Abstract
Flavin-binding LOV domains are broadly conserved in plants, fungi, archaea, and bacteria. These ≈100 residue photosensory modules are generally encoded within larger, multi-domain proteins that control a range of blue light-dependent physiologies. The bacterium Caulobacter crescentus encodes a soluble LOV-histidine kinase, LovK, that regulates the adhesive properties of the cell. Full-length LovK is dimeric as are a series of systematically truncated LovK constructs containing only the N-terminal LOV sensory domain. Non-conserved sequence flanking the LOV domain functions to tune the signaling lifetime of the protein. Size exclusion chromatography and small angle X-ray scattering (SAXS) demonstrate that the LOV sensor domain does not undergo a large conformational change in response to photon absorption. However, limited proteolysis identifies a sequence flanking the C-terminus of the LOV domain as a site of light-induced change in protein conformation/dynamics. Based on SAXS envelope reconstruction and bioinformatic prediction, we propose this dynamic region of structure is an extended C-terminal coiled-coil that links the LOV domain to the histidine kinase domain. To test the hypothesis that LOV domain signaling is affected by cellular redox state in addition to light, we measured the reduction potential of the LovK FMN cofactor. The measured potential of −258 mV is congruent with the redox potential of gram-negative cytoplasm during logarithmic growth (−260 to −280 mV). Thus a fraction of LovK in the cytosol may be in the reduced state under typical growth conditions. Chemical reduction of the FMN cofactor of LovK attenuates light-dependent ATPase activity of the protein in vitro, demonstrating that LovK can function as a conditional photosensor that is regulated by the oxidative state of the cellular environment.
Cellular adaptation requires protein domains that can perceive physical or chemical changes in the environment, and transduce detection of such events to downstream effectors. Flavin-binding light-, oxygen-, or voltage (LOV) domains, which were first identified in the phototropin (phot) plant photoreceptor family (1), are well-characterized protein photosensory domains. While phot LOV domains were initially postulated to serve as sensors of redox state (1) they were later shown to bind a flavin cofactor (2) and respond directly to the absorption of blue light via formation of a thermally-reversible cysteinyl-C(4a) adduct (3). Since these discoveries, LOV domains have generally become defined as a photosensory subset (4) of the larger Per-ARNT-Sim (PAS) domain superfamily (5). LOV domains are now known to be conserved across a diverse range of proteins in prokaryotes, eukaryotes, and archaea (4, 6).
Early studies on the structural mechanism of photon-dependent signaling in phot1 LOV domains revealed an allosteric switch, in which an amphipathic C-terminal α-helix packs against the LOV2 domain core in the dark state and unfolds and dissociates in the lit state (7). Later work revealed that N-terminal structural elements on phot1 LOV2 are also displaced upon photoexcitation (8). These solution and crystallographic analyses provided a framework for structural mechanisms of LOV signaling in phototropins, and insight from these studies have been successfully translated to the rational design of a chimeric LOV-Trp repressor that exhibits light-regulated DNA binding activity (9). However, recent examples of synthetic LOV-dihydrofolate reductase (10), LOV-Rac1 (11), and a LOV-histidine kinase (12) suggest multiple possible mechanisms through which LOV domains can regulate protein activity. Evidence for alternative modes of LOV signaling can be found in the fungal photoreceptor, VVD, in which cysteinyl-flavin adduct formation affects the equilibrium between monomer and dimer (13, 14). The structure of the LOV domain from the Bacillus subtilis σB regulator, YtvA (15–17) also suggests a mode of signaling that differs from phot LOV2, as YtvA does not have C-terminal Jα helix docked against the LOV domain but rather helices that extend away from LOV domain and connect to the C-terminal sulfate transporter anti-sigma factor antagonist (STAS) domain (18). An analogous, extended C-terminal helix is evident in the heme-binding PAS domain of the sensor histidine kinase, FixL (19).
While the biochemical and kinetic properties of isolated LOV domains from a variety of proteins have been broadly characterized, we generally do not understand the structural mechanisms by which LOV domains transduce signals within the context of the larger, multi-domain proteins in which they are encoded; recent studies on YtvA (20–22) and phototropin (23) have begun to address this question. Among the more prevalent classes of LOV proteins are the bacterial LOV histidine kinases (24). The Gram-negative aquatic bacterium, Caulobacter crescentus, encodes a soluble LOV-histidine kinase, LovK (Figure 1), that exhibits light-regulated autokinase activity and has been implicated in the regulation of cell adhesion (25). This simple, two-domain enzyme provides an excellent model to probe how LOV domains function in the context of multi-domain proteins. More generally, LovK provides a model to characterize the molecular basis of sensory transduction in histidine kinases.
Figure 1.
Conservation of LOV domains. CLUSTAL sequence alignment of a set of LOV domains for which kinetic properties have been characterized in vitro. Red bars and blue arrows above the alignment indicate the positions of α-helices and β-strands, respectively, in the Avena sativa phot1 LOV2 crystal structure (PDB ID: 2V1A). The box contains the signaling half-lives (i.e. half life of decay from the cysteinyl adduct state back to the unbonded dark state) of each of these constructs in vitro. The number of residues flanking the N- and C-termini of each characterized LOV protein construct are noted at the beginning and end of each sequence; these non-conserved flanking sequences are shown in Figure S1. An improved purification protocol for C. crescentus LovK (see Materials and Methods) has been implemented since our initial publication LovK recovery kinetics (25).
Here, we report a systematic analysis of the structural/oligomeric properties, signaling lifetime, and redox properties of LovK. Small angle X-ray scattering (SAXS) of the complete sensory domain of LovK provides evidence for a structure in which the C-terminal sequence linking the LOV domain to the kinase is extended away from the LOV domain core. This extended conformation is consistent with proposed models of coiled-coil-mediated signaling between the sensor domain and the C-terminal histidine kinase (12, 26). SAXS of the LovK sensor domain under constant blue-light illumination demonstrates that light-dependent regulation of LovK kinase activity does not require large changes in tertiary/quaternary structure of the protein, which is consistent with recent vibrational spectroscopic studies of full-length LovK (27). However, limited proteolysis under dark and illuminated conditions defines the extended C-terminal linker between the LOV domain and kinase as a site of conformational change upon photon absorption. Finally, we demonstrate that the reduction midpoint potential of the FMN cofactor of LovK is similar to that of the cytosol, and that the redox state of the FMN cofactor affects the capacity of LovK to function as a photosensor.
MATERIALS AND METHODS
Cloning and Expression Constructs
The gene encoding LovK (CC_0285) was PCR-amplified from C. crescentus genomic DNA and cloned into the overexpression vector pET28a (Novagen, Madison, WI) to produce pET28a-LovK (25). The C70A LovK mutant was generated using PCR-based site-directed mutagenesis as previously described (25). Truncated versions of the LovK LOV domains were PCR-amplified from pET28a-LovK and digested with NdeI and XhoI (NEB, Ipswich, MA). The LovK (1–138) construct was ligated into pET28a, while all other constructs were ligated into a pET28 variant in which the N-terminal His6 tag is followed by a TEV cleavage site (gift of R. Keenan) (see Table 1 for construct boundaries).
Table 1.
Primers used to construct the LovK truncation series, strain numbers of expression cell lines, and IPTG concentrations used to express proteins for purification.
| Construct | 5’ boundary | 3’ boundary | Strain | [IPTG] |
|---|---|---|---|---|
| LovK (1–368) |
GAATTCGATGGAAGACTATTCG GAATCGC |
AAGCTTTCAAAAGCGTCCTTGGC TAT |
FC410 | 1 mM |
| C70A LovK | TGATCGGCCGCAATGCCCGGTT CCTG |
CAGGAACCGGGCATTGCGGCCG ATCA |
FC543 | 1 mM |
| LovK (1–163) | CCCATATGTTGGAAGACTATTC G |
CTCGAGCTACTCCCTTGTCCGCT C |
FC722 | 1 mM |
| LovK (1–138) | CCCATATGTTGGAAGACTATTC G |
CTCGAGCTAGTCGGTCACGTCC AC |
FC931 | 2 mM |
| LovK (25- 163) |
CATATGGCCGCAGCGATCAGC GCC |
CTCGAGCTACTCCCTTGTCCGCT C |
FC598 | 1 mM |
| LovK (25- 138) |
CATATGGCCGCAGCGATCAGC GCC |
CTCGAGCTAGTCGGTCACGTCC AC |
FC596 | 1 mM |
Protein Expression and Purification
The pET28 expression vectors were transformed into Rosetta(DE3)pLysS cells to create overexpression strains. Expression strains were grown at 37°C to an OD600 of 0.1 (1 cm path length), at which point the temperature was lowered to 16°C and expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 16 hours. Cells were lysed by sonication in 20 mM Tris-HCl (pH 7.6), 500 mM NaCl, 20 mM imidazole, 1 mM β-mercaptoethanol, 5% glycerol. The recombinant protein was purified on Ni2+ Chelating Resin (GE Amersham Pharmacia, Piscataway, NJ) using a 20–500 mM imidazole gradient. Exposure to light during the purification led to loss of the flavin cofactor, so to avoid sample contamination with apo-LOV protein, affinity-purified sample was concentrated to 5 mL in the presence of excess FMN (Sigma-Aldrich, St. Louis, MO). Excess FMN and apo-LOV protein were removed via gel filtration chromatography; intact LovK was purified on a Hi Prep 26/60 Sephacryl S-200 column (GE Amersham Pharmacia) and the LOV domain truncations were purified on a 10/300 Superdex-75 column (GE Amersham Pharmacia). Gel filtration was performed in 20 mM Tris-HCl (7.6), 100 mM NaCl, 20 mM imidazole, 0.1 mM MgCl2. All further experiments were performed in this buffer unless otherwise specified. Thin-layer chromatography to identify LovK cofactor was performed as previously described (2).
UV/VIS Absorption Spectroscopy
Absorption spectroscopy was performed on a Shimadzu UV-1650 (Shimadzu Scientific Instruments, Columbia, MD) spectrophotometer using a 1 cm quartz cuvette. Purified LOV-proteins were kept in the dark for 24 hours prior to data collection, and photoexcited for 5 minutes with white light (1.1 mW/cm2). Spectra were collected at 120 second intervals to monitor the kinetics of thermal recovery. Recovery traces are the average of three independently-purified preparations of protein with error bars representing standard deviation. Recovery half-lives were calculated by fitting the kinetic absorption data in Prism (GraphPad Software, San Diego, CA) to the exponential function
| (1) |
where Ao is the initial absorbance at 446 nm, AF is the final absorbance at 446 nm (after complete recovery), k is the decay constant, and t is time.
Gel Filtration Chromatography
All LovK gel filtration was performed on a Hi Prep 26/60 Sephacryl S-200 column. All LOV domain gel filtration was performed on a 10/300 Superdex-75 column. Columns were calibrated with proteins from a Low Molecular Weight Gel Filtration Calibration kit (GE Life Sciences, Piscataway, NJ). For dark-state experiments, protein was loaded into an aluminum foil-covered sample injection loop under dim red light (1.7 µW/cm2), and automatically injected onto the column. The column was kept in the dark for the duration of the experiment. For lit-state experiments, protein was photoexcited with white light (1.1 mW/cm2) for 5 minutes prior to loading into the sample injection loop, and the column was illuminated by (11.9 µW/cm2) white light during the experiment. Dual-wavelength monitoring of the column elutant at 280 and 447 nm confirmed that the FMN cofactor in the lit-state proteins was still photobleached when the protein eluted from the column.
Small Angle X-ray Scattering
SAXS data were collected at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL) BioCAT beamline (Sector 18) on purified protein at concentrations of 0.4, 1.0, 2.0, and 4.0 mg/mL from a 1.5 mm flow capillary in which protein was flowed at 2 µl/s. Data were reduced in Igor Pro (WaveMetrics, Portland, OR). P(r) plots were calculated from the SAXS data using the indirect Fourier inversion algorithm of Svergun (28) in Gnom version 4.5a, and scattering envelopes were generated using DAMMIF (29). To measure SAXS in the lit state, protein samples were excited in the flow capillary with a blue AlGaInP LED (20° viewing angle, 8000 millicandela, 468 nm lambda max at 3.4 V; powered by a 3.3 volt, 4 amp AC adaptor) that was positioned orthogonal to the capillary throughout data collection as previously described (9). Complete adduct formation in the protein sample at this light intensity was confirmed spectroscopically prior to SAXS data collection.
Structural Modeling and Sequence Alignment
A cartoon model of dark-state LovK(1–163) was constructed in PyMol (MacPyMol, DeLano Scientific LLC) by appending the N-terminal 30 residues of the crystal structure of dark state N. crassa VVD (PDB 2PD7) (30) to the N-termini of the monomers within the dimeric crystal structure of B. subtilis YtvA (PDB 2PR5) (18). This model was superimposed on the scattering envelope of dark-state LovK(1–163) in PyMol. Sequences of LOV domain constructs characterized in vitro were aligned with ClustalW (31) and displayed with Boxshade (http://sourceforge.net/boxshade/).
Proteolytic Digests
LOV proteins were diluted to 20 µM and digested with concentrations varying from 0.1 µg/mL (1x) to 1 µg/mL (10x) of trypsin in 50 mM Na phosphate (pH 7.4). Dark digests were performed under dim red light (1.7 µW/cm2) on protein that had been allowed to photorecover for 24 hours. Lit digests were performed on protein photoexcited with white light (1.1 mW/cm2) for 5 minutes before the addition of trypsin, and illuminated throughout the reaction. Aliquots were removed from the reaction at 5 minute intervals. The reaction was stopped by adding the aliquots to equal volumes of 100 mM Tris-HCl (pH 6.8), 200 mM dithiothreitol, 4% SDS, 0.2% bromophenol blue, 20% glycerol and boiling for 5 minutes. Samples were run on 5%–12% SDS-polyacrylamide gradient gels and visualized by silver stain.
Redox Potential Measurements
Concentrated protein solutions were diluted to 20 µM in 100 mM potassium phosphate buffer (pH 7.0) in the presence of 200 µM xanthine and 1 µM benzyl viologen. Protein solutions were made anaerobic by repeated cycles of evacuation and equilibration with an atmosphere of purified argon in anaerobic glass cuvettes (32). Reduction potentials were measured using a xanthine/xanthine oxidase system to slowly reduce the sample in the presence and absence of an indicator dye (33). Experiments were monitored with a Shimadzu UV-2501PC spectrophotometer (Shimadzu Scientific Instruments, Columbia, MD) at 25°C, pH = 7.0. Phenosafranin (Em = −252 mV) and Safranin-T (Em = −290 mV) were used as reference dyes. Reduction potentials were calculated according to the Nernst equation
| (2) |
where A is absorption, η is the number of electrons transferred in the cell reaction, F is Faraday’s constant (23063 cal−1 V−1), R is the universal gas constant (1.987 cal K−1 mol−1), T is absolute temperature, and Em is the reduction potential.
ATPase Activity Assays
After purification and concentration, full-length LovK (1–368) was kept in the dark for 24 hours and diluted to 25 µM in kinase reaction buffer (50 mM Tris-HCl pH 7.6, 40 mM KCl, 10 mM MgCl2, 1 mM β-mercaptoethanol, 10% glycerol) in the presence or absence of 25 mM sodium dithionite and allowed to equilibrate in the dark for 30 minutes at room temperature. Reaction buffer was degassed for 48 hours prior to the addition of sodium dithionite, which was required for efficient reduction of the protein sample. Under these microaerobic conditions, the addition of sodium dithionite resulted in 50–60% reduction of the FMN cofactor. ‘Dark’ reactions were carried out under dim red light (1.7 µW/cm2), and ‘lit’ reactions were initiated after 2 minutes of illumination with white light (Hg fluorescent bulb; 1.1 mW/cm2). Reactions were initiated with the addition of 20 µM ATP, 5 µM [γ-32-P]ATP, allowed to proceed for 2 minutes, and were quenched by removing 4 µL aliquots and mixing with 2 µL 12 N formic acid. ‘Blank’ reactions containing no protein were performed in the presence and absence of sodium dithionite to ensure that the redox potential of the solution had no effect on the stability of the ATP. Aliquots were chilled on ice for 10 minutes, spun at maximum speed in a benchtop centrifuge for 2 minutes, spotted onto PEI Cellulose F plates TLC (Merck KgaA, Darmstadt, Germany), blotted with 50 µM ATP, and run for 7 minutes in 1 M formic acid, 1 M LiCl as previously described (25). Radioactive TLC plates were scanned using a Typhoon Trio Variable Mode Imager (GE Biosciences) and unhydrolyzed [γ-32P]ATP and free 32P were quantified with ImageQuant 5.2 software (GE Healthcare, Piscataway, NJ). ATP hydrolysis levels in the blank reactions were subtracted from the dark and lit-state reactions. Five replicates of each condition were performed for three independent protein purifications. All data were normalized to the average of the dark oxidized data. Statistical significance of ATPase activity differences were assessed by one-way ANOVA in Prism (GraphPad Software, San Diego, CA).
LovK/FMN Redox Exchange
20 µM solutions of LovK and free FMN were reduced by molar equivalent concentration of sodium dithionite in anaerobic glass cuvettes as described above. Redox exchange between free oxidized FMN and reduced LovK was initiated by adding oxidized FMN to reduced LovK at a 1:1 molar ratio.
RESULTS AND DISCUSSION
Conservation, diversity and signaling lifetime of LOV domains
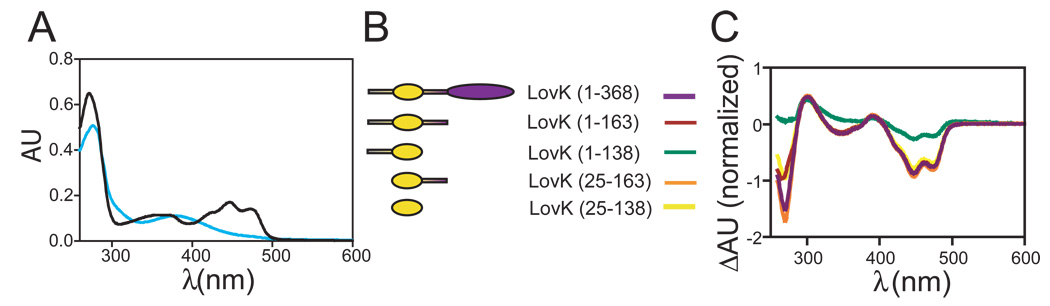
LovK has a dark-state flavin absorption maximum near 447 nm and vibronic bands near 425 and 475 nm (Figure 2A). Thin-layer chromatography of extract from denatured C. crescentus LovK demonstrates its cofactor is FMN (Table 2). As shown previously, LovK exhibits canonical LOV photochemistry (25): the 4a carbon of the FMN cofactor of LovK forms a covalent adduct with the conserved C70 residue, resulting in loss of 447 nm band and appearance of a band at 390 nm upon illumination (Figure 2C).
Figure 2.
Photochemistry of Caulobacter LovK. (A) Dark-state (black) and lit-state (blue) absorbance spectra of the isolated C. crescentus LOV domain (residues 25–138). (B) Schematic of the C. crescentus LovK truncation series. (C) Lit minus dark difference spectra of the LovK truncation series, normalized to the dark-state absorption at 447 nm.
Table 2.
Thin-layer chromatography of flavins. Retention factor (Rf) values of FMN and FAD standards, and the chromophore isolated from purified full-length LovK C70A (1–368).
| Species | Rf |
|---|---|
| FMN | 0.35 |
| FAD | 0.21 |
| LovK cofactor | 0.34 |
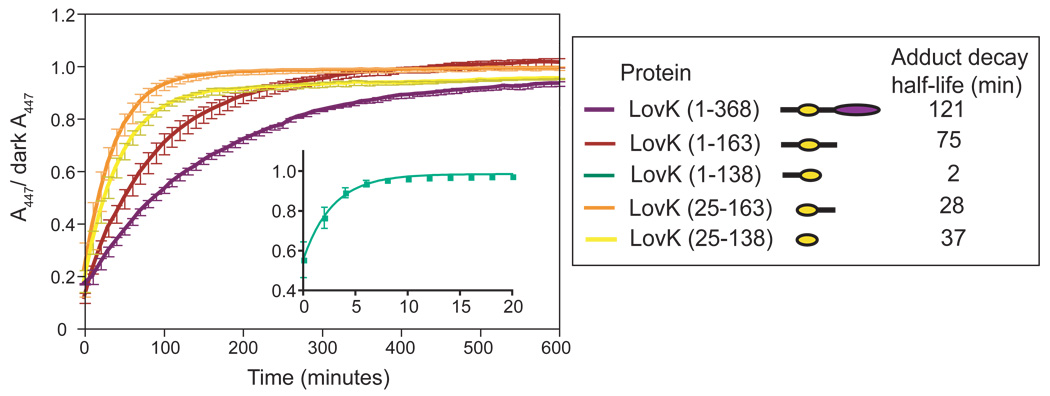
The rate constant at which the adduct species decays back to the ground state can be measured by monitoring the reappearance of the band at 447 nm (Figure 3); the reciprocal of this rate constant can effectively be considered the signaling lifetime of the protein. A number of prokaryotic and eukaryotic LOV domains have been characterized in vitro, and shown to exhibit signaling lifetimes (i.e. kinetics of cysteinyl-flavin adduct rupture) that range over five orders of magnitude (Figure 1). Some differences in the kinetics of adduct formation and rupture among LOV domains can be explained by variation in the residues that make up the binding pocket of the flavin cofactor (34–36), while other studies demonstrate that the signaling lifetime of LOV is modulated by non-conserved regions of structure outside the defined LOV domain core (37–39) (see Figures 1 and S1). To methodically test the effect of the non-conserved flanking sequence on the signaling lifetime of LovK, we expressed and purified full-length LovK and a series of systematic LovK truncations (see Figure 2B).
Figure 3.
The structural context of the LOV domain affects dark recovery kinetics. Recovery traces of the C. crescentus LovK truncation series. Legend box on right contains the recovery half-life of each construct calculated from the fit to Equation 1.
The kinetics of recovery from the adduct state depend not only on the buffer environment (40), but also on the structural context in which the LOV domain is contained (Figure 3). Full-length LovK (residues 1–368) has a long recovery, with a half-life of two hours (see Materials and Methods for purification protocols and buffer conditions under which kinetics were measured). The isolated LOV core (residues 25–138) recovers more quickly, with a half-life of 37 minutes. LovK (25–163), which contains the LOV core and the non-conserved linker sequence at its C-terminus, has a similar recovery half-life as LovK (25–138) at 28 minutes. A construct containing the LOV core and the non-conserved N-terminal flanking sequence, LovK (1–138), has dramatically faster recovery kinetics from the adduct state, with a half-life of only 2 minutes. However, it is not the case that this N-terminal region of structure has a general rate enhancement effect. LovK (1–163), a construct with fully intact N- and C-termini (but missing the kinase domain) is slower to recover than either LovK(25–138), LovK(1–138) or LovK(25–163) (Figure 3). Thus the effects of structure outside the LOV core on its signaling lifetime do not combine in a simple, additive way.
Steady-state lit minus dark difference spectra of these five constructs highlight the functional consequence of different adduct decay rates (Figure 2B). Namely, the maximum steady-state concentration of cysteinyl-flavin adduct is markedly decreased in LovK(1–138) relative to the other constructs under identical illumination conditions. This can be attributed to the fast rate (t1/2 = 2 minutes) of recovery from the adduct state in LovK(1–138). We also observe that the light-induced shift in absorbance at 280 nm, relative to 447 nm, varies between constructs (Figure 2B). We attribute this spectral difference to minor differences in the chemical environment surrounding buried aromatic amino acids and the flavin dimethylbenzene moiety in the five constructs.
Thermal recovery from the adduct state: a solvent accessibility model
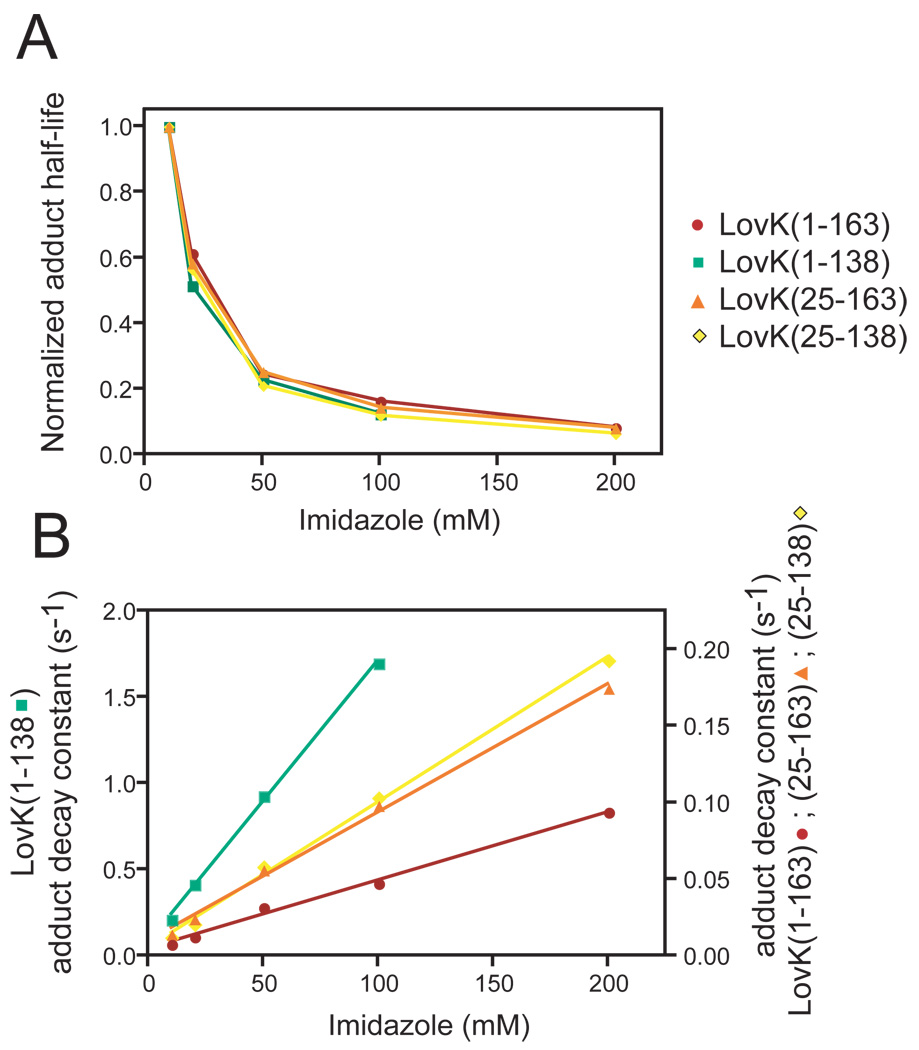
A possible mechanism underlying the different recovery rates among the C. crescentus LovK constructs is differential accessibility of the FMN adduct to the surrounding buffer environment. It is known that imidazole, a biological base, enhances the rate of thermal decay from the adduct state back to the non-bonded “dark” state (40). While the mechanism of base-mediated rate enhancement has not been determined definitively, the relationship between recovery rate and base concentration is both linear and non-saturable (40). This is consistent with a simple bimolecular reaction model involving a direct effect of base in bulk solvent on the stability of the cysteinyl-flavin adduct in the interior of the protein, such that
| (3) |
We observe that the relative (i.e. fold-change) rate enhancement is identical for all constructs (Figure 4A). This suggests the absolute difference in base-catalyzed adduct rupture can be modeled by a simple solvent accessibility term, a, such that
| (4) |
In this model, the slope of k versus concentration of base yields the solvent accessibility of the cysteinyl-flavin adduct, a, for each LOV domain construct (Figure 4B) (Table 3). The accessibility of the interior of the LovK LOV domain to solvent is similar in all of the truncation constructs except for LovK (1–138), which contains an intact N-terminus and no C-terminal linker. In the absence of sequence C-terminal to the LOV domain core, the 24 amino acids at the N-terminus of the LOV domain dramatically increase the rate of photorecovery (Figure 3, Figure 4B). This effect is attenuated by the addition of 25 residues C-terminal to the LOV domain core (Figure 3, Table 3). These data provide evidence that the two non-conserved regions flanking the LOV domain have opposing effects on protein signaling lifetime, perhaps by affecting the accessibility of solvent to the protein interior.
Figure 4.
The effect of imidazole on LOV domain recovery kinetics. (A) Normalized imidazole dependence of the recovery half-lives of the LOV truncation series (normalized to the half-life at 10 mM imidazole). (B) Absolute rate enhancement effect of imidazole on recovery from the adduct state in the four LovK LOV domain constructs. The recovery rate constant for each of the four constructs is graphed as a function of imidazole concentration.
Table 3.
Solvent accessibility constants of the LovK LOV domain truncation series. See Equation 4 for a definition the accessibility constant, a.
| Protein | Accessibility Constant (a) (104) |
|---|---|
| LovK (1–163) | 4.5 |
| LovK (1–138) | 163.6 |
| LovK (25–163) | 8.4 |
| LovK (25–138) | 9.5 |
Probing mechanism of LOV Domain signaling by size exclusion chromatography
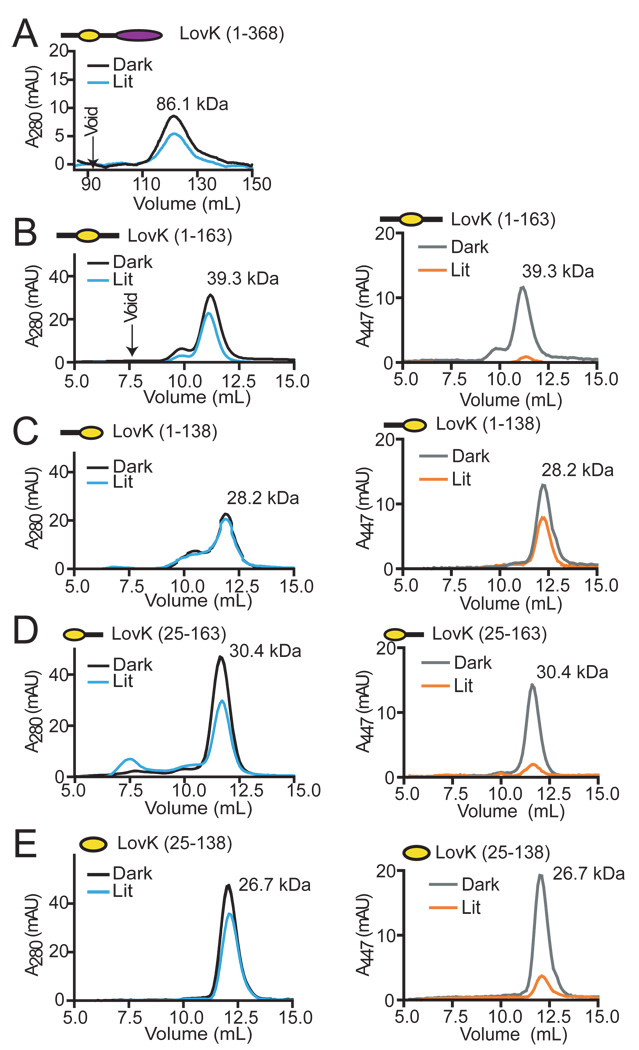
Signaling mechanisms have been proposed for LOV proteins that involve light-induced changes in oligomeric state (14), and/or localized unfolding of a region of structure (7, 41). An analysis of change in oligomeric state and hydrodynamic radius in C. crescentus LovK constructs (Figure 2B) in the dark and lit states by size exclusion chromatography reveals that LovK is principally dimeric, and does not undergo large structural changes upon illumination (Figure 5). Intact LovK(1–368) elutes at a volume consistent with a dimer in both the dark and lit states (Figure 5A), which does not support a model in which light-dependent changes in oligomeric state regulate activity of the kinase domain. The constructs containing the N-terminal flanking sequence (1–163 and 1–138) elute as apparent dimers; illumination of LovK(1–138) causes a slight shift in elution volume suggesting that this construct may undergo a modest change in structure in the lit state (Figure 5C). LovK(1–163) also elutes as a minor peak consistent with a trimer (Figure 5B). However, we do not believe this is a biologically-relevant oligomer, given that histidine kinases are known to function as dimers (42, 43), and that full-length LovK and other LOV constructs elute consistently as dimers in the light and dark. A possible explanation for this faster-eluting peak is the presence of a structural isoform in the population that is more elongated and less globular than the ‘true’ dimer. Together, these size exclusion data provide evidence that the signaling mechanism of LovK does not require a change in oligomeric state or large scale change in protein structure.
Figure 5.
Gel filtration of full-length LovK and systematically truncated LOV domains. Dark and lit-state elution profiles of (A) LovK(1–368), (B) LovK(1–163), (C) LovK(1–138), (D) LovK(25–163), and (E) LovK(25–138). Elution of full-length LovK was monitored at 280 nm; due to low concentration of this protein sample, signal at 447 nm could not be definitively interpreted. Elution profiles for the remaining LovK LOV domain constructs were monitored at 280 (left column) and 447 nm (right column) and show the expected loss of visible absorption signal in the lit state.
Assessing LovK structure in the light and dark by SAXS
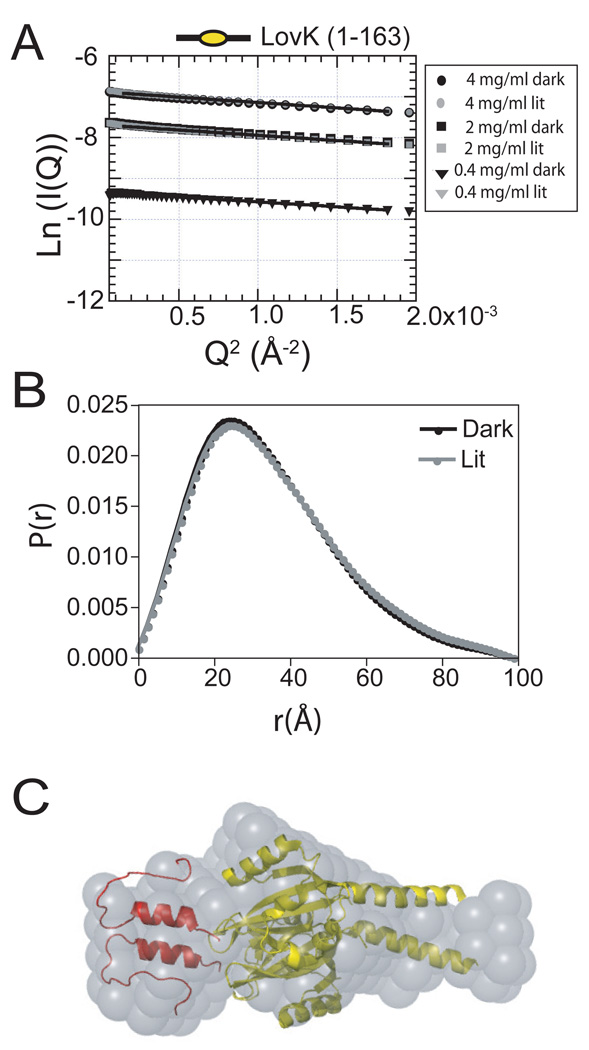
Analysis of LovK structure by small-angle X-ray scattering (SAXS) was confounded by the tendency of full-length LovK and 2 of the 4 truncated LOV domain constructs to form large aggregate species at the higher protein concentrations required for SAXS measurements (Figure S2). Preparations of LovK(1–163) yielded the highest quality SAXS data. Neither the Rg nor Io of LovK(1–163) change significantly upon saturating illumination of the protein with blue-light (Figure 6A and 6B), providing support for a model in which regulation of LovK kinase activity requires only minor structural rearrangements in the N-terminal sensory domain. A Guinier fit of LovK(1–163) data yielded a radius of gyration (Rg) of 27.6 Å (Figure 6B and Table 4). This result is consistent with dimeric LOV constructs from A. thaliana FKF1 (44) and phototropin 1 (41), which have radii of gyration in the range of 20–28 Å, and exhibit only minor increases (0.1–0.4 Å) in Rg upon photoexcitation.
Figure 6.
Small angle X-ray scattering of LovK (1–163). (A) Guinier plot of LovK (1–163) in the dark-state and lit-state. (B) P(r) plot of dark-state and lit-state LovK (1–163). (C) DAMMIF scattering envelope model (grey) of LovK (1–163) based on the SAXS data, superimposed with the crystal structures of the B. subtilis YtvA LOV domain (yellow; PDB ID 2PR5) and the LOV domain N-terminal cap from N. crassa VVD (red; 2PD7).
Table 4.
Dark- and lit-state radii of gyration of full-length LovK C70A and LovK (1–163) as determined by a Guinier fit to scattering data measured at a protein concentration of 2 mg/mL (see Figure 6).
| Construct | Light | Rg (Å) |
|---|---|---|
| LovK C70A | − | 42.7 ± 0.1 |
| LovK (1–163) | − | 27.6 ± 0.1 |
| LovK (1–163) | + | 27.8 ± 0.1 |
While wild-type full-length LovK(1–368) aggregated at the high protein concentrations necessary for SAXS analysis, the “blind” mutant, LovK C70A, had an identical gel filtration elution profile as wild type and was less prone to aggregation. A Guinier fit of the SAXS data from LovK C70A yielded an Rg=42.7 Å (Table 4). The closest histidine kinase homolog for which SAXS scattering data exist is a PAS-histidine kinase fragment of ThkA from Thermotoga maritima. This construct, which forms a 76 kDa dimer in solution, has an Rg of 37.3 Å (45). The larger Rhodopseudomonas palustris bacteriophytochrome 4 histidine kinase forms a 167 kDa dimer with an Rg of 52.5 Å (46). Based on SAXS data from these histidine kinases, the measured Rg for C. crescentus LovK C70A is consistent with the expected 87 kDa dimer of His6-LovK C70A.
Ab initio shape determination (29) of the LovK(1–163) sensor domain from its SAXS scattering profile produced an elongated envelope, providing evidence that the sensor domain dimer of LovK forms an extended structure. We fit this envelope with a crystal structure of the B. subtilis YtvA LOV domain (18), which contains an extended C-terminal helix. At the N-terminus of our low-resolution model, we placed the N-cap of the VVD LOV protein (30). The reconstructed scattering envelope of LovK(1–163) is best fit by a model in which both the N- and C-terminal flanking sequence extend away from the LOV domain core (Figure 6C).
In the context of bacterial sensor histidine kinases, helical coiled-coils linking the N-terminal sensor domain to the C-terminal kinase have long been postulated to serve a functional role in signaling (26). Recent studies on an engineered LOV histidine kinase provide experimental support for a mechanism involving signal transmission via rotation of extended coil-coil helices (12). Moreover, sequence analysis of naturally occurring PAS/LOV-histidine kinases has shown that the linker regions between the boundaries of the PAS sensor and histidine kinase domains contain (7n) or (7n+2) coiled-coil signatures with hydrophobic residues in a ‘heptad repeat’ pattern (12). The linker region between the histidine kinase and LOV domain of C. crescentus LovK (residues 150–175) also has a coiled-coil signature according to the Lupas/Stock algorithm (47), and the Paircoil algorithm of Berger and Keating (48). Our SAXS data on LovK(1–163) evidence a model in which these putative coiled-coil residues extend out and away from the C-terminus of the LOV domain core.
Photoexcitation destabilizes the LOV-HK linker sequence
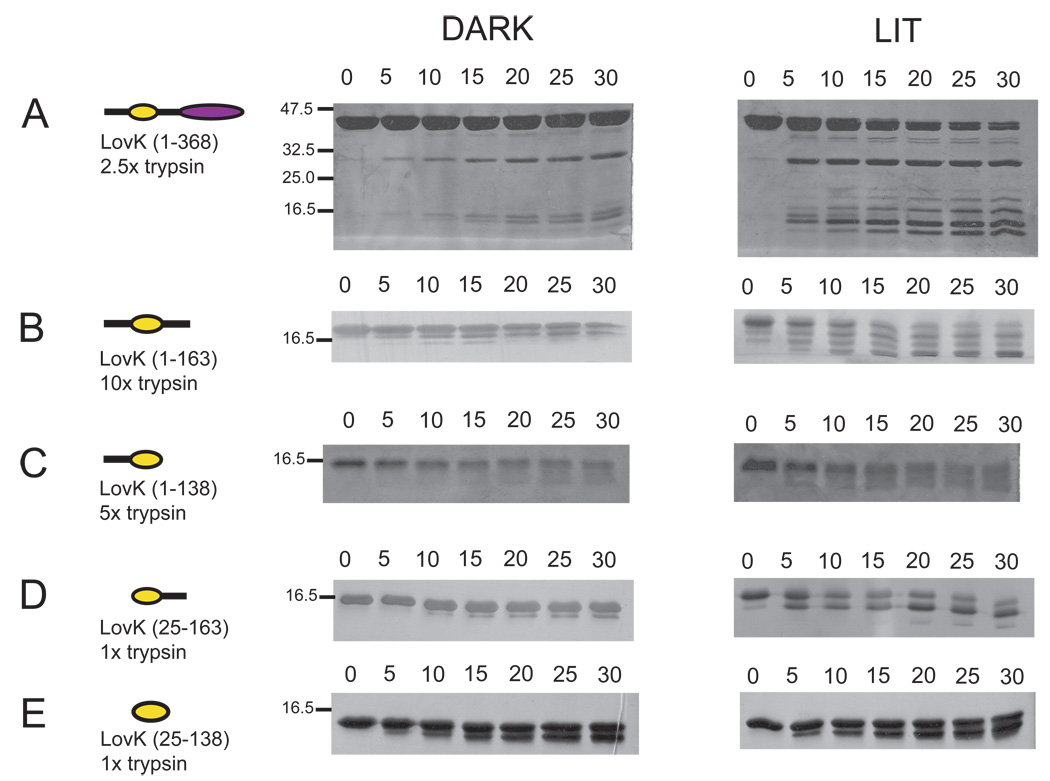
Limited proteolysis of proteins can provide information about how their structure changes upon perturbation. In particular, this method can detect changes in protein structural dynamics or stability that are not necessarily accompanied by a large-scale change in average structure (49). Thus, limited proteolysis provides information that is complementary to methods such as size exclusion chromatography and SAXS. Complete trypsin digestion of LovK would yield 49 fragments of molecular weights ranging from 0.1–2.6 kDa (50). Limited trypsin digestion of dark-state LovK(1–368) reveals two major cleavage products, of ~29 and ~15 kDa (Figure 7), which are consistent with the predicted sizes of the isolated histidine kinase and LOV domains, respectively. Proteolysis of LovK(1–368) after illumination with white light is more rapid and yields a greater number of cleavage products. These data suggest the protein is more conformationally flexible and thus more vulnerable to proteolysis in the lit state.
Figure 7.
Light-induced structural changes in C. crescentus LovK monitored by limited proteolysis. Tryptic digest time courses (in minutes) of dark and lit-state (A) LovK(1–368), (B) LovK(1–163), (C) LovK(1–138), (D) LovK(25–163), and (E) LovK(25–138). All protein samples are 20 µM; the trypsin concentration required to achieve similar levels of cleavage in 30 minutes varies from 0.1–1.0 µg/mL, depending upon the LOV domain construct. Protein molecular weight markers are noted on the sides of the dark-state gels.
Digestion of the four LovK truncation constructs reveals distinct patterns based on the presence or absence of different flanking sequences. The LovK(1–163) and (1–138) constructs, which include the N-terminal flanking sequence, are less susceptible to proteolysis than constructs without the N-terminus as evidenced by the higher trypsin:LOV ratio that is required to produce similar levels of cleavage per unit time (Figure 7). LovK(1–138), while relatively stable, does not show discrete proteolytic bands upon the addition of trypsin. Rather, this construct shows evidence of more non-specific degradation. The LovK(1–163) and (25–163) constructs, which include the C-terminal flanking sequence, show a pronounced increase in cleavage in response to illumination with visible light; constructs lacking this C-terminal extension do not show as pronounced a lit/dark difference in proteolysis (Figure 7). These data support a model in which structure at the N-terminus of the LOV domain is not significantly perturbed upon photoexcitation. Rather, it is the structure at the C-terminus, which links the LOV domain to the histidine kinase, that is the locus of the structural change in response to photon absorption and cysteinyl-C4(a) adduct formation. This result is analogous to the phototropin LOV2 signaling model in which illumination destabilizes a region of structure at the C-terminus of the LOV domain (7). However, the exact mechanism of signaling likely differs between LovK and phototropin, as the SAXS data support an extended conformation of the LOV C-terminus in LovK (see Figure 6).
Biochemical evidence that LovK can function as a sensor of cytoplasmic redox potential
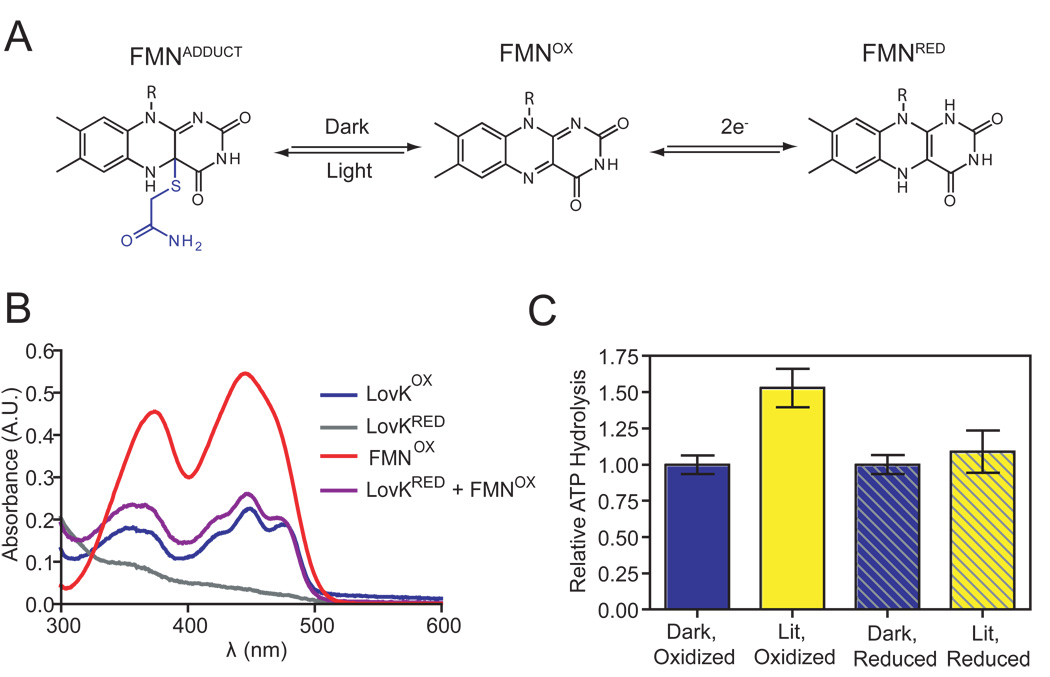
The LovK FMN chromophore is capable of undergoing chemical reactions other than light-dependent cysteinyl-C4(a) adduct formation. Flavoproteins are more commonly known for their capability to transfer electrons and detect changes in cellular redox state. Examples of flavin-binding PAS domains that detect changes in cellular redox include NifL and Aer (51, 52). The light-sensing role of the LOV domain in vivo is, in principle, influenced by the redox state of the FMN cofactor. The electron-poor, oxidized form of FMN is favored to accept an electron in the triplet excited state and form a cysteine-flavin radical pair that precedes cysteinyl-flavin adduct formation (53) (Figure 8A). While it is known that the redox potential of the buffer environment does not influence the kinetics of LOV photorecovery from the adduct state (54), to our knowledge, the effect of the oxidation state of LOV domains on their capacity to serve as photosensors remains unexplored.
Figure 8.
Integration of redox and light signals. (A) Oxidized FMN of a LOV domain forms a cysteinyl-C4a adduct upon illumination with blue light. The flavin cofactor of the LOV domain can also be reduced to form semiquinone (e−) or hydroquinone (2e−) species. (B) Chemical reduction of oxidized LovK FMN (blue) by sodium dithionite results in a loss of visible absorption signal (grey). Oxidized free FMN (red) and oxidized FMN bound to LovK (blue) are distinguished by fine structure in the LovK flavin spectrum. Mixing equimolar amounts of reduced LovK and oxidized free FMN results in the rapid transfer of electrons from LovKRED to FMNOX (t1/2 < 1 minute), restoring the absorption spectrum of oxidized LovK (purple). (C) Chemical reduction of the LovK FMN cofactor attenuates the lit/dark difference in LovK ATPase activity (mean +/− SEM). The lit, oxidized activity differs significantly from the other tested conditions (ANOVA, Tukey post test p = 0.014, n=15).
The cytoplasm of a bacterial cell is reducing, in the range of −260 to −280 mV under logarithmic growth conditions (55). We measured the potential for the two-electron reduction of full-length LovK at −258 mV, suggesting that the pool of cytoplasmic LovK will be at least partially reduced under normal growth conditions. With the exception of the LovK(1–138) construct (−303 mV), all of the truncated LovK constructs had two-electron reduction potentials in the −260 mV range (Table 5). As cysteinyl-flavin adduct formation in response to blue light absorption is only favored when FMN is in its oxidized form, increasing or decreasing the potential of the cytoplasm will, in theory, affect the responsiveness of a cell to blue light. In vitro ATPase activity assays of full-length LovK support a model in which a decrease in cytoplasmic reduction potential reduces light-dependent regulation of LovK. ATP hydrolysis activity of oxidized LovK was upregulated 1.6 fold upon illumination with white light (Figure 8B), which was consistent with our previous observation of an approximate 2-fold increase in ATPase activity in the lit state (25). Chemical reduction of LovK FMN (Figure 8B) (see Materials and Methods) to produce a mixed population of oxidized and reduced LovK did not affect its dark-state ATPase activity but attenuated the light-dependent increase in ATP hydrolysis. This result evidences a model in which the redox state of the cytoplasm can regulate LovK photoactivity (Figure 8C).
Table 5.
Reduction potentials of the FMN cofactor of all of the proteins in the LovK truncation series measured according to the method of Massey (33).
| Species | Potential (mV) |
|---|---|
| FMN | −207 |
| LovK (1–368) | −258 |
| LovK (1–163) | −259 |
| LovK (1–138) | −303 |
| LovK (25–163) | −263 |
| LovK (25–138) | −262 |
| cytoplasma | −260 to −280 |
Gram-negative cytoplasmic redox potential based on the measurements of Hwang et al. (55).
LovK has previously been implicated in the regulation of cell envelope composition (25), and envelope remodeling is known to be a common response to cellular stress (56). The cell envelope/adhesion phenotype of lovK mutants hints at a possible functional role for redox stress sensing by LovK. Indeed, extracellular stress can affect intracellular redox potential through a variety of mechanisms. Under normal growth conditions, cytoplasmic redox state is buffered mainly by the equilibria between the oxidized and reduced forms of thioredoxin and glutathione; these equilibria are affected by the availability of the reduced nicotinamide dinucleotides NADH and NADPH (57). Hypoxia can increase the cellular concentration of NADH, thereby lowering the cytoplasmic reduction potential. Oxidative stressors such as superoxide or nitric oxide can directly perturb the reduction potential of the cell or can affect it indirectly by altering the NADP/NADPH ratio (58, 59).
Thioredoxin, glutathione, and other redox-sensitive species in the cell display substrate specificity when participating in electron transfer reactions (57, 60). For example, ferric iron generated by oxidative stress is preferentially reduced by flavins in E. coli, rather than by NADH, thioredoxin, glutathione, or any of the other potential reducing agents in the cell (61). While molecules that may function as biological reductants of LovK in the cell are not known, we have in vitro evidence that NADH and NADPH do not likely to fill this role. Incubation of these reductants with equimolar concentrations of LovK failed to yield reduced protein over the course of several hours. However, the addition of one molar equivalent of oxidized FMN to a completely reduced sample of LovK resulted in a rapid (t1/2 < 1 minute) recovery of the absorption spectrum of oxidized LovK via reduction of free FMN (see Figure 8B). The ability of LovK to donate electrons to FMN suggests that flavins and other flavoproteins are candidates for such redox interactions in vivo.
The capacity of C. crescentus LovK to function as a conditional photosensor in the oxidized state may be a part of a more complex stress sensor pathway in which both light and cellular redox signals are coordinately integrated to affect adaptation to environmental stressors. Indeed, exposure to blue light is often coupled with exposure to UV light, a common cause of oxidative stress. We present a model in which LOV domains can have a dual role as sensors of light and cytosolic reduction potential, providing a possible explanation for the abundance of soluble LOV proteins encoded in the genomes of heterotrophic bacteria with no known photoresponse. LovK, and perhaps other LOV regulatory proteins, appear capable of integrating information from multiple environmental stimuli.
Supplementary Material
Acknowledgements
We thank members of the Crosson Lab for helpful discussions over the course of this study, and David Courson and Aretha Fiebig for criticism on drafts of the manuscript.
Abbreviations
- LOV
Light-oxygen-voltage
- PAS
Per-ARNT-Sim
- SAXS
small-angle X-ray scattering
- STAS
sulfate transporter anti-sigma factor antagonist
- phot
phototropin
- IPTG
isopropyl-β-D-thiogalactopyranoside
- Rg
radius of gyration
- Rf
retention factor
Footnotes
E.B.P was supported by National Institutes of Health Training Grant 5T32GM007183-34. S.C. acknowledges support for this project from NIH grant 1R01GM087353-2, the Arnold and Mabel Beckman Foundation (BYI), and the Mallinckrodt Foundation. Advanced Photon Source is supported by the DOE Office of Basic Energy Sciences (Contract No. DE-AC02-06CH11357). BioCAT is an NIH-supported Research Center (RR-08630). C.A.M. and B.A.P. are supported by NIH grant 2R01GM061087-8.
Supporting Information Available
An alignment of (A) N-terminal and (B) C-terminal LOV domain flanking sequence not presented in Figure 1 is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 2.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, Phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 4.Crosson S, Rajagopal S, Moffat K. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 5.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Losi A, Polverini E, Quest B, Gärtner W. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys J. 2002;82:2627–2634. doi: 10.1016/S0006-3495(02)75604-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 8.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 9.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci USA. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, Russ WP, Benkovic SJ, Ranganathan R. Surface sites for engineering allosteric control in proteins. Science. 2008;322:438–442. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Möglich A, Ayers RA, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb JS, Zoltowski BD, Pabit SA, Crane BR, Pollack L. Time-resolved dimerization of a PAS-LOV protein measured with photocoupled small angle X-ray scattering. J Am Chem Soc. 2008;130:12226–12227. doi: 10.1021/ja804236f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoltowski BD, Crane BR. Light activation of the LOV protein vivid generates a rapidly exchanging dimer. Biochemistry. 2008;47:7012–7019. doi: 10.1021/bi8007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akbar S, Gaidenko TA, Kang CM, O'Reilly M, Devine KM, Price CW. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor σB of Bacillus subtilis. J Bacteriol. 2001;183:1329–1338. doi: 10.1128/JB.183.4.1329-1338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avila-Pérez M, Hellingwerf KJ, Kort R. Blue light activates the σB-dependent stress response of Bacillus subtilis via YtvA. J Bacteriol. 2006;188:6411–6414. doi: 10.1128/JB.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaidenko TA, Kim TJ, Weigel AL, Brody MS, Price CW. The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J Bacteriol. 2006;188:6387–6395. doi: 10.1128/JB.00691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Möglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong WM, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK. Structure of a biological oxygen sensor: A new mechanism for heme-driven signal transduction. Proc Natl Acad Sci USA. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buttani V, Gärtner W, Losi A. NTP-binding properties of the blue-light receptor YtvA and effects of the E105L mutation. Eur Biophys J. 2007;36:831–839. doi: 10.1007/s00249-007-0155-1. [DOI] [PubMed] [Google Scholar]
- 21.Buttani V, Losi A, Eggert T, Krauss U, Jaeger KE, Cao Z, Gärtner W. Conformational analysis of the blue-light sensing protein YtvA reveals a competitive interface for LOV-LOV dimerization and interdomain interactions. Photochem Photobiol Sci. 2007;6:41–49. doi: 10.1039/b610375h. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y, Cao Z, Livoti E, Krauss U, Jaeger KE, Gärtner W, Losi A. Interdomain signalling in the blue-light sensing and GTP-binding protein YtvA: a mutagenesis study uncovering the importance of specific protein sites. Photochem Photobiol Sci. 2010;9:47–56. doi: 10.1039/b9pp00075e. [DOI] [PubMed] [Google Scholar]
- 23.Pfeifer A, Mathes T, Lu Y, Hegemann P, Kottke T. Blue light induces global and localized conformational changes in the kinase domain of full-length phototropin. Biochemistry. 2010;49:1024–1032. doi: 10.1021/bi9016044. [DOI] [PubMed] [Google Scholar]
- 24.Losi A. Flavin-based photoreceptors in bacteria. In: Silva E, Edwards AM, editors. Flavins - Photochemistry and Photobiology. Dorchester, UK: RCS Publishing; 2006. pp. 217–269. [Google Scholar]
- 25.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci USA. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh M, Berger B, Kim PS, Berger JM, Cochran AG. Computational learning reveals coiled coil-like motifs in histidine kinase linker domains. Proc Natl Acad Sci USA. 1998;95:2738–2743. doi: 10.1073/pnas.95.6.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandre MT, Purcell EB, van Grondelle R, Robert B, Kennis JT, Crosson S. Electronic and protein structural dynamics of a photosensory histidine kinase. Biochemistry. 2010;49:4752–4759. doi: 10.1021/bi100527a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 29.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;77 doi: 10.1016/S0006-3495(99)77443-6. 2896-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Palfey BA. Time-resolved spectral analysis. In: Johnson KA, editor. Kinetic Analysis of Macromolecules. New York: Oxford University Press; 2003. pp. 203–227. [Google Scholar]
- 33.Massey V. A simple method for the determination of redox potentials. In: Curti B, Ronchi S, Zanetti G, editors. Flavins and Flavoproteins. Berlin, Germany: Walter de Gruyter; 1990. [Google Scholar]
- 34.Christie JM, Corchnoy SB, Swartz TE, Hokenson M, Han IS, Briggs WR, Bogomolni RA. Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry. 2007;46:9310–9319. doi: 10.1021/bi700852w. [DOI] [PubMed] [Google Scholar]
- 35.Nash AI, Ko WH, Harper SM, Gardner KH. A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochemistry. 2008;47:13842–13849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jentzsch K, Wirtz A, Circolone F, Drepper T, Losi A, Gärtner W, Jaeger KE, Krauss U. Mutual exchange of kinetic properties by extended mutagenesis in two short LOV domain proteins from Pseudomonas putida. Biochemistry. 2009;48:10321–10333. doi: 10.1021/bi901115z. [DOI] [PubMed] [Google Scholar]
- 38.Kasahara M, Swartz TE, Olney MA, Onodera A, Mochizuki N, Fukuzawa H, Asamizu E, Tabata S, Kanegae H, Takano M, Christie JM, Nagatani A, Briggs WR. Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 2002;129:762–773. doi: 10.1104/pp.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Losi A, Quest B, Gärtner W. Listening to the blue: the time-resolved thermodynamics of the bacterial blue-light photoreceptor YtvA and its isolated LOV domain. Photochem Photobiol Sci. 2003;2:759–766. doi: 10.1039/b301782f. [DOI] [PubMed] [Google Scholar]
- 40.Alexandre MTA, Arents JC, van Grondelle R, Hellingwerf KJ, Kennis JTM. A base-catalyzed mechanism for dark state recovery in the Avena sativa phototropin-1 LOV2 domain. Biochemistry. 2007;46:3129–3137. doi: 10.1021/bi062074e. [DOI] [PubMed] [Google Scholar]
- 41.Nakasako M, Iwata T, Matsuoka D, Tokutomi S. Light-induced structural changes of LOV domain-containing polypeptides from Arabidopsis phototropin 1 and 2 studied by small-angle X-ray scattering. Biochemistry. 2004;43:14881–14890. doi: 10.1021/bi0485530. [DOI] [PubMed] [Google Scholar]
- 42.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surette MG, Levit M, Liu Y, Lukat G, Ninfa EG, Ninfa A, Stock JB. Dimerization is required for the activity of the protein histidine kinase CheA that mediates signal transduction in bacterial chemotaxis. J Biol Chem. 1996;271:939–945. doi: 10.1074/jbc.271.2.939. [DOI] [PubMed] [Google Scholar]
- 44.Nakasako M, Matsuoka D, Zikihara K, Tokutomi S. Quaternary structure of LOV-domain containing polypeptide of Arabidopsis FKF1 protein. FEBS Lett. 2005;579:1067–1071. doi: 10.1016/j.febslet.2004.12.078. [DOI] [PubMed] [Google Scholar]
- 45.Yamada S, Akiyama S, Sugimoto H, Kumita H, Ito K, Fujisawa T, Nakamura H, Shiro Y. The signaling pathway in histidine kinase and the response regulator complex revealed by X-ray crystallography and solution scattering. J Mol Biol. 2006;362:123–139. doi: 10.1016/j.jmb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Evans K, Grossmann JG, Fordham-Skelton AP, Papiz MZ. Small-angle X-ray scattering reveals the solution structure of a bacteriophytochrome in the catalytically active Pr state. J Mol Biol. 2006;364:655–666. doi: 10.1016/j.jmb.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 47.Lupas A, Van Dyke M, Stock J. Predicting Coiled Coils from Protein Sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 48.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 49.Fontana A, Polverino de Laureto P, De Filippis V, Scaramella E, Zambonin M. Probing the partly folded states of proteins by limited proteolysis. Fold Des. 1997;2:R17–R26. doi: 10.1016/S1359-0278(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 50.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucl. Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez-Argudo I, Little R, Shearer N, Johnson P, Dixon R. The NifL-NifA System: a multidomain transcriptional regulatory complex that integrates environmental signals. J Bacteriol. 2004;186:601–610. doi: 10.1128/JB.186.3.601-610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor BL, Rebbapragada A, Johnson MS. The FAD-PAS domain as a sensor for behavioral responses in Escherichia coli. Antioxid Redox Signal. 2001;3:867–879. doi: 10.1089/15230860152665037. [DOI] [PubMed] [Google Scholar]
- 53.Schleicher E, Kowalczyk RM, Kay CW, Hegemann P, Bacher A, Fischer M, Bittl R, Richter G, Weber S. On the reaction mechanism of adduct formation in LOV domains of the plant blue-light receptor phototropin. J Am Chem Soc. 2004;126:11067–11076. doi: 10.1021/ja049553q. [DOI] [PubMed] [Google Scholar]
- 54.Noll G, Hauska G, Hegemann P, Lanzl K, Noll T, von Sanden-Flohe M, Dick B. Redox properties of LOV domains: chemical versus photochemical reduction, and influence on the photocycle. Chembiochem. 2007;8:2256–2264. doi: 10.1002/cbic.200700304. [DOI] [PubMed] [Google Scholar]
- 55.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 56.MacRitchie DM, Buelow DR, Price NL, Raivio TL. Two-component signaling and gram negative envelope stress response systems. Adv Exp Med Biol. 2008;631:80–110. doi: 10.1007/978-0-387-78885-2_6. [DOI] [PubMed] [Google Scholar]
- 57.Ritz D, Beckwith J. Roles of thiol-redox pathways in bacteria. Annu Rev Microbiol. 2001;55:21–48. doi: 10.1146/annurev.micro.55.1.21. [DOI] [PubMed] [Google Scholar]
- 58.Eklow L, Moldeus P, Orrenius S. Oxidation of glutathione during hydroperoxide metabolism. A study using isolated hepatocytes and the glutathione reductase inhibitor 1,3-bis(2-chloroethyl)-1-nitrosourea. Eur J Biochem. 1984;138:459–463. doi: 10.1111/j.1432-1033.1984.tb07938.x. [DOI] [PubMed] [Google Scholar]
- 59.Gilbert HF. Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol. 1990;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- 60.Jones DP. Redefining oxidative stress. Antioxid Redox Signal. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 61.Woodmansee AN, Imlay JA. Reduced flavins promote oxidative DNA damage in non-respiring Escherichia coli by delivering electrons to intracellular free iron. J Biol Chem. 2002;277:34055–34066. doi: 10.1074/jbc.M203977200. [DOI] [PubMed] [Google Scholar]
- 62.Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, Goldbaum FA, Briggs WR, Bogomolni RA. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 63.Pathak GP, Ehrenreich A, Losi A, Streit WR, Gärtner W. Novel blue light-sensitive proteins from a metagenomic approach. Environ Microbiol. 2009;11:2388–2899. doi: 10.1111/j.1462-2920.2009.01967.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.