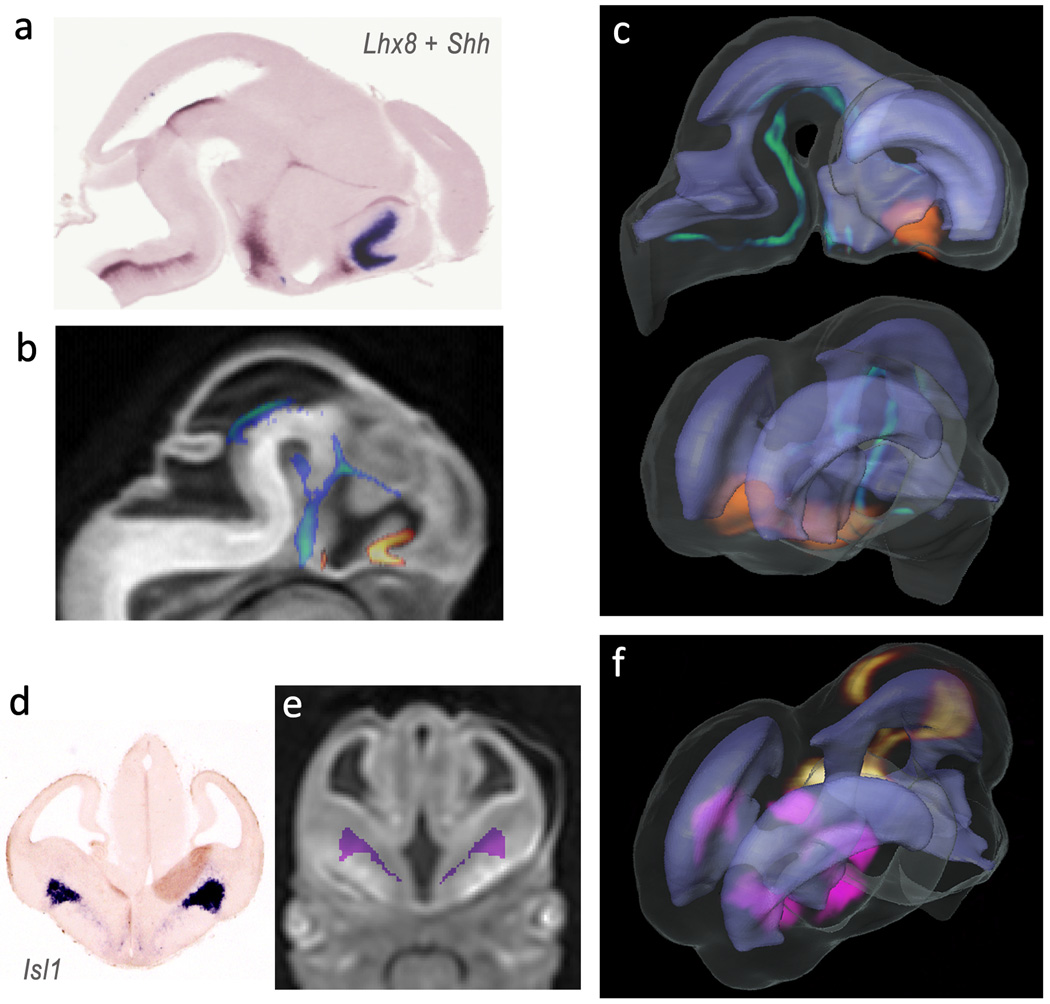
DTMI of the E12 mouse brain was used to establish an anatomical reference for mapping of gene expression patterns from serial 2D ISH sections. Fig. 7 illustrates the results of mapping gene expression data for four different transcription factors expressed in the mouse brain at E12. Expression data from sagittal two-color ISH sections co-stained for expression of Lhx8 and Shh genes, and coronal ISH sections stained for expression of Isl1 and Pitx2 were mapped to high resolution MR images. In Fig. 7 (b,e), the expression patterns of Lhx8 (orange), Shh (green) and Isl1 (pink) are overlaid on corresponding 2D slices from MR images after intensity and landmark based registration. From the original datasets of 2D ISH sections, it was not possible to determine the spatial relationships or possible overlaps between expression regions of the different genes. Mapping of gene expression data enabled simultaneous visualization of 3D expression patterns and relative spatial distributions of different genes within the E12 mouse brain (Fig. 7, c and f).
Figure 7.
Mapping of gene expression data to high resolution 3D MR images in an E12 mouse embryo. Lhx8 + Shh expression data from two-color sagittal ISH sections (a), and Isl1 (d) and Pitx2 expression data from serial coronal ISH sections were mapped to the MR images using intensity and landmark based registration. The expression patterns after coregistration are overlaid on corresponding 2D sections from the MR images (b,f). After the mapping procedures, the gene expression patterns of Shh (green), Lhx8 (orange), Isl1 (pink) and Pitx2 (yellow) are visualized in 3D (c,f) to reveal their relative spatial distributions within the brain and spatial correlations with surrounding anatomical structures such as the ventricles (rendered in light purple).
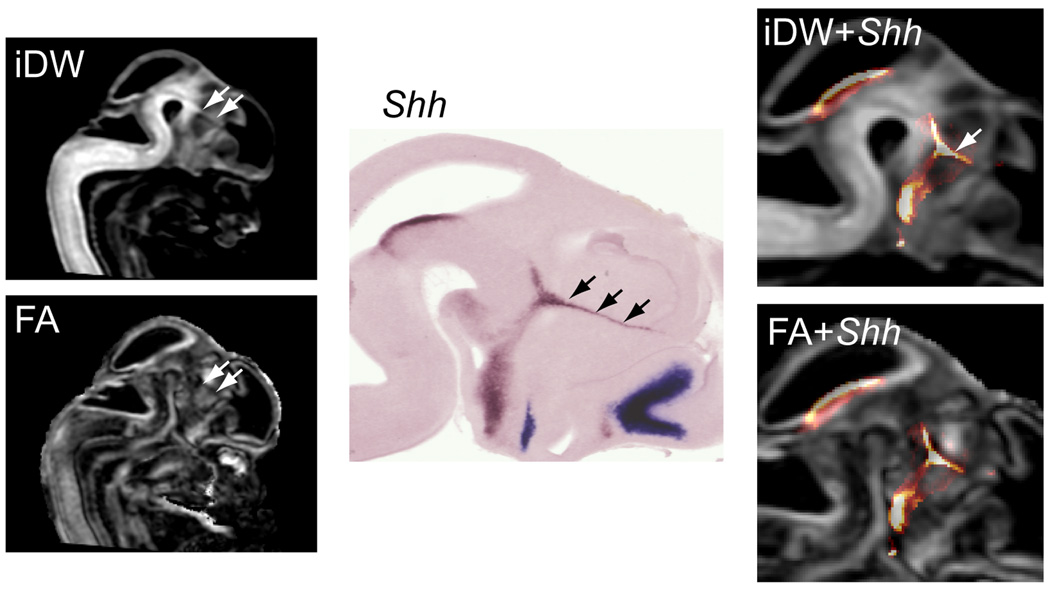
With the coregistered gene expression and DTMI data, it was possible to use structure-specific gene expression patterns to guide the delineation of structures in the DTMI data. In Fig. 8, an example of structural identification based on mapping of gene expression patterns is illustrated. There are developmental boundaries in the embryonic brain, that besides morphological characteristics, also serve as signaling centers and can be identified by the specific genes they express. The zona limitans intrathalamica (ZLI), that separates the dorsal and ventral thalamus in the embryonic brain, is also the only structure in the alar plate that expresses the Shh gene. In the DTMI data, a structure that resembles the ZLI based on its location relative to the brain and ventricles could be identified in the iDW and FA images based on differences in contrast with the surrounding tissue (white arrows in Fig. 8), while the ZLI could be very clearly demarcated in the ISH section stained for Shh expression (black arrows in Fig. 8). By mapping the Shh gene expression pattern to the MR images, the exact location of the ZLI in the MR images could be confirmed. These results demonstrate the possibility of combining gene expression information with high resolution anatomical DTI to study the evolution of structural and developmental boundaries during embryonic development.
Figure 8.
Mapping the Shh gene expression data to the MR images in an E12 mouse brain helped to locate the zona limitans interthalamica (ZLI) in the early thalamus. In high resolution diffusion-weighted (iDW) and fractional anisotropy (FA) images of an E12 mouse embryo (left panel), the region indicated by the white arrows can be differentiated from surrounding tissues, and may correspond to the ZLI. In the sagittal ISH section stained for Shh expression (middle), the ZLI is clearly visible (indicated by black arrows). After mapping the Shh expression data to the MR images, the location of ZLI in MR images is confirmed (right panel).
Discussion
This study demonstrates the feasibility and efficacy of fast diffusion tensor microimaging, which is important for anatomical characterization of the mouse brain at microscopic levels. The theoretical resolution limit of DTI is approximately 10–20 µm, dictated by the echo time and diffusion constant of free water at room temperature. In the present study, a spatial resolution of up to 50 µm was achieved on an 11.7 T system. The DW-GRASE based fast acquisition scheme was able to significantly reduce the scan time compared to conventional spin echo acquisitions, and therefore allow us to acquire high resolution within a given scan time. The use of double refocusing diffusion gradients and twin navigator echo phase correction further increased the robustness of the acquisition scheme. Compared to EPI based diffusion acquisition, the DW-GRASE based method is less sensitive to B0 field inhomogeneity. DW-GRASE based DTI showed no noticeable degradation in image quality in terms of the measured fractional anisotropy and potential distortion artifacts (Fig. 3). We also demonstrated the feasibility of acquiring in vivo mouse brain DTI using the DW-GRASE sequence. At the level of resolution used in this study for ex vivo DTMI, finer anatomical details of the adult mouse brain could be revealed as compared to our previous DTI results at lower resolutions. We were able to reconstruct early white matter tracts in the E12 mouse brain, which has not been reported previously. Because of the well-known limitation of the six-element tensor estimation and the deterministic tractography approach employed in this study (40), the tract reconstruction was limited to well-defined core regions. With more sophisticated diffusion sampling schemes (31,41,42) and tract reconstruction methods (43,44), more structural and connectivity information may be revealed. These results underscore the importance of imaging at higher resolutions for studying the neuroanatomy of the mouse brain at microscopic levels. With the availability of higher field strengths (~17.5 T), higher SNR and thus finer resolution (~40 µm) should be achievable. In this study, we implemented the GRASE readout scheme with acquisition of three echoes per RF refocusing pulse. With the employment of improved shimming techniques such as FAST(EST)MAP (45), it may be possible to increase the number of gradient echoes per refocusing pulse, thus leading to even faster acquisition than the current method and further reduction in scan time.
In this study, the phase correction scheme for the DW-GRASE sequence was based on an extension of the twin-navigator echo scheme proposed in (25). For diffusion-weighted FSE acquisitions, alternative pulse sequence modifications to minimize artifacts from breakdown of the CPMG condition have been reported in previous studies (46,47), and the use of an image-based method for correction of eddy-current induced artifacts has been demonstrated for ex vivo applications (48). The twin-navigator echo based approach was used in the present study, since in addition to breakdown of the CPMG condition, the k-space data acquired using DW-GRASE also suffer from phase modulation due to off-resonance effects that cause phase differences between gradient and spin echoes, and the phase differences mapped by the navigator echoes can be used to correct for artifacts from both these sources (Fig. 2). The navigator echoes are also useful to correct for phase differences due to subject motion for in vivo imaging, as demonstrated in Fig. 3b. It is necessary to note that several factors such as the amount of T2- and T2*-induced signal attenuation, gradient linearity and gradient calibration may adversely affect the quality of the GRASE images, and should therefore be carefully evaluated.
At present, our knowledge of mouse brain anatomy during embryonic development is far from complete. There are only a limited number of 2D histological atlases for murine brains at these ages. Although the resolution of DTMI is still not comparable to that of OPT (5 ~ 10 µm) (5) or histology (<1 µm), DTMI can provide superior 3D contrasts within the CNS. As shown in this study, it is possible to delineate fine neuroanatomy of the embryonic brain and trace early developing axonal tracts as early as E12 in the mouse, which is not possible with OPT. However, despite the rich anatomical contrasts available from DTI, accurate identification and labeling of embryonic brain structures is still difficult due to our limited knowledge of mouse brain anatomy at these stages. The presence of developing and transient structures and lack of existing brain atlases in early embryonic stages makes it difficult to interpret and correlate the DTI contrasts with embryonic neuroanatomy for accurate assignment of structures. In the adult stages, the DTI contrasts are relatively well-understood, since the anatomy of the adult mouse brain has been well characterized. In future studies, high resolution DTI of subsequent embryonic and neonatal stages may make it possible to retrogradely trace and identify the early embryonic precursors of structures in the adult brain by examining the changes in DTI contrasts with development.
This study also demonstrated the mapping of gene expression information from 2D ISH sections to anatomical DTI images in the embryonic brain. Analysis of temporal and spatial patterns of gene expression in the context of neuroanatomy is important for understanding gene interactions and functions during embryonic development. While the current resolution is still lower than the in-plane resolution of the ISH data (< 1 µm), it is approaching the through-plane resolution (40 µm), and therefore can be used for mapping and visualization of the 3D patterns of gene expression. Effective matching of gene expression data to DTI images can provide complementary information for understanding embryonic CNS development, with delineation of anatomical structures from DTI, and developmental boundaries defined by patterns of gene expression. Gene mapping procedures will also make it possible to inspect the 3D expression patterns of different genes, examine possible overlaps between the regions of expression and study the interactions of these genes in the context of neuroanatomy. Current efforts on integrating gene expression data with MRI data of the mouse brain mainly focus on the adult brain because conventional MRI can provide satisfactory contrast for structural delineation in the adult brain. The techniques described in this paper will enable us to extend current studies to the embryonic and neonatal mouse brains and study the gene expression patterns in four dimensions.
To summarize, this study demonstrated the potential of high resolution DTI for microanatomical characterization in the mouse brain. At the level of resolution currently achievable, the structural details provided by DTI will be important for potential applications in anatomical phenotyping and development of atlases of the mouse brain, and to further our understanding of the developmental processes in the embryonic CNS.
Acknowledgement
This study was supported by National Institutes of Health (NIH) grants R01AG20012-01, R01EB003543, P41RR15241-01, R21NS059529 and R21NS065306.
References
- 1.Sunkin SM. Towards the integration of spatially and temporally resolved murine gene expression databases. Trends in Genetics. 2006;22(4):211–217. doi: 10.1016/j.tig.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Paxinos G, Halliday GH, Watson C, Koutcherov Y, Wang H. Atlas of the Developing Mouse Brain at E17.5, P0 and P6. Academic Press; 2006. [Google Scholar]
- 3.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 4.Venkataraman S, Stevenson P, Yang Y, Richardson L, Burton N, Perry TP, Smith P, Baldock RA, Davidson DR, Christiansen JH. EMAGE--Edinburgh Mouse Atlas of Gene Expression: 2008 update. Nucleic Acids Res. 2008;36(Database issue):D860–D865. doi: 10.1093/nar/gkm938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296(5567):541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs RE, Ahrens ET, Dickinson ME, Laidlaw D. Towards a microMRI atlas of mouse development. Comput Med Imaging Graph. 1999;23(1):15–24. doi: 10.1016/s0895-6111(98)00059-7. [DOI] [PubMed] [Google Scholar]
- 7.Benveniste H, Kim K, Zhang L, Johnson GA. Magnetic resonance microscopy of the C57BL mouse brain. Neuroimage. 2000;11(6 Pt 1):601–611. doi: 10.1006/nimg.2000.0567. [DOI] [PubMed] [Google Scholar]
- 8.Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66(1):259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basser PJ, Pierpaoli C. A simplified method to measure the diffusion tensor from seven MR images. Magn Reson Med. 1998;39(6):928–934. doi: 10.1002/mrm.1910390610. [DOI] [PubMed] [Google Scholar]
- 10.Mori S, Zhang J. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron. 2006;51(5):527–539. doi: 10.1016/j.neuron.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Ng L, Pathak SD, Kuan C, Lau C, Dong H, Sodt A, Dang C, Avants B, Yushkevich P, Gee JC, Haynor D, Lein E, Jones A, Hawrylycz M. Neuroinformatics for genome-wide 3D gene expression mapping in the mouse brain. IEEE/ACM Trans Comput Biol Bioinform. 2007;4(3):382–393. doi: 10.1109/tcbb.2007.1035. [DOI] [PubMed] [Google Scholar]
- 12.MacKenzie-Graham A, Jones ES, Shattuck DW, Dinov ID, Bota M, Toga AW. The informatics of a C57BL/6J mouse brain atlas. Neuroinformatics. 2003;1(4):397–410. doi: 10.1385/NI:1:4:397. [DOI] [PubMed] [Google Scholar]
- 13.MacKenzie-Graham A, Lee EF, Dinov ID, Bota M, Shattuck DW, Ruffins S, Yuan H, Konstantinidis F, Pitiot A, Ding Y, Hu G, Jacobs RE, Toga AW. A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J Anat. 2004;204(2):93–102. doi: 10.1111/j.1469-7580.2004.00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Richards LJ, Yarowsky P, Huang H, van Zijl PC, Mori S. Three-dimensional anatomical characterization of the developing mouse brain by diffusion tensor microimaging. Neuroimage. 2003;20(3):1639–1648. doi: 10.1016/s1053-8119(03)00410-5. [DOI] [PubMed] [Google Scholar]
- 15.Aggarwal M, Zhang J, Miller MI, Sidman RL, Mori S. Magnetic resonance imaging and micro-computed tomography combined atlas of developing and adult mouse brains for stereotaxic surgery. Neuroscience. 2009;162(4):1339–1350. doi: 10.1016/j.neuroscience.2009.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Miller MI, Plachez C, Richards LJ, Yarowsky P, van Zijl P, Mori S. Mapping postnatal mouse brain development with diffusion tensor microimaging. Neuroimage. 2005;26(4):1042–1051. doi: 10.1016/j.neuroimage.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Song SK, Yoshino J, Le TQ, Lin SJ, Sun SW, Cross AH, Armstrong RC. Demyelination increases radial diffusivity in corpus callosum of mouse brain. Neuroimage. 2005;26(1):132–140. doi: 10.1016/j.neuroimage.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 18.Tyszka JM, Readhead C, Bearer EL, Pautler RG, Jacobs RE. Statistical diffusion tensor histology reveals regional dysmyelination effects in the shiverer mouse mutant. Neuroimage. 2006;29(4):1058–1065. doi: 10.1016/j.neuroimage.2005.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, van Zijl PC, Mori S. Three dimensional diffusion tensor magnetic resonance micro-imaging of adult mouse brain and hippocampus. NeuroImage. 2002;15:892–901. doi: 10.1006/nimg.2001.1012. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Zhang J, Mori S, Nathans J. Axonal growth and guidance defects in Frizzled3 knock-out mice: a comparison of diffusion tensor magnetic resonance imaging, neurofilament staining, and genetically directed cell labeling. J Neurosci. 2006;26(2):355–364. doi: 10.1523/JNEUROSCI.3221-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schambra U, Lauder JM, Silver J. Atlas of the Prenatal Mouse Brain. Academic Press; 1991. [DOI] [PubMed] [Google Scholar]
- 22.Bulfone A, Puelles L, Porteus MH, Frohman MA, Martin GR, Rubenstein JL. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13(7):3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vue TY, Aaker J, Taniguchi A, Kazemzadeh C, Skidmore JM, Martin DM, Martin JF, Treier M, Nakagawa Y. Characterization of progenitor domains in the developing mouse thalamus. J Comp Neurol. 2007;505(1):73–91. doi: 10.1002/cne.21467. [DOI] [PubMed] [Google Scholar]
- 24.Hoekman MF, Jacobs FM, Smidt MP, Burbach JP. Spatial and temporal expression of FoxO transcription factors in the developing and adult murine brain. Gene Expr Patterns. 2006;6(2):134–140. doi: 10.1016/j.modgep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Mori S, van Zijl PCM. A motion correction scheme by twin-echo navigation for diffusion-weighted magnetic resonance imaging with multiple RF echo acquisition. Magn Reson Med. 1998;40:511–516. doi: 10.1002/mrm.1910400403. [DOI] [PubMed] [Google Scholar]
- 26.Oshio K, Feinberg D. GRASE (gradient- and spin-echo) imaging: a novel fast MRI technique. Magn Reson Med. 1991;20:344–349. doi: 10.1002/mrm.1910200219. [DOI] [PubMed] [Google Scholar]
- 27.D'Arceuil HE, Westmoreland S, de Crespigny AJ. An approach to high resolution diffusion tensor imaging in the fixed primate brain. NeuroImage. 2006;35(2):553–565. doi: 10.1016/j.neuroimage.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 28.Oshio K. vGRASE: separating phase and T2 modulations in 2D. Magn Reson Med. 2000;44:383–386. doi: 10.1002/1522-2594(200009)44:3<383::aid-mrm7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 29.Mugler JP. Improved three-dimensional GRASE imaging with the SORT phase-encoding strategy. Journal of Magnetic Resonance Imaging. 1999;9:604–612. doi: 10.1002/(sici)1522-2586(199904)9:4<604::aid-jmri15>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 30.Reese TG, Heid O, Weistoff RM, Wedeen VJ. Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo. Magn Reson Med. 2003;49:177–182. doi: 10.1002/mrm.10308. [DOI] [PubMed] [Google Scholar]
- 31.Jones DK. The effect of gradient sampling schemes on measures derived from diffusion tensor MRI: A Monte Carlo study. Magn Reson Med. 2004;51:807–815. doi: 10.1002/mrm.20033. [DOI] [PubMed] [Google Scholar]
- 32.Jiang H, van Zijl PCM, Kim J, Pearlson GD, Mori S. DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. Computer Methods and Programs in Biomedicine. 2006;81(2):106–116. doi: 10.1016/j.cmpb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Mori S, Crain BJ, Chacko VP, van Zijl PCM. Three dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annals of Neurology. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 34.Kataoka A, Shimogori T. Fgf8 controls regional identity in the developing thalamus. Development. 2008;135:2865–2871. doi: 10.1242/dev.021618. [DOI] [PubMed] [Google Scholar]
- 35.Woods RP, Grafton ST, Watson JD, Sicotte NL, Mazziotta JC. Automated image registration: II. Intersubject validation of linear and nonlinear models. Journal of Computer Assisted Tomography. 1998;22(1):153–165. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]
- 36.Joshi SC, Miller MI. Landmark matching via large deformation diffeomorphisms. IEEE Trans Image Process. 2000;9(8):1357–1370. doi: 10.1109/83.855431. [DOI] [PubMed] [Google Scholar]
- 37.van Dusschoten D, Moonen CT, de Jager PA, Van As H. Unraveling diffusion constants in biological tissue by combining Carr-Purcell-Meiboom-Gill imaging and pulse field gradient NMR. Magn Reson Med. 1996;36:907–913. doi: 10.1002/mrm.1910360613. [DOI] [PubMed] [Google Scholar]
- 38.Chahboune H, Ment LR, Stewart WB, Ma X, Rothman DL, Hyder F. Neurodevelopment of C57B/L6 mouse brain assessed by in vivo diffusion tensor imaging. NMR Biomed. 2007;20(3):375–382. doi: 10.1002/nbm.1130. [DOI] [PubMed] [Google Scholar]
- 39.Sun SW, Neil JJ, Liang HF, He YY, Schmidt RE, Hsu CY, Song SK. Formalin fixation alters water diffusion coefficient magnitude but not anisotropy in infarcted brain. Magn Reson Med. 2005;53(6):1447–1451. doi: 10.1002/mrm.20488. [DOI] [PubMed] [Google Scholar]
- 40.Mori S, van Zijl PC. Fiber tracking: principles and strategies - a technical review. NMR Biomed. 2002;15(7–8):468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- 41.Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40(5):885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 42.Frank LR. Characterization of anisotropy in high angular resolution diffusion-weighted MRI. Magn Reson Med. 2002;47(6):1083–1099. doi: 10.1002/mrm.10156. [DOI] [PubMed] [Google Scholar]
- 43.Koch MA, Norris DG, Hund-Georgiadis M. An investigation of functional and anatomical connectivity using magnetic resonance imaging. Neuroimage. 2002;16(1):241–250. doi: 10.1006/nimg.2001.1052. [DOI] [PubMed] [Google Scholar]
- 44.Behrens TE, Woolrich MW, Jenkinson M, Johansen-Berg H, Nunes RG, Clare S, Matthews PM, Brady JM, Smith SM. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med. 2003;50(5):1077–1088. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- 45.Gruetter R, Tkac I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn Reson Med. 2000;43(2):319–323. doi: 10.1002/(sici)1522-2594(200002)43:2<319::aid-mrm22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 46.Pipe JG, Farthing VG, Forbes KP. Multishot diffusion-weighted FSE using PROPELLER MRI. Magn Reson Med. 2002;47:42–52. doi: 10.1002/mrm.10014. [DOI] [PubMed] [Google Scholar]
- 47.Alsop DC. Phase insensitive preparation of single-shot RARE: Application to diffusion imaging in humans. Magn Reson Med. 1997;38:527–533. doi: 10.1002/mrm.1910380404. [DOI] [PubMed] [Google Scholar]
- 48.Tyszka JM, Frank LR. High-field diffusion MR histology: Image-based correction of eddy-current ghosts in diffusion-weighted rapid acquisition with relaxation enhancement (DW-RARE) Magn Reson Med. 2009;61:728–733. doi: 10.1002/mrm.21876. [DOI] [PMC free article] [PubMed] [Google Scholar]