Abstract
Epistasis could be an important source of risk for disease. How interacting loci might be discovered is an open question for genome-wide association studies (GWAS). Most researchers limit their statistical analyses to testing individual pairwise interactions (i.e., marginal tests for association). A more effective means of identifying important predictors is to fit models that include many predictors simultaneously (i.e., higher dimensional models).
We explore a procedure called screen and clean (SC) for identifying liability loci, including interactions, by using the lasso procedure, which is a model selection tool for high dimensional regression. We approach the problem by using a varying dictionary consisting of terms to include in the model. In the first step the lasso dictionary includes only main effects. The most promising SNPs are identified using a screening procedure. Next the lasso dictionary is adjusted to include these main effects and the corresponding interaction terms. Again, promising terms are identified using lasso screening. Then significant terms are identified through the cleaning process. Implementation of SC for GWAS requires algorithms to explore the complex model space induced by the many SNPs genotyped and their interactions. We propose and explore a set of algorithms and find that SC successfully controls Type I error while yielding good power to identify risk loci and their interactions. When the method is applied to data obtained from the Wellcome Trust Case Control Consortium study of Type 1 Diabetes it uncovers evidence supporting interaction within the HLA class II region as well as within Chromosome 12q24.
Keywords: association test, gene-gene interaction, lasso, model selection
Introduction
With the advent of relatively-inexpensive molecular methods for genotyping, genome-wide association studies (GWAS) have been carried out with notable success. Although the primary interest in GWAS is to identify single nucleotide polymorphisms (SNPs) that are directly associated with a disease, there is growing evidence supporting the occurrence of epistasis and its contribution to risk for complex disease [Evans et al., 2006; Manolio and Collins, 2007]. Consequently, there is much interest in searching for interactions between two or more SNPs [Cordell, 2009]. The search for loci that interact is typically conducted in a candidate gene study or a genome-wide association study. Several strategies are available, including exhaustive searches [Marchini et al., 2005], data mining [Strobl et al., 2007], and Bayesian model selection [Zhang and Liu, 2007].
Due to the large number of potential comparisons for interactions, however, an exhaustive search involving all combinations of two or more markers across the genome is daunting [Cordell, 2009]. Exploring models fitting main effect and interactions simultaneously in the setting of GWAS are impractical or even impossible, depending on the complexity of the models evaluated. One natural way to reduce the computational load is to adopt two-stage strategies [Marchini et al., 2005; Kooperberg and LeBlanc, 2008; Hoh et al., 2000], in which a small number of promising SNPs are selected at the first stage (henceforth candidate SNPs) and the higher order interactions are only considered among these candidate SNPs. With these strategies, the question of how to choose the promising SNPs at the first stage is crucial. Evans et al. [2006] investigate complex epistatic models and determine conditions for which it is challenging to capture the right terms to include in the second stage of a two-stage approach. It is not clear how often these conditions arise in practice because many genetic interactions demonstrate substantial marginal effects. We explore the potential of two-stage searches using a new statistical approach.
Our aim is to find a parsimonious model including SNPs, pairs of SNPs, and even higher order interactions that best explains the phenotype. This multivariate model selection approach can improve performance over tests for individual SNPs because it decreases the unexplained variance in the model. It has long been recognized that failing to account for these sources of heterogeneity can reduce the power to detect genetic factors in both linkage and association studies [Chatterjee et al., 2006; Hoggart et al., 2008]. In addition, a model selection approach will tend to include fewer spurious results because a SNP or multiple SNP interaction will only be included in the model if it substantially improves prediction beyond that obtained from the terms already included. A computationally efficient method for model selection is the lasso method, which is a tool for high dimensional regression [Tibshirani, 1996].
A good model identifies a set of SNPs and interactions between SNPs (covariates) that predict the phenotype. A parsimonious model tends to err on the side of simplicity, including only a subset of predictive SNPs, while a complex model tends to include too many SNPs, some having no impact on risk. Like stepwise regression, the lasso can explore models with more covariates than observations, but the lasso is a “less greedy” procedure than stepwise regression in that it tends to find less complex models. As it searches the model space it can both drop and add covariates. Using a computationally efficient procedure, the algorithm returns a suite of solutions, ranging from parsimonious to complex, indexed by a complexity parameter. Thus, using the lasso, the problem of identifying genetic variants associated with the phenotype is equivalent to selecting a complexity parameter between 0 and 1. The chosen parameter identifies a single solution from among the range of solutions suggested by the algorithm. A good choice corresponds to a model that controls the Type I error rate and yet attains good power. The complexity parameter is typically chosen by statistical sampling procedures, such as cross-validation. This approach yields a model with good predictive power, but the model often includes extra terms and thus it has a high Type I error rate [Devlin et al., 2003].
Due to limited research funds, or as a result of how the research unfolds, GWAS are conducted in stages. These multistage designs help to identify SNPs truly affecting risk by winnowing, at each stage, the list of associated SNPs. The same design feature can be used to improve the Type I error rate of the lasso.
A lasso-based procedure called Screen and Clean (SC) incorporates multiple stage experimental designs into the lasso procedure, attaining good power and yet controlling for spurious findings for models with only main effects [Wasserman and Roeder, 2009]. The SC procedure first screens for an inclusive model among the immense class of possibilities using the lasso, then it cleans the lasso-solution, removing terms from the model, using a traditional hypotheses testing approach. Screening is performed on stage 1 data and cleaning on stage 2 data. By exploiting the two-stage design, an optimal model can be discovered, overcoming a serious hurdle to using lasso for GWAS. SC has the important statistical feature that it finds a consistent model that controls the Type I error for main effects [Wasserman and Roeder, 2009]; if all of the SNPs are genotyped at each stage of the study, a multi-split refinement of SC is available [Meinshausen et al., 2008]. We examine the properties of this alternative.
In this paper, we follow the idea of incorporating multiple stage experimental design into the lasso procedure and expand the SC procedure to select optimal models with main and interacting effects. We focus on pairwise interactions, however, the principal of hierarchical model selection extends naturally to higher order interactions. The extended SC procedure is able to control the Type I error and attain good power for models with interactions, just as it does for models with only main effects. SC is extended in two important ways: first, we incorporate SNP-SNP interactions; and second, we devise a computationally efficient approach to the problem that scales successfully to GWAS. The set of SNPs or SNP-SNP interactions considered at a given step of the lasso regression model is called the dictionary (D). We use a dictionary that expands and contracts at each step of development. The method is a powerful alternative to marginal methods that test each SNP or pair of SNPs individually [Kooperberg and LeBlanc, 2008; Lin, 2006]. The method builds on published multivariate regression ideas [Wu et al., 2009]. It differs in that the proposed approach controls Type I error. Competing lasso procedures do not provide valid p-values. We illustrate SC using the publicly available genome-wide data on Type 1 diabetes data from Welcome Trust Case Control Consortium [The Welcome Trust Case Control Consortium, 2007].
Methods
Stages and Dictionaries
For a two-stage study design N1 subjects are genotyped at L SNPs in stage 1 and N2 subjects are genotyped at stage 2. The purpose of the second stage is to validate the findings of stage 1. As genome-wide platforms become more cost effective both stages are likely to yield genotypes for the whole genome. In this scenario we can use all of the data efficiently by performing multiple-splits of the data, repeating the screen and clean procedure.
For simplicity we assume that the L measured SNPs are coded for the additive model (X = 0, 1 or 2), but our results extend naturally to other genetic models.
In the interest of parsimony we use statistical interactions synonymously with SNP-SNP interactions, even though epistasis can be considerably more complex in reality [Phillips, 2008; Cordell, 2002]. Let Y be a phenotype which can be either binary or quantitative. We consider main effect models with
| (1) |
where g is an appropriate link function. Likewise we consider interaction models with
| (2) |
Let S = {j : βj ≠ 0, j ∈ 1, … , L} ∪ {(i, j) : βij ≠ 0, (i, j) ∈ 1, … , L} be the set of terms associated with the phenotype either as main effects or interactions. We assume that the number of terms associated with the phenotype is small. Our goal is to identify these terms.
In a traditional GWAS, each SNP is tested for association and hence the dictionary (D) is the full set of SNPs that passes quality control criterion. In a multi-step statistical procedure, the dictionary can contract if we discard terms that show little evidence of association in a previous step. Or it can also expand, if we add additional terms, such as interactions. We indicate the set of covariates in the dictionary after contraction or expansion by and , respectively. In this manner the many promising avenues of a huge dictionary can be explored without directly investigating the whole space. Naturally this approach works better if the true model is hierarchical (i.e., associated interactions are accompanied by main effects). Even when the true model is not hierarchical, however, models with strong interactions often demonstrate weak main effects and hence are approximately hierarchical.
Screen and Clean
Screen and Clean Illustration
A variable dictionary is critical when exploring the model given in equation (2) because it is usually not possible to fit the full model simultaneously due to the large number of covariates. To accommodate the high-dimensional challenge, we consider a statistical procedure that employs a hierarchical search. At step 1, the dictionary consists of all SNPs D = {X1, X2, … , XL} entered as main effects. Those that exceed a threshold for inclusion based on lasso screening are recorded as the candidate SNPs. At step 2, the dictionary consists of the candidate SNPs identified in step 1, plus all pairwise interactions of these terms. In summary, the dictionary contracts () in step 1, and based on these results the dictionary expands to include interactions,
Finally, terms in the resulting dictionary are tested for association.
To illustrate the concept of a contracting and expanding dictionary in action we preview SC in two selected simulation data sets (A and B). We chose these two data sets to contrast what happens when screen happens to be too liberal (A) versus too strict (B). The algorithm employed in this example, which is subsequently described in detail, is SC for interactions. Two data sets are drawn from the model g(E[Y∣X]) = β(X5X6 + X10X11). For each model, we generate 400 individuals, each with 15 SNPs coded using an additive model X1, …, X15.
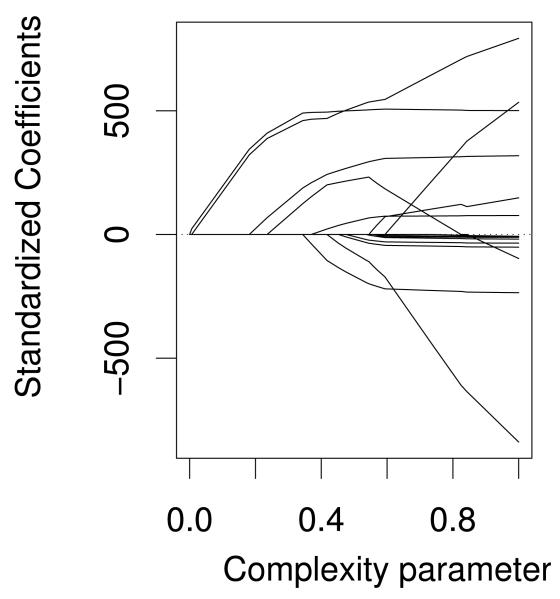
The lasso plot (Fig. 1) displays the family of solutions provided by the lasso algorithm for data set A in the initial screen of the dictionary consisting of all 15 SNPs. Let and denote the coefficient of the j’th term in the dictionary, estimated by lasso and least squares, respectively. The complexity parameter, or tuning parameter, γ is defined as . As the complexity parameter moves from 0 to 1, new terms are introduced to or dropped from the model. Typically the model moves from parsimonious to rich as this parameter increases. The plotted curves depict the standardized regression coefficients as a function of the complexity parameter γ. Each of them starts at 0, indicating that for γ = 0 the model includes no covariates. As γ increases, 2 covariates enter the model with large positive coefficients indicating that these terms have a strong positive association with the phenotype. As γ increases to 0.24, 2 more terms enter the model with positive coefficients. In fact, any choice of the complexity parameter between 0.24 and 0.34 will yield only the 4 terms actually needed to form the 2 interactions in the true model (5,6,10,11). At γ = 0.83 the fourth SNP is dropped from the model; but this term eventually re-enters the model with a negative coefficient. As γ increases to 1, the remaining 11 terms enter the model, but the pattern of coefficients is illogical and indicative of a model that includes several correlated terms, many of which are uninterpretable. Cross validation yields a complexity parameter of 0.45. With this choice the model identifies 7 candidate SNPs (4 true, 3 spurious). It is typical for cross-validated screening to yield an overly rich model [Devlin et al., 2003; Wasserman and Roeder, 2009]. This is why we need the Clean step of the procedure.
Figure 1.
Family of Solutions from the Lasso Algorithm. As the complexity parameter increases, SNPs are enter into (or drop out of) the model, one by one. Likewise, the complexity parameter determines the attenuation factor. At 0, all coefficients are 0. As the complexity parameter increases, the lasso coefficients approach the least squares solution. The traces plot each standardized coefficient as it enters the model and becomes less attenuated. Using cross-validation, a complexity parameter is selected that corresponds to a particular solution chosen from the family of solutions.
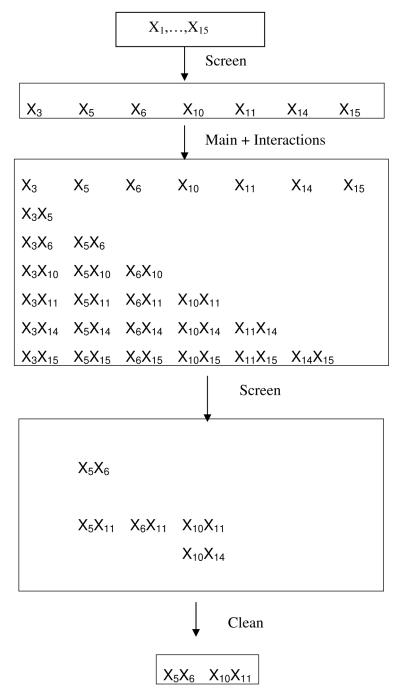
The flow chart (Fig. 2) depicts the expansion and contraction of the dictionary at each step of the SC analysis for data set A. After the initial screen, we contract the dictionary by removing all main effects not identified by the cross-validated model. We also expand the dictionary by adding all 21 pairwise interactions derived from the main effects discovered in the initial screen (Fig. 2). Applied to this dictionary, screen identifies a dictionary consisting of 5 interactions. Finally, using an independent sample of simulated data, the clean step of the procedure removes the 3 spurious interactions. The final model discovers the truth, even though the initial screen, based on cross-validation, included three spurious terms.
Figure 2.
Screen and Clean Flowchart from Simulation A. Step 1: all 15 SNPs are in the model dictionary. Step 2: Screening removes all but 7 terms. Step 3: Dictionary includes these 7 main effects, plus pairwise interactions. Step 4: Screening removes all but 5 interactions. Step 5: Cleaning removes all but 2 interaction. This is the estimated model, which is also the true simulation model.
By chance, for set B, screen prunes the initial 15 SNP dictionary to 3 of the 4 causal SNPs, missing SNP 5 due to a lack of power. At the next step, the dictionary adds the 3 pairwise interaction terms corresponding to these main effects. With this dictionary of 6 terms, screen drops the main effects, but retains all 3 interactions. Finally, in the clean step, the model settles on one effect (X10X11). This is a true effect; the other true effect can not be discovered because the model did not identify X5 in the screen step for main effects.
Screen and Clean Procedure
To describe the SC procedure we require notation for the number of variables under consideration. Thus we let #A denote the number of variables in a set A. The SC procedure (SCm) designed for the main effects model given in equation (1) corresponds with a two stage experimental design. Step 1: set the upper limit of the number of covariates to enter the screen process, Lu. This helps us to deal with the computational load; we generally set Lu=5000 (see more discussion in simulations). If #D > Lu, perform marginal tests on each effect in D and select the Lu effects with the smallest p-values. Include only these terms in the revised dictionary. Step 2: using data from stage 1, the model g(E[Y|X]) = β0 + ∑j∈D βjXj is applied to the dictionary. The lasso identifies a set of indices for each value of the complexity parameter γ. The complexity parameter is selected by cross-validation. The resulting dictionary, , includes all the terms for which when . Step 3: using data from stage 2, find the least squares estimate for the terms in . From this analysis, obtain Tj, the traditional t-statistic obtained from the least squares analysis of the screened model, which includes terms. Clean the model of superfluous terms by selecting , in which c = Zα/(2m).
To discover interactions as in equation (2), we extend the SCm algorithm to handle dictionaries with interactions. We call the procedure SCi. Repeat Steps 1 and 2 as for SCm. Step 3: obtain . If , perform marginal tests on each term in and update to include only the Lu terms with the smallest p-values. Again using data from stage 1, fit a model including all of the main effects and interactions delineated by these Lu best terms. For each γ ∈ (0, 1) we obtain a contracted dictionary including . Select by leave-one-out cross-validation, and use to define the dictionary, , to be used for the final step; let m* = #S. Step 4: using the second stage data, clean the model as follows. Find the least squares estimate for the model defined by S. The chosen model is {j : ∣Tj∣ > c} ∪ {(i, j) : ∣Tij∣ > c}, where c = Zα/(2m*), and Tj and Tij are the t-statistics for main effects and interactions, respectively. The resulting procedure is designed to control Type I error at level α.
Genome Wide Association
Multivariate methods such as SC do not scale directly to the immense computational burden imposed by a GWAS (results not shown). SC is computationally challenged by large numbers of covariates (p) and large numbers of subjects (n). With n=400 and p=1000, the procedure takes less than one minute to perform; but as the number of covariates increases to 5,000 and n increases to 1,000, the procedure requires about an hour. Approached directly, the computational challenge for hundreds of thousands of SNPs is prohibitive; however, this does not prevent us from employing SC in a GWAS. When the dimension of the problem is large we adjust the algorithm to obtain the results in a reasonable time. The adjustments include pre-selection of SNPs to those with promising marginal signals, and reducing the effort involved to perform cross-validation. These adjustments can be combined together or used individually.
Prescreening can be used to limit the number of SNPs in the dictionary. Based on a marginal test of association, most of the SNPs can be eliminated from consideration. We suggest prescreening the dictionary to include only those SNPs with a marginal p-value less than p0. Because SC is based on a 2-stage process, prescreening has no impact on the Type I error of the procedure. In addition, the number of SNPs entered in SC can be reduced by restricting the analysis to tag SNPs [de Bakker et al., 2005; Rinaldo et al., 2005].
The computational effort increases quadratically as a function of n and p. Consequently we view p ≈ 5000 as a practical upper limit on the number of covariates for n ≈ 2000. This is due to two computational features in SC. Like a stepwise procedure, the lasso searches the covariate sets by adding and dropping covariates sequentially. The default maximum number of steps taken by the lasso algorithm increases with the number of samples and the number of variables in the model. This default value can be lowered to obtain an approximate solution; however, the algorithm might then fail to discover some subtle signals in the data. Second, the computational cost of the cross-validation increases linearly in the number of samples when using leave-one-out cross validation. For large n we suggest k-fold cross-validation, which leaves n/k observations out in each step of the algorithm instead of leaving one out [Hastie et al., 2001]. We obtain good results using k of 30 to 40.
Here we summarize the SCi algorithm, with adaptations that facilitate analysis of GWAS.
SCi Algorithm
- Create a dictionary D including all SNPs with minor allele frequency (MAF) > 0.01. To ensure that #D < Lu, restrict this set by including only
- those SNPs with marginal p-values < p0m,
- tag SNPs.
Using stage 1 data, screen D for main effects to obtain . In cross-validation, restrict the class of models to those with R1 or fewer terms.
- Obtain by including pairwise interactions. Optionally restrict this set by
- including only those interactions with marginal p-value < p0i.
Screen to obtain . Again, in cross-validation, restrict the class of models to those with R2 or fewer terms.
Using stage 2 data, clean S.
The final model includes those terms with cleaned p-values < α. These p-values have been corrected for multiple testing.
Multiple-split SC
When genotypes for the full panel of SNPs are available for every individual in the data there is no obvious split of the data into one set for screening and another set for cleaning. In this scenario, the SC analysis results vary depending on how this single-split is chosen. For this scenario, Meinshausen et al. [2008] extended the single-split SC procedure to a multi-split procedure. The analysis involves randomly splitting the data repeatedly, running SC for each split to obtain a set of p-values for each covariate, and then obtaining a single composite p-value from the sample of p-values.
For b = 1, … , B,
randomly split the data into two portions: for screen and for clean;
using , screen to find the variables, S(b), with ;
- clean using ;
- based on the results of Clean, compute the p-values for variables in S(b);
- set for variables not in S(b);
- obtain a p-value that is corrected for multiple testing
.
Thus far, the algorithm is the usual SC procedure applied repeatedly over B splits of the data. Typically a variant associated to the phenotype will produce a distribution of , b = 1, … , B including several small p-values and several 1’s. Thus we cannot obtain a single p-value for each variant by taking the mean of the ’s. The alternative is to examine the distribution of p-values, from which a summary p-value can be obtained. Meinshausen et al. [2008] recommend the following algorithm which provides a conservative overall p-value:
obtain the empirical quantile function qδ for δ in the interval [0.05, 1];
find δ* to minimize the function qδ/δ;
set Pj = min (4 × qδ* /δ*, 1).
The multiplication by 4 accounts for selecting the quantile that yields the smallest p-value [Meinshausen et al., 2008].
Other Features
To control for confounding effects of population structure we suggest including eigenvectors estimated using either principal component analysis [Price et al., 2006] or spectral analysis [Lee et al., 2009]. For case-control data there is also the option of matching cases and controls by estimated ancestry and using the conditional logit model on the matched strata [Luca et al., 2008].
The lasso is designed for linear regression and quantitative traits. For dichotomous traits, logistic regression replaces linear regression naturally at a number of steps in the algorithm. This works conveniently for univariate p-values and cleaning of the data. When screening a large model space the computational challenge is greater for logistic regression. Wu et al. (2009) describe an approach that they call lasso penalized logistic regression. Following the classification literature [Hastie et al., 2001], we have found that linear regression provides a practical alternative to logistic regression even when the response variable is binary.
Results
Simulation Results with a Moderate Number of SNPs
We generate 400 individuals, each with 1000 SNPs with genotypes encoded by having X = 0, 1, or 2 minor alleles. We use half of the samples to screen (stage 1) and the remaining half to clean (stage 2). The SNPs are block-wise dependent with 200 blocks of size 5. Linkage disequilibrium within blocks is set low (Pearson’s correlation coefficient [Devlin and Risch, 1995] ρ = 0.25) and high (ρ = 0.75). We generate a quantitative phenotype Y according to four models, with random error ∊ ~ N(0, 1). Models M1, M2 and M3 each contain multiplicative interaction terms with varying numbers of SNP-SNP pairs involved in the interaction. For ease of exposition, the coefficient β is constant for each term in all models, but for model M3 the strength of the association decreases by a multiplier for each successive SNP pair.
For computational efficiency we performed our simulations using a relatively small sample size (400) and a large genetic signal (β ranged from 0.25 to 2.0.) In regard to their power, these choices are statistically equivalent to a more realistic scenario with sample size 1500 and genetic heritability attributable to each interaction ranging from 0.1% to 7%. We performed 1000 simulation for each combination of model, β, and ρ.
Define power as the fraction of discoveries of interactions over the total number of interactions in the model; the false discovery rate (FDR) as the fraction of false discoveries of interactions among the total number of discoveries of interactions; and the Type I error rate as the fraction of the simulations with at least one false discovery over the total number of simulations.
We evaluate the Type I error of SCm and SCi, using data simulated based on model M0 (Table I). SCm successfully controls the Type I error for each condition explored. When the marginal effects become more substantial, the Type I error of SCi increases slightly over the nominal level.
Table I.
Type I error in model M0 for Screen and Clean for main effects only (SCm), Screen and Clean for main effects and interactions (SCi). Levels of disequilibrium (ρ) and strength of association (β) vary as indicated.
| β = 0.5 | β = 1 | β = 2 | ||||
|---|---|---|---|---|---|---|
| Type I | SCm | SCi | SCm | SCi | SCm | SCi |
| ρ = 0 | 0.06 | 0.06 | 0.06 | 0.07 | 0.05 | 0.11 |
| ρ = 0.25 | 0.05 | 0.04 | 0.04 | 0.09 | 0.05 | 0.10 |
| ρ = 0.75 | 0.06 | 0.04 | 0.07 | 0.05 | 0.05 | 0.09 |
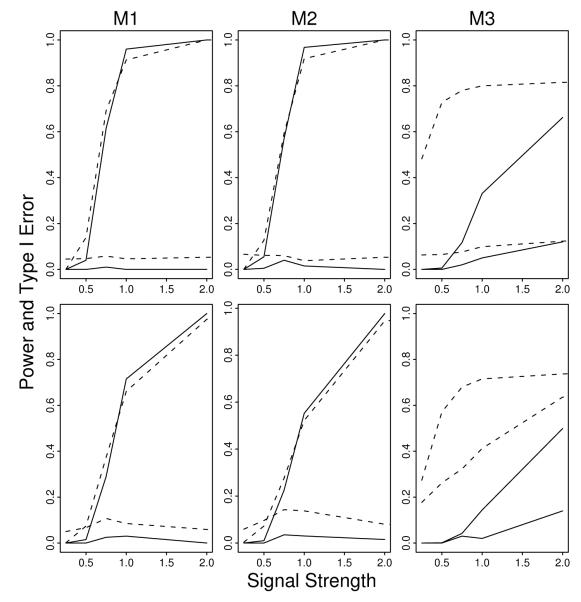
For most configurations of parameters in models M1-M3, SCi controls Type I error well (Fig. 3; dashed lines). The procedure has low power for small β, but power increases rapidly as the signal grows. Comparing panels moving left to right it is apparent that more complex models have lower power than simpler ones. Poor performance for SCi occurs when both the block correlation and the model complexity are high (bottom-right panel). This suggests that better performance might be obtained if the analysis were performed using only tag SNPs.
Figure 3.
Power and Type I error rates for the (one-split) SC method and the multi-split SC method. Plotted against the strength of the signal (β) are power (top two lines) and Type I error (bottom two lines) for one-split SCi (dashed lines) and the multi-split SCi (solid line). Top (bottom) row is low (high) correlation within blocks. Columns correspond to models of increasing complexity (M1, M2, and M3).
Next, for the same conditions, we compare the performance of the method using a single-split versus the multi-split procedure (Fig. 3; dashed versus solid lines). For this scenario, we performed 200 simulations with SCi repeated 5 times. For all three models the Type I error improved substantially with the multi-split procedure. For model M3 controlling the Type I error, came at the cost of a substantial loss in power, especially when the SNPs had a higher correlation. Moreover, the Type I error is still slightly inflated when the correlation is high.
To combat this problem we repeated the experiment on model M3 using tag SNPs (ρ < 0.1). The power is essentially unchanged from when all SNPs were included, but Type I error is successfully controlled (results not shown).
Simulations with Large Numbers of SNPs
To demonstrate the application of SC to genome wide association studies, we simulated data sets with 1500 samples and 100,000 SNPs. The SNPs are generated to simulate tag SNPs that possess LD structure similar to a Markov chain: nearest neighbor SNPs have correlation ρ = 0.3. We set minor allele frequency at 0.3; in practice SNPs with a smaller minor allele frequency will require a bigger signal to yield the same power. We use two-thirds of the samples to screen (stage 1) and the remaining one-third to clean (stage 2). (This is just one option for splitting the data. Using a greater fraction of the data for cleaning might be advantageous.)
To assess the power of SC in GWAS we simulated 100 data sets for two more complex models. For model M4, 100 causal SNPs fall into sets of 10 SNPs of equal signal strength, with 10 levels ranging from low to high signal. For model M5, 25 pairs of SNPs are grouped into sets of 5, with strength of interaction signal set at 5 levels ranging from low to high. We use simulations from model M4 to evaluate tests for main effects and model M5 for interaction effects.
For model M4, let X{Sj} = {Xj1, … , Xj10}, j = 1, … , 10, represent 10 sets of 10 randomly selected SNPs for each simulation. For each j, the effect size is j × β, so that, in total, 100 SNPs affect Y , including 10 SNPs at each level, with β = 0.3, i.e.,
Assignment of the 100 causal SNPs vary by simulation.
For model M5, let , j = 1, … , 5, represent 5 sets of 5 randomly selected pairs of SNPs for each simulation. For each j, the effect size is j × β, j = 1, … , 5, so that, in total, 25 SNPs affect Y , including 5 pairs of SNPs for each level, with β = 0.9, i.e.,
Again the 25 pairs of causal SNPs vary by simulation.
For each simulated data set, using stage 1 data, we first pre-screen the SNPs and include only those SNPs with a marginal p value less than 0.05, effectively reducing the size of the dictionary to approximately 5000. In screen, we use the default parameters in lasso and leave-one-out cross-validation and otherwise follow the described procedure for SC in GWAS.
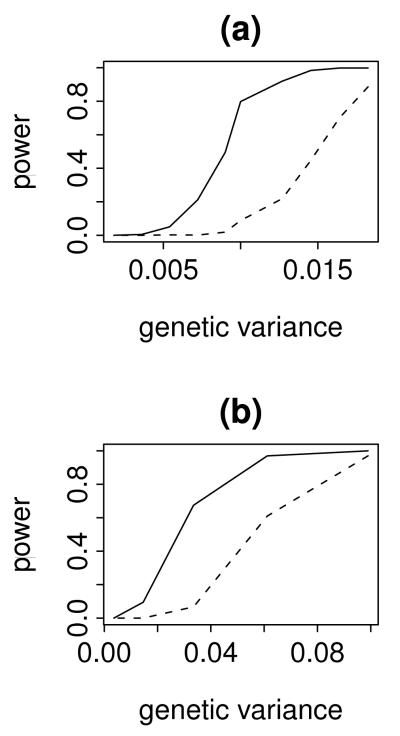
For main effects SCm achieves reasonable control of both Type I error (0.077) and the FDR (0.0016). For interactions, SCi has a higher than desired Type I error (0.13), but these errors are fairly uncommon compared to true discoveries, as evidenced by the well controlled FDR (0.014). To assess power, we used SCm on Model M4 and SCi on Model M5 (Fig. 4). Notice that the power to detect main effects is much greater than the power to detect interactions.
Figure 4.
Power for SCm (a) and SCi (b) methods. Genetic variation is the average fraction of the total genetic variance attributable to a SNP (a) or a SNP-SNP interaction (b).
To assess the advantages of multivariate model selection we compared SC with methods designed for marginal testing of main effects [Lin, 2006] and interactions [Kooperberg and LeBlanc, 2008]. Both marginal methods use the first stage data to screen for important main effects. The second stage data is then combined with the first stage data for a test of each effect selected in the screening process. In a test for main effects, Lin’s method had false positive rates comparable to SCm (Type I error = 0.088, FDR = 0.003), but substantially lower power (Fig. 4, top panel). Kooperberg and LeBlanc’s (KL) method tests for interactions formed from all pairwise combinations of screened main effects. This method showed false positive rates equivalent to SCi (Type I = 0.18, FDR = 0.022), but considerably lower power (Fig. 4, bottom panel).
We also explored some of the simpler non-additive models investigated by Evans et al. [2006]. We chose these models because they are derived from pairwise combinations of recessive and dominant single SNP models. The recessive-recessive (RR) only has an effect if all four minor alleles are present; the recessive-dominant (RD) has an effect if SNP one has both minor alleles and SNP two has at least one minor allele; the dominant-dominant (DD) has an effect if both SNPs have at least one minor allele; and the dominant-recessive-dominant (DRD), has the effect if at least three minor alleles are present. A two-step analysis is likely to fail if the main effects explain an insufficient fraction of the variance contained in the interaction. For this set of models, the fraction of the variance attributable to main effects and epistatic effects for each varies (Table II). For comparison we include a model with the core element we used for most of our simulations Y = βX1X2 + ∊; we label this model M, because it is based on a multiplicative interaction. Model M is most similar to models DD and DRD, hence we expect SC to be most challenged by the recessive and partially recessive models. In our simulations, as expected, power is similar to model M for models DD and DRD, but not promising for models RR and RD.
Table II.
The fraction of the genetic variance attributable to each main effect and the epistatic effect in five genetic models. The models are a selection of those explored in Evans et al. (2006): recessive-recessive (RR), recessive-dominant (RD), dominant-dominant (DD) and dominant-dominant, except that the double heterozygote does not have the effect (DRD) and multiplicative interaction (M).
| Model | RR | RD | DRD | DD | M |
|---|---|---|---|---|---|
| lucus 1 | 0.083 | 0.046 | 0.26 | 0.34 | 0.32 |
| locus 2 | 0.083 | 0.49 | 0.26 | 0.34 | 0.32 |
| epistasis | 0.83 | 0.47 | 0.48 | 0.32 | 0.37 |
Next we try a simulation of case and control data. We simulate 600 cases and 600 controls from a population of SNPs with first-order Markov dependency ρ = 0.3, MAF = 0.3. The data were generated using the following model with 5 pair-wise interactions of randomly selected SNPs.
We simulated two types of data sets, one with 5000 SNPs and the other with 50000 SNPs. In the situation of 50000 SNPs, we first select top 5000 SNPs by marginal test using logistic regression. Then, for both types of the data sets, we apply the SCi procedure. The power was essentially equivalent for both scenarios. The Type I error increased from approximately 0.05 to 0.15.
Analysis of Type 1 Diabetes (T1D) data
The Wellcome Trust Case Control Consortium [WTCCC, 2007] data includes 1963 cases with T1D and 2938 controls (post QC) obtained from people living in Great Britain who self-identified as white Europeans; see WTCCC [2007] for details about the sample and quality control procedures. The samples were genotyped with the GeneChip 500K Mapping Array Set (Affymetric chip).
At least twelve regions in the genome have strong statistical support in the literature for association with T1D (i.e., 1p13.2, PTPN22; 2p24.2, IFIH1; 2q33.2, CTLA4; 6p21.32, HLA class II; 6q15, BACH2; 10p15.1, PRKCQ; 11p15.5, INS; 12q12.2, ERBB3; 12q24.13, SH2B3/PTPN11; 15q25.1, CTSH; 16q13.13, CLEC16A; 18p11.21 PTPN2) with reported p-values less than 10−8 [Hindor et al., 2009]. Univariate analyses of the WTCCC data show genome-wide significance for four of these (i.e., PTPN22, HLA class II, ERBB3, and the SH2B3/PTPN11 region). In addition rs12708716 (CLEC16A) on Chromosome 16 is borderline significant. The INS gene, which is not tagged well by this array, does not show evidence of association in these data [WTCCC 2007].
We reanalyzed these data using the multi-split SC approach with 56 random splits of the data (1/3 screen and 2/3 clean). Of the 469,612 SNPs passing WTCCC QC, we also removed 594 SNPs with poor genotype clustering patterns and all SNPs on chromosome X. Next we restricted the dictionary to those with univariate p-value less than 0.017. From the remaining 10,000 SNPS, we chose SNPs using H-clust, set to pick tag SNPs with squared correlation less than 0.04 and minor allele frequency greater than 0.01; for a cluster of SNPs in LD, H-clust used preference scores based on the univariate p-values for association of each SNP [Rinaldo et al., 2005]. In this way, our tag SNP selection process includes the SNP most likely to be associated with T1D within each LD block. After this process, we further ensured that the SNP dictionary included no SNPs with squared correlation greater than 0.045 on the same chromosome. The resulting dictionary included 3437 SNPs. We recoded the genotype data for the additive model. For the SCi algorithm, to keep the model size computationally manageable we used R1 = 250 and R2 = 2000.
Our results are similar to the WTCCC’s univariate analysis (Table III). All five of their best signals also appeared as significant effects in our model. In addition, on Chromosome 4, SNP rs17388568, which was borderline significant (5.7 × 10−7) using conditional logistic regression, is also borderline in our analysis (multiple testing corrected p-value = 0.35). Our model also identified 4 additional SNPs in the HLA region. Because we restricted our analysis to tag SNPs the LD between these SNPs is minor, suggesting the signal in the MHC is due to multiple variants.
Table III.
Best SNP main effects found via SCm and corrected for multiple testing. Nominal univariate p-values, not corrected for multiple testing, are obtained using logistic regression.
| Chr | (bp) | Position SNP |
SC P-value |
Univariate P-value |
|---|---|---|---|---|
| 1 | 114105331 | rs6679677 | 5.6 × 10−14 | 5.1 × 10−25 |
| 4 | 123548812 | rs17388568 | 0.35 | 5.7 × 10−7 |
| 6 | 31735428 | rs2242655 | 1.8 × 10−2 | 5.4 × 10−6 |
| 6 | 32297010 | rs415929 | 1.8 × 10−2 | 2.9 × 10−5 |
| 6 | 32712350 | rs9272346 | 1.1 × 10−76 | 8.9 × 10−122 |
| 6 | 32910181 | rs241432 | 3.5 × 10−4 | 1.7 × 10−6 |
| 6 | 33111665 | rs448733 | 2.7 × 10−2 | 1.1 × 10−5 |
| 12 | 54756892 | rs11171739 | 2.6 × 10−5 | 1.3 × 10−11 |
| 12 | 110971201 | rs17696736 | 1.3 × 10−2 | 1.0 × 10−11 |
| 16 | 11115395 | rs9746695 | 1.4 × 10−3 | 9.6 × 10−9 |
Applying SCi we identified three SNP-SNP interactions as significant (Table IV), corresponding to univariate SNP-SNP p-values that would not have been sufficient to attain genome-wide significance in a standard analysis. This suggests that the SCi procedure can indeed be more powerful than a series of univariate tests, especially when searching through the vast model space of SNP-SNP interactions.
Table IV.
Best SNP interaction effects found via SCi and corrected for multiple testing. Nominal univariate p-values, not corrected for multiple testing, are obtained using logistic regression.
| Chr | Position (bp) |
SNP | Chr | Position (bp) |
SNP | SC P-value |
P-value | Univariate P-value |
r2 |
|---|---|---|---|---|---|---|---|---|---|
| 6 | 32911818 | rs241429 | 6 | 32910181 | rs241432 | 3.5 × 10−66 | 8.2 × 10−5 | 1.7 × 10−6 | 0.02 |
| 6 | 32911818 | rs241429 | 6 | 32712350 | rs9272346 | 1.9 × 10−25 | 8.2 × 10−5 | 9.0 × 10−122 | < 0.001 |
| 12 | 110918887 | rs11066119 | 12 | 110971201 | rs17696736 | 2.3 × 10−15 | 9.6 × 10−5 | 1.0 × 10−11 | < 0.001 |
Two of the pairs involve SNPs in the MHC region. Both of these include a SNP that was identified as main effects paired with another that did not demonstrate significant main effects (rs241429, univariate p-value of 8.2 × 10−5). The remaining interaction involves a pair of SNPs on Chromosome 12, one discovered as a main effect (rs17696736) that tags the SH2B3/PTPN11 region, paired with (rs11066119, univariate p-value of 9.6 × 10−5). Pairs of SNPs are not in linkage disequilibrium (Table IV). Moreover, because each of these variants is significant in the multivariate model, we can conclude that each variant exhibits a significant association with the phenotype, after accounting for all of the other variants in the model. The genotype by genotype counts support our findings (Supplementary Table A).
Our initial analyses of these data, conducted after removal of SNPs that failed standard QC measures but prior to removal of SNPs that failed visual QC inspection, identified several additional SNP-SNP interactions (results not shown). Unfortunately, these SNPs did not pass the visual QC inspection performed by WTCCC. From this we conclude that interactions are much more sensitive to poor genotype quality than tests of main effects, and care must be taken to thoroughly inspect the data for problems with genotype calling. Our reported results were conducted after removal of all SNPs with small univariate p-values that showed poor genotype clustering.
Discussion
A multivariate regression model can have greater power than a series of marginal tests to detect signals when multiple variables affect the outcome, as seen here and in other research [Chatterjee et al., 2006; Hoggart et al., 2008; Longmate, 2001; Ritchie et al., 2003; Millstein et al., 2006; Zhang and Liu, 2007]. Building on this idea we propose the SC algorithm, which identifies the most promising SNPs and interactions simultaneously using the lasso regression procedure. Because the class of models that includes all potential interactions is too large to be practical, we vary the dictionary of SNPs and SNP-SNP interactions considered at each step of the analysis. First only main effects are considered. Next we include interactions corresponding to SNPs that exhibit at least a weak main effect. Using an independent source of data, in the final step we look for replication of those terms that look most promising in the first step analysis. This approach lies somewhere between classical replication analysis and joint analysis of the data [Skol et al., 2006]. Contrary to joint analyses, only the replication data are used in the validation study, but SNPs and SNP-SNP interactions that go on to the second stage for validation need not exceed the threshold for genome wide significance in stage one.
We applied our procedure on data simulated with linkage disequilibrium and data similar to a GWAS. Because SC is designed to model the effects of a multiple gene system it provides good control of the type I error and false discovery rate even when the SNPs are in LD. Although many marginal tests are available in the literature, we compared our results with two methods that work well with data collected in two stages using a joint analysis approach [Kooperberg and LeBlanc, 2008; Lin, 2006]. In data simulated to mimic a GWAS with many SNP and SNP-SNP interactions present, the marginal methods lacked power relative to the SC approach. It is not surprising that a multiple regression approach has greater potential to handle these complex models than a sequential approach based on marginal tests because it reduces the variance and allows the causal SNPs, or SNPs highly correlated with causal SNPs, to compete for variance prediction against other SNPs having no impact.
From statistical theory and practice we know that regression models that include highly correlated predictors have myriad undesirable properties. Consistent with the theory our simulation results show that SC works better when linkage disequilibrium among SNPs is small. Moreover, including correlated SNPs increases the computational burden severely without adding substantial information. Thus, we recommend always using tag SNPs for SC. To enhance the chance that associated SNPs are included in the set of tag SNPs we suggest chosing the SNP with the smallest marginal p-value among any correlated set of SNPs [Rinaldo et al., 2005]. After applying the SC procedure to the tag SNPs, one can investigate all of the SNPs genotyped in the vicinity of the tag SNPs identified in the initial analyses.
Contrary to some methods in the literature, SC identifies promising main effects, followed by pursuit of epistatic effects. Of course, some kinds of epistasis do not lend themselves to a two-step approach because the majority of the genetic variance resides strictly in the interactions and hence the SNPs cannot be identified via single-locus tests of association [Evans et al., 2006]. For other epistatic models, however, the power to detect each locus using a single-locus strategy is high enough that a two-step strategy does have advantages. When using the lasso, success in finding individual SNPs in step 1 is further enhanced because the model space is multivariate. Nevertheless, if a model has low variance attributable to either of the two SNPs involved, there is little chance that the epistatic effect will be discovered with a two-step approach.
Our approach differed from that of Evans et al. in a number of ways. Regardless of the true model, we used an additive model for the SC. With this approach we gain power when the model is approximately additive by using fewer degrees of freedom, but we lose power when the model is far from additive. In contrast, Evans et al. use a genotype model which allows for multiple degrees of freedom for single locus and two-locus tests. Our simulations were designed for data collected from a two-stage experimental design. With this design it is assumed that only the most promising SNPs are evaluated at stage 2. Thus, SC is not limited to an additive model, but can be used for any genetic model or family of models. With SC the second set of data is utilized to clean the model of superfluous terms. The other approaches include an implicit correction for joint analysis of data from both stages of data. This correction is more powerful than the one utilized by Evans et al. in their simulations.
An option, which we did not pursue in this paper, is SC applied directly to a dictionary including all main effects and interactions without the benefit of screening. This approach is not computationally feasible if the number of SNPs is large. To bypass the hierarchical search and yet avoid severe computational hurdles one could combine the best features of marginal testing and the multivariate approach: define the preliminary dictionary to be the set of all possible pairwise interactions; contract this huge dictionary by testing for main effects and interactions using a marginal test to obtain a dictionary corresponding to terms with smallest p-values; and screen this dictionary using stage 1 data and clean using stage 2 data. This approach is not limited by the assumed hierarchical structure, but it has the disadvantage of being very computationally intensive [Purcell et al., 2007; Marchini et al., 2005].
In principle we would like to create even richer interaction dictionaries. One way to consider more SNPs, and yet achieve the advantages of the multivariate analysis, is to split the SNPs into subcategories that are more likely to be involved in interactions. For instance, we might chose subsets of SNPs from different pathways and create pathway dependent dictionaries [Bochdanovits et al., 2008; Emily et al., 2009]. With this approach, each dictionary is less likely to exceed the computational limit, and yet likely interactions are included in the screening process. This approach could be successful in discovering complex epistatic models.
Application of the method to data obtained from the Wellcome Trust Case Control Consortium study of Type 1 Diabetes cases and controls uncovered evidence supporting multiple HLA class II independent T1D associations within the HLA class I regions occurring at HLA-B and HLA-A [Valdes et al., 2005; Nejentsev et al., 2007; Howson et al., 2009]. Analyses of interacting SNP pairs discovered association occurring within HLA class II as well as within the Chromosome 12q24 region. The HLA region represents the largest genetic risk element for T1D as well as other autoimmune diseases [Klein and Sato, 2000]. A likely mechanism by which certain HLA alleles influence T1D susceptibility is related to their ability to bind and present autoantigens to autoreactive T-lymphocytes in the thymus [Todd et al., 1987; Morel et al., 1988; Nepom and Erlich, 1991]. Likewise, the Chromosome 12q24 region has been confirmed as associated with T1D [Todd et al., 2007; Barrett et al., 2009]. These studies have identified a large LD block, estimated at greater than 1.2Mb, harboring at least two genes with possible functional relevance to T1D, such as PTPN11 and SH2B3 [Todd et al., 2007; Smyth et al., 2008].
SNP pairs that interact to influence disease susceptibility may do so by mechanisms involving transcriptional control, mRNA processing, changes in amino acid sequence, or a combination of mechanisms. For example, HLA loci polymorphisms in the promoter region have been described that result in changes in expression [Beaty et al., 1995]. Non-HLA loci that influence T1D risk have also been linked to changes in expression (i.e., INS) as well as altered RNA processing (i.e., CTLA4) [Ounissi-Benkalha and Polychronakos, 2008]. In these examples altered expression has been proposed to affect autoimmunity by influencing negative selection of autoreactive T-lymphocytes [Fan et al., 2009]. The SNP pairs identified by our analyses may also impact gene expression, however, additional experiments will be needed to sufficiently characterize these elements in order to elucidate their mechanism of interaction with T1D risk.
Supplementary Material
Supplementary Table A. Genotype by genotype counts (expected counts) for those SNP-SNP pairs with significant interactions.
Acknowledgements
This work was funded by the National Institutes of Health grant MH057881 awarded to B.D. and K.R. and by the Department of Defense (grant W81XWH-07-1-0619) awarded to M.T.
Electronic References
- The R code is available at http://www.stat.cmu/~jwu/screenNclean/
- Hclust http://www.wpic.pitt.edu/WPICCompGen/hclust/hclust.htm.
References
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS, The Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty JS, West KA, Nepom GT. Functional effects of a natural polymorphism in the transcriptional regulatory sequence of hla-dqb1. Mol Cell Biol. 1995;15:4771–4782. doi: 10.1128/mcb.15.9.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochdanovits Z, Sondervan D, Perillous S, van Beijsterveldt T, Boomsma D, Heutink P. Genome-wide prediction of functional gene-gene interactions inferred from patterns of genetic differentiation in mice and men. PLoS ONE. 2008;3:e1593. doi: 10.1371/journal.pone.0001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Kalaylioglu Z, Moslehi R, Peters U, Wacholder S. Powerful multilocus tests of genetic association in the presence of gene-gene and gene-environment interactions. Am J Hum Genet. 2006;79:1002–1016. doi: 10.1086/509704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PIW, Yelensky R, Peér I, Gabriel SB, Daly JJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Wasserman L. Analysis of multilocus models of associaiton. Genet Epiodemiol. 2003;25:36–47. doi: 10.1002/gepi.10237. [DOI] [PubMed] [Google Scholar]
- Emily M, Mailund T, Schauser L, Schierup MH. Using biological networks to search for interacting loci in genomewide association studies. Eur J Hum Genet. 2009;11 doi: 10.1038/ejhg.2009.15. (doi: 10.1038/ejhg.2009.15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DM, Marchini J, Morris AP, Cardon LR. Two-stage two-locus models in genome-wide association. PLoS Genet. 2006;2:e157. doi: 10.1371/journal.pgen.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Rudert WA, Grupillo M, He J, Sisino G, Trucco M. Thymus-specific deletion of insulin induces autoimmune diabetes. The EMBO Journal. 2009;00:00–00. doi: 10.1038/emboj.2009.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The elements of statistical learning. Springer; New York: 2001. [Google Scholar]
- Hindorff LA, Junkins HA, Mehta JP, Manolio TA. A Catalog of Published Genome-Wide Association Studies. 2009 Available at: www.genome.gov/26525384.
- Hoggart CJ, Whittaker JC, De Iorio M, Balding DJ. Simultaneous analysis of all snps in genome-wide and re-sequencing association studies. PLoS Genet. 2008;4:e1000130. doi: 10.1371/journal.pgen.1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh J, Wille A, Zee R, Cheng S, Reynolds R, Lindpaintner K, Ott J. Selecting snps in two-stage analysis of disease association data: a model-free approach. Ann Hum Genet. 2000;64:413–417. doi: 10.1046/j.1469-1809.2000.6450413.x. [DOI] [PubMed] [Google Scholar]
- Howson JM, Walker NM, Clayton D, Todd JA, Diabetes Genetics Consortium Confirmation of hla class ii independent type 1 diabetes associations in the major histocompatibility complex including hla-b and hla-a. Diabetes Obes Metab. 2009;11(Suppl 1):31–45. doi: 10.1111/j.1463-1326.2008.01001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Sato A. The hla system. N Engl J Med. 2000;343:782–786. doi: 10.1056/NEJM200009143431106. Second of two parts. [DOI] [PubMed] [Google Scholar]
- Kooperberg C, LeBlanc M. Increasing the power of identifying gene x gene interactions in genome-wide association studies. Genet Epidemiol. 2008;32:255–263. doi: 10.1002/gepi.20300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AB, Luca D, Klei L, Devlin B, Roeder K. Discovering genetic ancestry using spectral graph theory. Genet Epidemiol. 2009 doi: 10.1002/gepi.20434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY. Evaluating statistical significance in two-stage genomewide association studies. Am J Hum Genet. 2006;78:505–509. doi: 10.1086/500812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmate JA. Complexity and power in case-control association studies. Am J Hum Genet. 2001;68:1229–1237. doi: 10.1086/320106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca D, Ringquist S, Klei L, Lee AB, Gieger C, Wichmann HE, Schreiber S, Krawczak M, Lu Y, Styche A, Devlin B, Roeder K, Trucco M. On the use of general control samples for genome-wide association studies: Genetic matching highlights causal variants. American Journal of Human Genetics. 2008;82:453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins F. Genes, environment, health, and disease: facing up to complexity. Hum Hered. 2007;63:63–66. doi: 10.1159/000099178. [DOI] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci influencing complex diseases. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- Meinshausen N, Meier L, Buhlmann P. P-values for high dimensional regression. 2008. arXiv:0811.2177v2.
- Millstein J, Conti DV, Gilliland FD, Gauderman WJ. A testing framework for identifying susceptibility genes in the presence of epistasis. Am J Hum Genet. 2006;78:15–27. doi: 10.1086/498850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel PA, Dorman JS, Todd JA, McDevitt HO, Trucco M. Aspartic acid at position 57 of the hla-dq beta chain protects against type i diabetes: a family study. Proc Natl Acad Sci USA. 1988;85:8111–8115. doi: 10.1073/pnas.85.21.8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejentsev S, Howson JM, M WN, Szeszko J, Field SF, Stevens HE, Reynolds P, Hardy M, King E, Masters J, Hulme J, Maier LM, Smyth D, Bailey R, Cooper JD, Ribas G, Campbell RD, Clayton DG, Todd JA, Wellcome Trust Case Control Consortium Localization of type 1 diabetes susceptibility to the mhc class i genes hla-b and hla-a. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepom GT, Erlich H. Mhc class-ii molecules and autoimmunity. Annu Rev Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- Ounissi-Benkalha H, Polychronakos C. The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends Mol Med. 2008;14:268–275. doi: 10.1016/j.molmed.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Phillips PC. Epistasis - the essential role of gene interactions in the structure and evolution of genetic systems. Nature Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, de Bakker PIW, Daly MJ, Sham PC. Plink: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo A, Bacanu SA, Devlin B, Sonpar V, Wasserman L, Roeder K. Characterization of multilocus linkage disequilibrium. Genet Epiodemiol. 2005;28:193–206. doi: 10.1002/gepi.20056. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epiodemiol. 2003;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Smyth DJ, Cooper JD, Howson JM, Walker NM, Plagnol V, Stevens H, Clayton DG, Todd JA. Ptpn22 trp620 explains the association of chromosome 1p13 with type 1 diabetes and shows a statistical interaction with hla class ii genotypes. Diabetes. 2008;57:1730–1737. doi: 10.2337/db07-1131. [DOI] [PubMed] [Google Scholar]
- Strobl C, Boulesteix AL, Zeileis A, T H. Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinformatics. 2007;8 doi: 10.1186/1471-2105-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Welcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statist Soc B. 1996;58:267–288. [Google Scholar]
- Todd JA, Bell JI, McDevitt HO. Hla-dq beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgovirte C, Genetics of Type 1 Diabetes in Finland. Simmonds MJ, Heward JM, Gough SC, Wellcome Trust Case Control Consortium. Dunger DB, Wicker LS, Clayton DG. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes AM, Erlich HA, Noble JA. Human leukocyte antigen class i b and c loci contribute to type 1 diabetes (t1d) susceptibility and age at t1d onset. Hum Immunol. 2005;66:301–313. doi: 10.1016/j.humimm.2004.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman L, Roeder K. High dimensional variable selection. Annal Stat. 2009 doi: 10.1214/08-aos646. To appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Chen YF, Hastie T, Sobel E, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25:714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu JS. Bayesian inference of epistatic interactions in case-control studies. Nat Genet. 2007;39:1167–1173. doi: 10.1038/ng2110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table A. Genotype by genotype counts (expected counts) for those SNP-SNP pairs with significant interactions.