Abstract
Many proteins, especially transporters, are thought to undergo large conformational alterations during their functional cycle. Since x-ray crystallography usually gives only the most stable conformation, other methods are needed to probe this conformational change. Site-directed disulfide cross-linking is often very useful for this purpose. We illustrate this by using the Escherichia coli AcrB, a proton-motive-force-dependent multidrug efflux transporter. Crystallographic studies of the asymmetric trimer of AcrB suggest that each protomer in the trimeric assembly goes through a cycle of conformational changes during drug export (functional rotation hypothesis). Site-directed disulfide cross-linking between those residues that come close to each other in only one stage in the cycle inactivated the transporter, showing that the conformational changes indeed occurred in vivo and that they are required for drug transport. A dsbA strain, which has a diminished activity to form disulfide bonds in the periplasm, was used to verify the conclusion by showing a restored transport activity in this strain. Furthermore, we describe “a real-time cross-linking experiment,” in which rapidly reacting, sulfhydryl-specific cross-linkers, methanethiosulfonates, inactivate the AcrB double-cysteine mutant expressed in dsbA cells instantaneously.
Keywords: Site-directed cross-linking, conformational change, disulfide bond, sulfhydryl-specific cross-linker, methanethiosulfonate, dsbA, multidrug efflux transporter, AcrB, Escherichia coli
1. Introduction
Disulfide cross-linking is an attractive method for probing conformational changes in proteins, as was shown more than 20 years ago (1). When this method is applied to intact cells of Escherichia coli, however, it is important to realize that the formation of disulfide bonds is facilitated by an elaborate system in the periplasm (2), whereas such a system does not exist within the membrane bilayer or in the cytosol. Thus the addition of oxidizing agents (1) or cross-linkers (3) is often necessary to cover the cysteine residues in the latter locations. It may also be possible to use genetically altered strains that are able to form disulfide bonds in the cytosol (4, 5). In the example to be described, cross-linking occurs in the periplasm, and we also describe the manipulation of the disulfide-forming machinery in this compartment.
Multidrug efflux transporters of the RND (resistance-nodulation-division) superfamily, such as AcrB of Escherichia coli, play an important role in both intrinsic and elevated multidrug resistance of Gram-negative bacteria (6). These transporters associate with two other classes of proteins, the outer membrane channel such as TolC and the periplasmic membrane fusion protein such as AcrA (6). This tripartite complex spans the entire thickness of the cell envelope, containing the inner and outer membranes as well as the periplasm between them (7) and expels, driven by a proton-motive force, the drugs directly into the external medium. This process must obviously involve large conformational alterations of AcrB.
The AcrB exists as a homotrimer in which each subunit provides 12 transmembrane helices and a large periplasmic headpiece (8). The newly elucidated asymmetric crystal structure of AcrB trimer (9–11) shows that each protomer has a conformation significantly different from that of its neighbors, presumably representing each of the steps (open access, drug binding, and drug extrusion) in the conformational change cycle for drug export and suggests a functionally rotating mechanism.
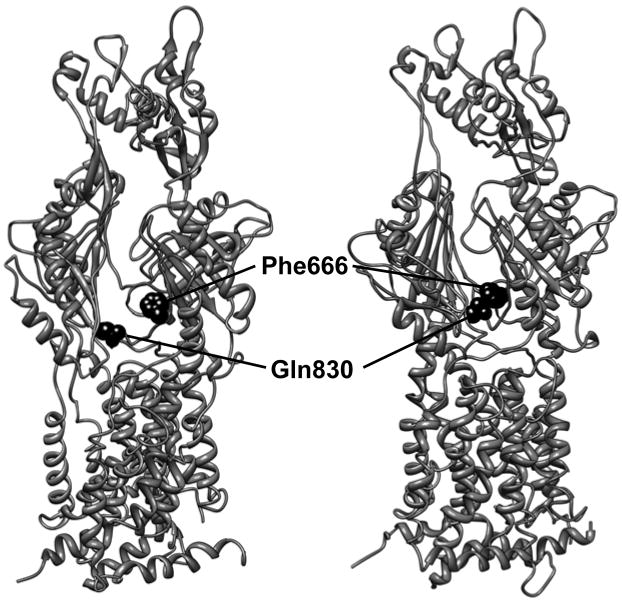
We provided biochemical evidence supporting this hypothesis by site-directed disulfide cross-linking experiments (12), taking advantage of the observation that the cyclic conformational change of AcrB involves the opening and closing of the large external cleft in the periplasmic domain. The AcrB proteins with double-Cys mutations on both sides of the cleft, where a disulfide bond would be produced only in the extrusion protomer (Fig. 1), are inactive. The fact that the inactivation is due to the formation of disulfide bonds has been supported by the restoration of transport activity in a dsbA strain, which has a defective DsbA, a main enzyme that catalyzes disulfide bond formation in the periplasm (2). However, dsbA did not restore the activity of some mutant pairs, and formation of large aggregates of AcrB suggested that cross-linking may have occurred during the assembly of the trimeric complex. In view of these potential problems relying on the activity of naturally cross-linked proteins, it is important that the cross-linking and inactivation of AcrB could be observed in “real rime” by using methanethiosulfonate (MTS) cross-linkers, which are known to act much more rapidly than the usual cross-linkers (13, 14). The addition of such cross-linkers to dsbA cells expressing double-Cys mutant AcrB proteins that are pumping out a fluorescent dye, ethidium, stops the function of the pump instantaneously. (An alternative approach is the reduction of disulfide bonds with externally added dithiothreitol, which resulted in the recovery of the transport function [15]). All these results show that when the external cleft in the periplasmic domain of AcrB becomes closed to allow the formation of a disulfide bridge or a cross-linked structure, then the resultant loss of flexibility in the conformation inactivates the efflux pump.
Fig. 1.
Predicted conformational change of AcrB protomer during drug transport and the choice of the residues to introduce Cys mutations. The figure shows two of the protomers (the access protomer on the left and the extrusion protomer on the right) of the asymmetric AcrB structure (9), viewed from outside of the trimer. The large cleft in the periplasmic domain, wide open in the access protomer (left), is nearly completely closed in the extrusion protomer (right). The distance between the two residues, Phe666 and Gln830 (shown as dark space-filling models), on the opposing walls of the cleft becomes close enough to produce a disulfide bond only in the extrusion protomer (right, 4.4 Å), as opposed to the access (left) and binding (not shown) protomers (>9 Å). These two residues were therefore chosen as a pair of the positions to introduce the Cys mutations in order to test the conformational change of AcrB protomer during transport function. The figure was drawn by using the UCSF Chimera package (18), by using the PDB file 2DRD.
2. Materials
2.1. Site-Directed Mutagenesis and Mutant Plasmids Preparation
For manipulation of recombinant DNA, we follow the standard methods (16).
Template plasmid (see Note 1): pSCLBH (12). This pSPORT1 (medium copy-number vector with an amp marker; Gibco BRL, Invitrogen Corp) based plasmid expresses the cysteineless and hexahistidine-tagged AcrB protein, CL-AcrBHis, under the control of the lac promoter. The codons for two intrinsic cysteines of the acrB gene have been converted to serine codons. The CL-AcrBHis is fully functional.
PfuUltra high-fidelity (HF) DNA polymerase (Stratagene).
10x PfuUltra HF reaction buffer (Stratagene).
dNTP mix: 2.5 mM each dNTP, store at −20°C.
Oligonucleotide primers to introduce Cys mutations (see Note 2).
DpnI restriction enzyme (New England Biolabs).
Competent cells of E. coli DH5α (16). Store at −80°C.
Luria-Bertani (LB:Tryptone 1%; Yeast Extract 0.5%; NaCl 0.5%) agar plates, supplemented with ampicillin (100 μg/mL).
LB liquid medium, supplemented with ampicillin (100 μg/mL).
Plasmid miniprep kit (Fermentas Inc or Qiagen Inc).
Tris-EDTA (TE) buffer: 10 mM Tris-HCl, 1 mM EDTA, pH 8.0. Autoclaved, store at room temperature.
Autoclaved distilled water. Store at room temperature.
2.2. Functional Characterization of Single- and Double-Cys Mutants and Effect of dsbA Mutation on Transport Activities
E. coli AG100YB (12): acrB deletion mutant (ΔacrB::Spcr), generated from a K-12 strain AG100. In this strain, 90% of the acrB gene is deleted and replaced with the spectinomycin resistance (Spcr) gene, aadA. Prepare as competent cells, and store at −80°C. (Bacterial strains and plasmids mentioned in this chapter are available from the authors.)
E. coli AG100YBD (12): ΔacrB dsbA mutant (dsbA1::kan), derived from AG100YB. Prepare as competent cells, and store at −80°C.
LB agar plates, supplemented with ampicillin (100 μg/mL).
LB liquid medium supplemented with ampicillin (100 μg/mL).
LB agar, autoclaved and kept at 50°C.
Cholic acid sodium salt: hydrate, from ox or sheep bile, ≥99% (Sigma)
Square dishes with grid, 100 × 100 × 15 mm (Fisher Scientific)
Disposable loops, 1 μL (Fisher Scientific).
2.3. Real-Time Inactivation of AcrB Double-Cys Mutant by the Use of Fast-Acting MTS Cross-Linker
AG100YBD (ΔacrB dsbA) strains transformed by the plasmids expressing wild-type cysteineless AcrB, or single- or double-Cys mutant.
LB liquid medium with ampicillin (100 μg/mL).
Assay buffer: 50 mM sodium phosphate, 0.1 M NaCl, 0.1% (vol/vol) glycerol, pH 7.0. Autoclaved and stored at room temperature.
Shimadzu RF-5301PC spectrofluorometer (Shimadzu Scientific Instruments, Inc.).
Quartz cuvette for fluorometry, 10 mm path length (Starna Cells, Inc.)
Micro stirring bar, 7 mm length × 2 mm diameter (Fisher Scientific).
Ethidium bromide, 2 mM stock solution in distilled water. Store at −20°C.
MTS reagents: 5 mM 1,2-ethanediyl bismethanethiosulfonate (MTS-2-MTS, approximately 5.2 Å spacer) and 10 mM pentyl MTS (5-MTS) (Toronto Research Chemicals (see Note 3). Freshly dissolved in dimethyl sulfoxide (DMSO)-ethyl acetate (3:1, vol/vol) (see Note 4), and kept on ice until use (see Note 5).
Carbonyl cyanide m-chlorophenylhydrazone (CCCP), as a proton conductor: 10 mM stock solution in ethanol. Store at −20°C. The solution is stable at least for a month.
3. Methods
3.1. Site-Directed Mutagenesis and Preparation of Mutant Plasmids
The method for introduction of Cys mutations follows the QuickChange site-directed mutagenesis protocol from Stratagene. A detailed instruction manual is available at their website.
Combine 5 μL 10x PfuUltra HF reaction buffer, 5–20 ng template plasmid, 4 μL dNTP mix, 2 μL each of two complementary oligonucleotide primers (10 pmol/μL in TE buffer). Adjust the volume to 49.2 μL with distilled water, then add 0.8 μL PfuUltra HF DNA polymerase (2.5 U/μL) (see Note 6) and mix gently.
Overlay the reaction mixture with approximately 30 μL of mineral oil if necessary.
Cycle the reaction using the PCR program of 3 min at 95°C followed by 16 cycles of amplification, and an appropriate time for the final extension at 68°C. An amplification cycle consists of: 45 s at 95°C, 45 s at an appropriate annealing temperature, and 1 min/kb of plasmid length at 68°C.
Remove the mineral oil, and treat the reaction mixture with 10 U of DpnI at 37°C for 1 h to digest the template DNA.
Transform E. coli DH5α with the DpnI-treated DNA. Up to 8 μL of the reaction can be used to 100 μL of the competent cells prepared with the CaCl2 procedure. Incubate the transformation plates overnight at 37°C.
Prepare the plasmids from the culture of colonies using Plasmid miniprep kit, and sequence the acrB gene to ensure the presence of the desired mutation and also to confirm that there are no unintended nucleotide sequence alterations.
3.2. Functional Characterization of Single- and Double-Cys Mutants and Effect of dsbA Mutation on Transport Activities
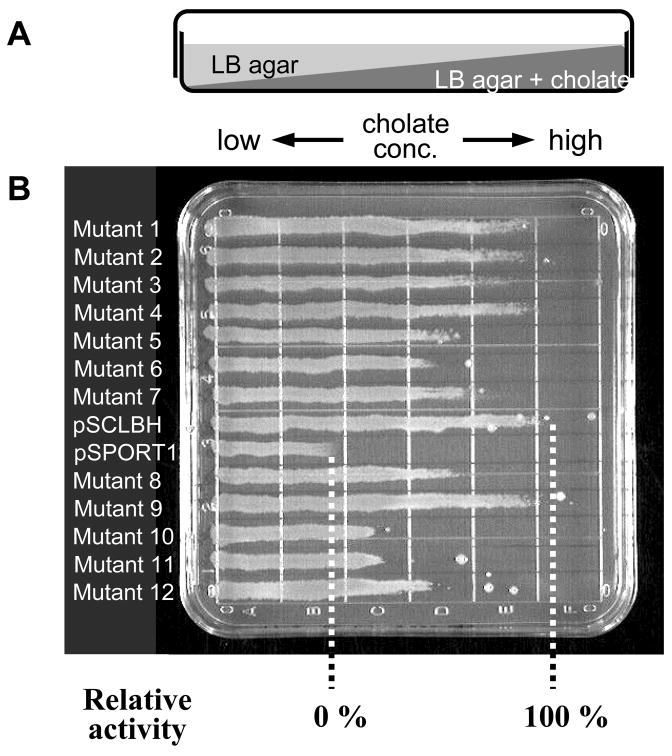
Activity of each mutated AcrB protein can be evaluated by the drug susceptibility of the Δ acrB strain expressing each mutant protein. We use the gradient plate technique (17) (Fig. 2A and B), with a plate containing two agar layers that allow a gradual increase of the drug concentration along one horizontal axis.
Fig. 2.
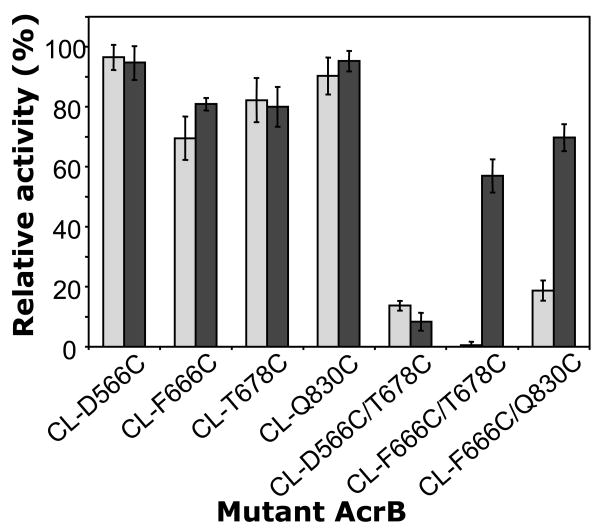
Effect of dsbA mutation on the activities of AcrB Cys mutants evaluated by drug susceptibility of the host strains. (A) Side view of a gradient plate containing cholate in the lower layer. (B) An example of measurement of bacterial drug susceptibility using the gradient plate. Fully functional control, the ΔacrB strain harboring wild-type AcrB-expressing-plasmid, pSCLBH, grew well up to the region containing cholate at high concentrations, while the nonfunctional control, the host with vector only (pSPORT1), grew only for a short length. The relative activity of each mutant was calculated from the length of its growth compared to the two controls, pSCLBH (100%) and pSPORT1 (0%). (C) Activities of AcrB single- and double-Cys mutants and the effect of dsbA mutation. AcrB proteins containing double-Cys mutations on both sides of the cleft are strongly compromised in function in dsbA+ host strain AG100YB (gray bars, three columns on the right). In contrast in the dsbA host AG100YBD (black bars), the CL-F666C/T678C and CL-F666C/Q830C double mutants largely retain their transport activities, indicating that the inactivation of these two double-Cys mutants in the dsbA+ host is due to the formation of disulfide bonds. This is one of the biochemical evidences that the conformational changes, including the closure of the cleft suggested from the asymmetrical trimer structure, indeed occur in vivo and that they are required for AcrB function. (Reproduced from ref. 12.)
This method, which relies on the formation of disulfide bonds between nearby Cys residues in the periplasm, works for many pairs of Cys residues. However, there is a possibility that disulfide bonds are formed during the formation of the multi-protein assembly, that is, in a way that has nothing to do with the conformational alterations during the functional cycle of the transporter (12). In order to exclude such a possibility, the real-time cross-linking of already assembled AcrB, described below, is very useful.
Transform competent cells of a pair of isogenic strains, AG100YB (Δ acrB) and AG100YBD (Δ acrB dsbA), with plasmids containing mutated acrB. As fully functional and nonfunctional AcrB controls, cells are also transformed with plasmids containing the wild-type cycteineless acrB and vector only. Incubate the transformation plates overnight at 37°C.
Pick a single colony from the fresh transformation plates (see Note 7), and grow in LB liquid medium with ampicillin until an optical density at 660 nm (OD660) reaches approximately 0.6 (mid-exponential-phase) (see Note 8).
To prepare a linear concentration gradient of cholic acid in LB agar plates, add cholic acid to LB agar kept at 50°C, dissolve and mix well, and pour 20 mL into the square dishes. Slant the plates sufficiently so that the entire bottom is just covered. Wait until the agar hardens. It is necessary to adjust the concentration of cholic acid in the bottom layer depending on the susceptibility levels of the strains used (see Note 9).
Place the dishes in the normal horizontal position, and overlay ~28 mL LB agar without drugs to cover the bottom layer (adjust the volume so that the bottom layer is covered sufficiently as shown in Fig. 2A). Wait until the agar hardens, and the surface becomes dry. Use the plates in the same day.
Dilute mid-exponential-phase cultures to an OD660 of 0.1 with LB broth with ampicillin.
Streak the diluted cultures using Fisherbrand disposable 1 μL loops as a line across the plate, in parallel with the drug gradient (Fig. 2B, see Note 10). Each plate should also be inoculated with control strains harboring wild-type pSCLBH and the vector pSPORT 1 (see Note 11). Incubate the plates at 37°C for 24 h.
Measure bacterial growth across the plates, from low to high drug concentrations.
Calculate the relative activity of each mutated AcrB protein as follows. Divide the length of growth of each mutant strain minus that of the strain carrying the vector alone, by the length of growth of the strain containing pSCLBH minus that of the vector-containing strain, and multiply the result by 100. Thus, the full efflux activity and no activity should produce values of 100 and 0%, respectively (Fig. 2B). An example of the results obtained is shown in Fig. 2C.
3.2. Real-Time Inactivation of AcrB Double-Cys Mutant by the Use of Fast Acting MTS Cross-Linker
Pick fresh single colonies of AG100YBD transformants containing pSCLBH, single-and double-Cys mutant plasmids, and grow them in 2 mL LB with ampicillin overnight without shaking at room temperature (see Note 12).
Dilute the cultures into 10 mL of LB medium with ampicillin to an OD660 ~0.08, and grow with shaking at 37°C to an OD660 of 0.7–0.9 without IPTG induction.
Harvest the cells at room temperature, wash once with 10 mL of assay buffer, and resuspend in 10 mL of assay buffer. Measure the OD660.
Dilute the cells with assay buffer to an OD660 of 0.2 (see Note 13), place 2 mL of cells into the quartz 1 × 1 cm cuvette with micro stirring bar. Set the cuvette in the spectrofluorometer, and stir gently.
Add 5 μL of 2 mM ethidium bromide (final concentration, 5 μM, see Note 13) into the cell suspension, and monitor the ethidium influx rate into the cells at room temperature with fluorescence increase at excitation and emission wavelengths of 520 nm and 590 nm, respectively.
After a few minutes of preincubation with ethidium bromide, add to the cells one of the following: 8 μL of solvent alone (DMSO-ethyl acetate [3:1, vol/vol]), 8 μL of 5 mM MTS-2-MTS (final concentration, 20 μM), or 8 μL of 10 mM 5-MTS (final concentration, 40 μM) (see Note 14). Follow the accumulation of ethidium for several minutes more.
As a positive control of AcrB inactivation, add 8 μL of 10 mM CCCP (final concentration, 40 μM) to the cells instead of MTS reagents.
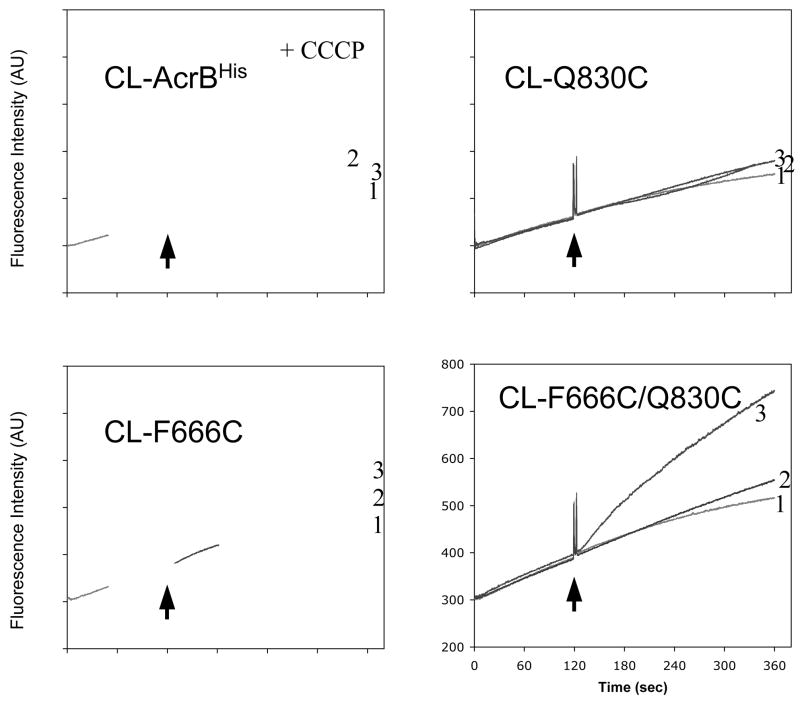
An example of the data is shown in Fig. 3. It is seen that only in the mutant protein containing the double Cys residues, an instantaneous inactivation of the AcrB pump occurred, causing the increased influx of ethidium (the bottom right panel).
Fig. 3.
Real-time inactivation of AcrB double-Cys mutant by cross-linking with MTS reagent. Cellular accumulation of ethidium by the ΔacrB dsbA host AG100YBD cells expressing each AcrB protein was monitored continuously by measuring the fluorescence of the ethidium-nucleic acid complex. After 2 min of incubation with ethidium bromide (arrow), solvent alone (curve 1) or MTS reagents (curve 2, 5-MTS; curve 3, MTS-2-MTS) was added to cell suspension. The AcrB with double-Cys mutation in the cleft, CL-F666C/Q830C, was active in the dsbA host in the efflux of ethidium, so that only very slow entry of ethidium was seen initially (bottom right panel). However, upon addition of MTS-2-MTS, cross-linking apparently occurred, so that AcrB became inactivated, inducing a rapid ethidium influx and increased fluorescence (curve 3, bottom right panel). In contrast, MTS-2-MTS had little effect on the ethidium entry rate in cells expressing no-Cys (CL-AcrBHis) or a single-Cys (CL-F666C and CL-Q830C) AcrB. A control reagent, 5-MTS (a non-cross-linker), produced no AcrB inactivation in any of the mutants (curves 2). As a positive control, a proton conductor (CCCP), was added to the cells expressing CL-AcrBHis, resulting in a rapid influx of ethidium because of inactivation of AcrB due to the loss of the proton-motive force (top left panel). (Reproduced from ref. 12.)
Acknowledgments
This work was supported by research grant AI-09644 from U.S. Public Health Service.
Footnotes
The template plasmid should be isolated from a dam+ E. coli strain for the following removal step by DpnI digestion. The majority of the commonly used E. coli strains are dam+. We use E. coli DH5α or DH10B (Invitrogen) for plasmid preparation.
We designed most of the primers in the length of 20–30 bases, with the calculated melting temperature of about 60°C or higher, and checked that there is no significant matching sequences in other parts of acrB.
We chose the MTS reagents that are likely to cross the outer membrane. MTS cross-linkers with different size spacers are available from approximately 3.9 Å (MTS-1-MTS) to approximately 10.4 Å (MTS-6-MTS). We also examined MTS-5-MTS (approximately 9.1 Å spacer) and obtained similar results as MTS-2-MTS. As a non-cross-linker control reagent, we used 5-MTS whose length is similar to that of MTS-2-MTS.
MTS-2-MTS is soluble in DMSO, but 5-MTS is not. We found that ethyl acetate (good solvent for 5-MTS) alone affected significantly the ethidium influx rate into the cells. To minimize this effect and to obtain a reasonable solubility for both MTS-2-MTS and 5-MTS, DMSO-ethyl acetate (3:1, vol/vol) was the best solvent we found so far.
MTS reagents are sensitive to moisture and are hydrolyzed in water over a period of time. The reagents should be stored in a desiccator at −20°C and warmed up to room temperature before opening of the vial. We made up the solutions in organic solvent immediately prior to use and the solutions, kept on ice, were used within ~1.5 h.
The protocol from Stratagene recommends the use of 1 μL (2.5 U) of PfuUltra HF DNA polymerase. We found that 0.5–0.8 μL enzyme also provided enough products for obtaining plasmids with point mutation.
We observed that the levels of resistance of the Δ acrB cells expressing AcrB mutants from pSPORT1 derivatives often varied after storage of the transformed strains for several days at 4°C. Therefore, for these experiments we only used freshly transformed cells.
To avoid the nonreproducible drug susceptibility patterns caused by strong overexpression of AcrB alone, acrB is expressed without isopropyl-β-D-thiogalactopyranoside (IPTG) induction.
To calculate the relative activity of each mutated AcrB, both controls (fully functional pSCLBH and nonfunctional vector only) should grow and show large differences on the lengths of growth zone on the same plate (see Fig. 2B). We tested several AcrB substrates with gradient plates and found that cholic acid satisfied this requirement. With some compounds such as novobiocin and rhodamine 6G, it was difficult to find the appropriate concentration in the bottom layer, because the difference in susceptibility to these compounds was too large between the two controls. Thus at concentrations at which the fully active control grew well, no growth was seen for the nonfunctional control, whereas at other concentrations that gave a short growth zone for the vector only control, cells expressing the fully active transporter gave growth covering the entire length of the plate. We also adjusted the concentration of cholic acid in the lower layer to 10,000 μg/ml for AG100YB and to 16,000 or 18,000 μg/ml for AG100YBD, since the resistance levels of these strains to cholate were different.
We streak the cells twice on the same line without refilling the loop, from high to low drug concentrations using one side of the loop and then low to high concentrations using the other side of the loop. Dry the surface of the plate well to avoid flooding of the plate.
We assay all mutants at least four times with controls, and change the position of strains on each plate.
Overgrowth of the preculture sometimes caused non-reproducible results in ethidium influx assay. To avoid this, the preculture was prepared from a fresh transformant colony without shaking, either at room temperature or at 30°C. We also tested the frozen glycerol stock, which was made with a culture in mid-exponential phase (OD660~0.5) from a fresh colony and stored in aliquots at −80°C, as the inoculum for the preculture. This method produced similar results until up to two weeks of storage of the stock.
The concentrations of cell suspension and ethidium bromide can be modified depending on the conditions of experiments. Generally, higher concentrations of the cells and ethidium bromide show faster increase of the fluorescence. For our conditions (host strain and the expression levels of AcrB), the cell suspension of OD660 of 0.2 and 5 μM ethidium bromide produced good results.
MTS reagents at these final concentrations produced the best results for these strains. MTS reagents at higher concentrations caused increased influx of ethidium even with the cells expressing the wild-type cysteineless AcrB. On the other hand, MTS-2-MTS at lower concentrations sometimes failed to cause a detectable inactivation of AcrB double-Cys mutants. We also note that a very low concentration of MTS reagents (4 μM) gave the best results for another host strain derived from BL21.
References
- 1.Falke, Koshland DE., Jr Global flexibility in a sensory receptor: a site-directed cross-linking approach. Science. 1987;237:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- 2.Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 3.Moore KJ, Fillingame RH. Structural interactions between transmembrane helices 4 and 5 of subunit a and the subunit c ring of Escherichia coli ATP synthase. J Biol Chem. 2008;283:31726–31735. doi: 10.1074/jbc.M803848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart EJ, Åslund F, Beckwith J. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masip L, Pan JL, Haldar S, et al. An engineered pathway for the formation of protein disulfide bonds. Science. 2004;303:1185–1189. doi: 10.1126/science.1092612. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Three’s company: component structures bring a closer view of tripartite drug efflux pumps. Curr Opin Struct Biol. 2004;14:741–747. doi: 10.1016/j.sbi.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 9.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structure of a multidrug transporter reveals a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 10.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 11.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takatsuka Y, Nikaido H. Site-directed disulfide cross-linking shows that cleft flexibility in the periplasmic domain is needed for the multidrug efflux pump AcrB of Escherichia coli. J Bacteriol. 2007;189:8677–8684. doi: 10.1128/JB.01127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akabas MH, Stauffer DA, Xu M, Karlin A. Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science. 1992;258:307–310. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- 14.Kenyon GL, Bruice TW. Novel sulfhydryl reagents. Methods Enzymol. 1977;47:407–430. doi: 10.1016/0076-6879(77)47042-3. [DOI] [PubMed] [Google Scholar]
- 15.Seeger MA, von Ballmoos C, Eicher T, et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 17.Bryson V, Szybalzski W. Microbial selection. Science. 1952;116:45–51. [PubMed] [Google Scholar]
- 18.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]