Summary
Cell division in Gram-negative organisms requires coordinated invagination of the multi-layered cell envelope such that each daughter receives an intact inner membrane (IM), peptidoglycan (PG) layer, and outer membrane (OM). Here, we identify DipM, a putative LytM endopeptidase in Caulobacter crescentus, and show that it plays a critical role in maintaining cell envelope architecture during growth and division. DipM localized to the division site in an FtsZ-dependent manner via its peptidoglycan binding LysM domains. Although not essential for viability, ΔdipM cells exhibited gross morphological defects, including cell widening and filamentation, indicating a role in cell shape maintenance and division that we show requires its LytM domain. Strikingly, cells lacking DipM also showed OM blebbing at the division site, at cell poles, and along the cell body. Cryo electron tomography of sacculi isolated from cells depleted of DipM revealed marked thickening of the peptidoglycan as compared to wild type, which we hypothesize leads to loss of trans-envelope contacts between components of the Tol-Pal complex. We conclude that DipM is required for normal envelope invagination during division and to maintain a sacculus of constant thickness that allows for maintenance of OM connections throughout the cell envelope.
Keywords: divisome, DipM, outer membrane, M23 peptidase, LytM, LysM
Introduction
The bacterial cell envelope serves to maintain the cell’s shape and provide a selective barrier between the contents of the cytoplasm and the extracellular environment. Though they serve a structural purpose, the membranes and peptidoglycan cell wall that constitute the envelope are subject to constant remodeling as the cell grows, divides, and, in some cases, differentiates. Peptidoglycan (PG) is the polymeric network of strands of alternating N-acetyl-glucosamine and N-acetyl-muramic acid sugars connected by peptide crosslinks that imparts mechanical strength and shape to the envelope. Its dynamic structure is regulated by an array of enzymes that catalyze formation and breakage of bonds within the network in a manner that is directed by the bacterial cytoskeleton (den Blaauwen et al., 2008, Vollmer and Bertsche, 2008). The inner and outer membranes (IM and OM) of Gram-negative species are linked to the PG and to each other by membrane-protein-PG interactions to maintain remarkably constant cell envelope geometry as the cell grows and divides. The coordination of PG growth and remodeling with membrane remodeling is particularly important during cell division, when invagination of the multi-layered cell envelope culminates in daughter cell separation and formation of new cell poles.
Cell division in most bacteria is accomplished by the action of ~20 proteins that assemble into a molecular machine (the divisome) at the incipient division site (Harry et al., 2006). The site of division is established by localized polymerization of the tubulin-like FtsZ GTPase into a structure (the Z-ring) that acts as a scaffold for assembly of other divisome components and was recently shown to generate constrictive forces that may drive invagination (Osawa et al., 2008). FtsZ is required for the mid cell recruitment of all other cell division proteins, which arrive in several waves as the cell cycle progresses (Aarsman et al., 2005, Gamba et al., 2009). Although the mechanisms underlying bacterial division remain to be elucidated, it is hypothesized that FtsZ and associated factors generate constrictive forces that invaginate the inner membrane and that may be transduced to PG synthetic machinery to bias new cell wall growth inward. Repeated rounds of FtsZ-driven constriction and inward-directed cell wall synthesis eventually lead to IM fission, complete invagination and separation of PG, and OM fission.
The efficiency of division requires a balance in the activities of PG synthetic and hydrolytic enzymes, as well as dynamic membrane-PG connections that allow the IM, PG, and OM layers of the envelope to invaginate coordinately and eventually separate. Among the proteins reported to localize to the division site in the Gram-negative organisms Escherichia coli and/or Caulobacter crescentus are PG biosynthetic enzymes, including FtsI (Weiss et al., 1997, Wang et al., 1998, Costa et al., 2008), MurG (Aaron et al., 2007, Mohammadi et al., 2007), PBP2 (Vats et al., 2009, Den Blaauwen et al., 2003) and PBP1B (Bertsche et al., 2006), which mediate construction of new cell wall at the division site. Of these, only the FtsI transpeptidase is specific to PG synthesis during division (Spratt, 1977); the others play more general roles in both the elongation and division modes of PG synthesis. In addition to synthesis, cleavage of existing PG at the division site is necessary for OM invagination and separation of daughter PG (sacculi). The division site localized hydrolytic amidase AmiC helps to fulfill this function, as deletion of amiC (alone or with its homologs amiA and amiB) in Escherichia coli leads to chaining of cells with compartmentalized cytoplasms that are connected by constricted PG linkages (Bernhardt and de Boer, 2003, Heidrich et al., 2001, Heidrich et al., 2002). AmiC may also collaborate with the endopeptidases PBP4 and PBP7 and the carboxypeptidase PBP5 during septal splitting, as combining mutations in these factors in E. coli exacerbates cell separation failure (Priyadarshini et al., 2006). Two putative PG endopeptidases of the LytM/M23 family, EnvC and NlpD, were also recently shown to localize to the division site in E. coli and to contribute to PG splitting and cell separation (Uehara et al., 2009, Bernhardt and de Boer, 2004, Hara et al., 2002, Ichimura et al., 2002). Similar to the case of amiC, when combinations of genes encoding LytM-domain containing proteins in E. coli were deleted, cell chaining was observed.
In Gram-negative organisms, cell wall remodeling is linked to outer membrane invagination at the division site primarily by the trans-envelope Tol-Pal complex, which localizes to the division site and establishes IM-PG-OM contacts that bring the OM inward during division (Gerding et al., 2007). When components of this five-member protein complex were mutated in E. coli, OM blebbing at division sites and poles was observed under normal growth conditions and cells failed to separate efficiently when grown in low salt (Gerding et al., 2007). More remarkably, in Caulobacter crescentus TolA, TolB, and Pal are essential for viability and cell division, with dramatic cell separation defects and OM blebbing occurring when they are depleted (Y.-C. Yeh, LS, and H. McAdams, unpublished).
The temporal coordination of envelope invagination varies among bacteria, leading to morphological differences at the division site and in the resulting cell poles. This likely reflects mechanistic differences in the progression of division in different organisms. In Gram-positive organisms like the spore-forming bacterium Bacillus subtilis, which lack an OM, daughter cells with independent IMs are connected by a wall of PG perpendicular to the long axis of the cell (the septum) that is cleaved apart after a significant time has passed since its synthesis (Burdett and Higgins, 1978). In E. coli, IM invagination, PG synthesis and cleavage, and OM ingression are more tightly coordinated in time, such that OM invagination follows closely behind in-growth and cleavage of the cell wall (Burdett and Murray, 1974a, Burdett and Murray, 1974b). A third scenario exists in Caulobacter, where IM invagination occurs synchronously with PG synthesis and cleavage, but OM invagination lags behind significantly (Judd et al., 2005). Although extensively studied in E. coli, in particular, the process of coordinated PG synthesis and invagination of the cell membranes at the division site is still not well understood. Moreover, little characterization of this process has been undertaken in Caulobacter, a morphologically and phylogenetically distinct α-proteobacterium. In this study, we identified and characterized DipM, a putative endopeptidase of the LytM family in Caulobacter and determined that it plays a critical role in maintaining proper cell envelope architecture during growth and division.
Results
DipM, a putative LytM endopeptidase, is localized to the site of cell division
In a microscopy-based screen for cell division proteins in Caulobacter (E.D.G. and L.S., unpublished), we identified a novel protein encoded by CC1996 that we named division involved peptidase carrying LysM domains, or DipM. Sequence analysis of DipM indicates that it contains an N-terminal signal peptide (with cleavage predicted between residues 24 and 25 (Juncker et al., 2003)), several peptidoglycan-binding Lysin Motifs (LysM domains) in the N-terminal and central regions of the protein (Buist et al., 2008), and a C-terminal LytM/M23 peptidase domain (see Fig. 3A). This domain organization is similar to that of the NlpD protein in E. coli, which is functionally implicated in PG hydrolysis during division to allow cell separation (Uehara et al., 2009). Morover, dipM resides adjacent to the surE and pcm stationary phase survival genes in Caulobacter, a genomic position occupied by the nlpD gene in E. coli (Ichikawa et al., 1994, Lange and Hengge-Aronis, 1994), further suggesting that these proteins may be orthologs. However, whereas NlpD is OM-linked lipoprotein, DipM is predicted to be soluble in the periplasm. The similarities between NlpD and DipM prompted us to investigate a role for DipM in Caulobacter cytokinesis.
Figure 3.
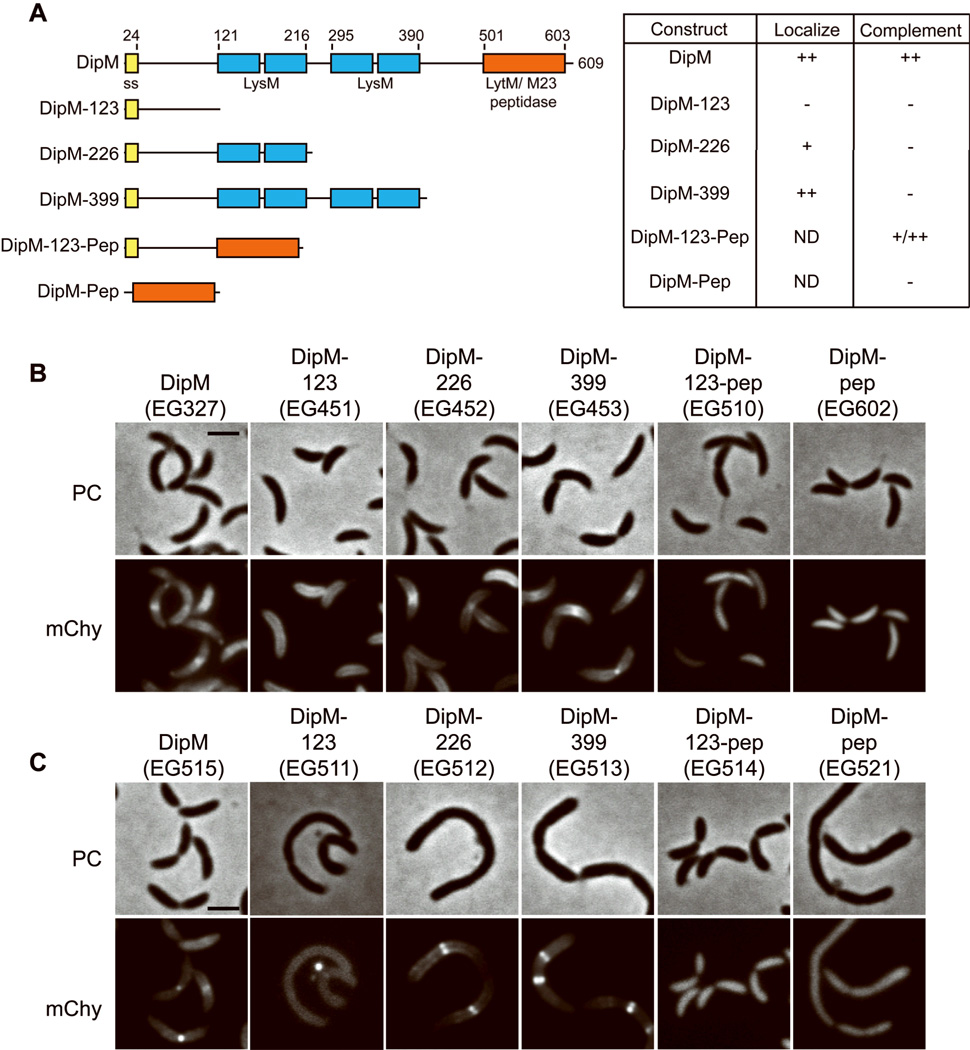
The LysM domains of DipM mediate its mid cell targeting, while the LytM domain is critical for its function. A. Graphic representation of domain organization of DipM and DipM truncations showing relative location of signal sequence (ss), LysM domains, and LytM domain. Numbers indicate amino acid position. Table summarizes localization and ability to complement loss of wild type DipM for each. ND, not determined. B. Localization of mCherry (mChy) fusions to the indicated DipM truncation derivatives in otherwise wild type cells. PC = phase contrast. Cells were grown in PYE with 0.5 mM vanillate for 2 h prior to imaging. C. Morphology of cells expressing the indicated DipM truncation derivatives in the absence of DipM. Cells were grown in PYE with 0.2% glucose to deplete DipM and 0.5 mM vanillate to express the indicated mCherry fusion protein for 17 h prior to imaging. Bars = 2 µM.
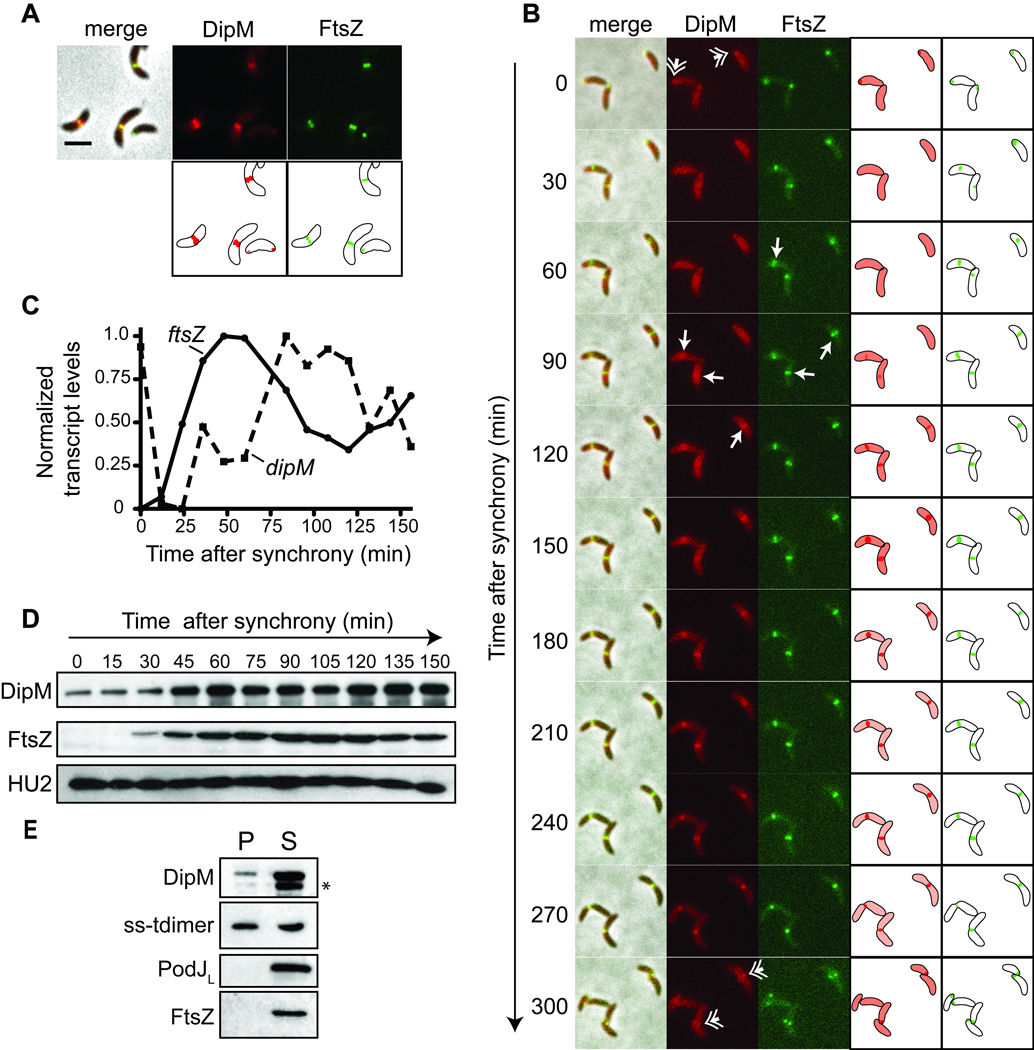
We asked if, like NlpD and EnvC, DipM localizes with FtsZ at the cell division site. To test this idea, a strain bearing xylose-inducible dipM-mCherry at the chromosomal xylX locus as well as vanillate-inducible ftsZ-cfp at the chromosomal vanA locus was generated. In the presence of both inducers, DipM-mCherry was found to colocalize with FtsZ-CFP at the division site (Fig 1A), consistent with a possible role in cell division. DipM-mCherry was also frequently observed in a focus that co-localized with FtsZ at the new pole in cells without a mid cell band of DipM-mCherry (Figs 1A,B) and, occasionally, at the opposite pole (Figs 1A, 3B). We followed the localization of DipM-mCherry and FtsZ-CFP over the course of the cell cycle by isolating swarmer cells and imaging them as they grew synchronously on agarose pads (Fig. 1B). As reported previously (Thanbichler and Shapiro, 2006), FtsZ-CFP began as a focus at the new pole of swarmer cells, and appeared at the incipient division site in a loose pattern that was focused into a stable ring at 60 to 90 minutes post-synchrony. DipM-mCherry first co-localized with FtsZ at the new pole, then began to accumulate in a mid cell band shortly after a distinct, focused ring of FtsZ-CFP was observed (90 to 120 min post-synchrony), indicating that DipM is recruited to the division site early in the assembly of the divisome.
Figure 1.
DipM is a cell cycle regulated division site localized protein. A. Co-localization of DipM-mCherry and FtsZ-CFP in strain EG281. Cartoon representations of localized protein are depicted below fluorescent panels. Cells were grown in PYE media with 0.3% xylose for 2 h and 0.5 mM vanillate for 1 h prior to imaging. Bar = 2 µm. B. Localization of DipM-mCherry and FtsZ-CFP in strain EG281 over the cell cycle. Cartoon representations of DipM (red) and FtsZ (green) localizations are also depicted. Cells were grown as in A prior to synchrony, then applied to M2G agarose pads for imaging. Hatched arrows indicate polar DipM. Arrows indicate time at which FtsZ or DipM is observed in a stable mid cell band in each cell. C. Normalized abundance of ftsZ and dipM transcript levels over the cell cycle in wild type cells (CB15N). D. Immunoblots using antibodies against the indicated proteins of cell lysates from synchronized wild type cells (CB15N) at the indicated times post-synchrony. Cells were grown in M2G. E. Immunoblots of periplasmic (P) and spheroplast (S) fractions using antibodies that recognize the indicated proteins after fractionation of strain LS4032. * indicates putative DipM cleavage product below full-length DipM band.
A number of cell division proteins in Caulobacter are transcriptionally and post-translationally regulated over the course of the cell cycle and peak in abundance and/or activity at their time of function (Laub et al., 2000, Quardokus et al., 1996, Sackett et al., 1998). To gain insight into the timing of DipM accumulation, we assessed dipM transcript and DipM protein levels over the cell cycle. Analysis of Affymetrix data from synchronously growing Caulobacter cells (McGrath et al., 2007) indicated that dipM transcript levels were high in swarmer cells, dropped precipitously, then increased in stalked cells and peaked in pre-divisional cells (Fig. 1C). The accumulation of dipM mRNA followed the peak of ftsZ transcription (Fig. 1C), but occurred close to the time at which other cell division genes, for example ftsA, are transcribed (Sackett et al., 1998). In order to assess DipM protein levels over the course of the cell cycle, polyclonal antibodies were raised against purified recombinant DipM and used to probe Caulobacter cell lysates taken at various times post-synchronization. Similar to FtsZ, DipM protein levels were low in swarmer cells but increased in stalked cells and were high through the remainder of the cell cycle, consistent with it acting at the time of cell division (Fig. 1D).
To determine if DipM resides in the periplasm, as predicted computationally (Gardy et al., 2005), cells bearing xylose inducible tdimer2 that is targeted to the periplasm by fusion to the signal sequence from E. coli TorA (ss-tdimer) (Judd et al., 2005) were fractionated into periplasmic and spheroplast fractions and probed with anti-DipM antibodies. The fractions were also probed with antibodies that recognize periplasmic ss-tdimer, cytoplasmic FtsZ, and membrane-associated PodJ. As predicted, DipM and ss-tdimer were both detected in the periplasmic fraction (Fig. 1E). A significant fraction of both DipM and ss-tdimer was also observed in the spheroplast fraction, probably owing to incomplete release of periplasmic contents during fractionation. DipM may also be retained in spheroplasts by binding to residual peptidoglycan in the spheroplast fraction (see below). Nevertheless, cytoplasmic FtsZ and membrane-bound PodJ controls were found exclusively in the spheroplast fraction. In further support of a periplasmic localization for DipM, an N-terminal mCherry fusion to CC1996, subsequently identified as DipM, did not exhibit any subcellular localization (Werner et al., 2009), presumably owing to release of the fluorescent protein moiety upon cleavage of the signal sequence. Moreover, when we attempted to fuse the GFP derivative Cerulean to the C-terminus of DipM, we failed to detect a fluorescent signal (data not shown). This result is consistent with the expectation that DipM is exported to the periplasm by the canonical Sec pathway in an unfolded state, whereby GFP derivatives do not fold in a manner that supports maturation of the fluorophore. Collectively, these data argue that DipM is, indeed, found in the periplasm.
DipM is required for normal cell morphology
Having established that DipM is a new cell cycle regulated division-site localized protein, we investigated its cellular function by perturbing the levels of the protein by overexpression, deletion and depletion. Overexpression of dipM was achieved by placing its expression under the control of the xylose-inducible PxylX promoter on a high copy number plasmid. In the presence of xylose-inducer, cells first elongated and eventually became rounded and lysed (Fig. 2A). Cell lysis was also observed upon overexpression of LytM-domain containing proteins in E. coli (Hara et al., 2002, Lange and Hengge-Aronis, 1994) and is presumably a result of excess PG-hydrolytic activity weakening the cell wall.
Figure 2.
DipM is required for efficient cell division and normal cell morphology. A. Morphology of cells from a strain bearing PxylX-dipM on a high copy number plasmid (EG301) grown in PYE with 0.2% glucose or 0.3% xylose for 17 h. Arrows indicate rounded cells. B. Genomic context of dipM and strategy for generating deletion and depletion strains. C. Spot dilutions of the indicated strains after 3 days of growth at 30°C. Three independent ΔdipM isolates are shown. D. Morphology of ΔdipM cells (EG590). Smooth filaments (asterisk); shorter, wide cells (arrow); and cells with envelope roughening (hatched arrow) were observed. E. Cells from DipM depletion strain EG353 grown for the indicated amount of time in PYE without xylose inducer. F. Immunoblots using anti-DipM antibodies or anti-HU2 control antibodies of CB15N (WT), ΔdipM (Δ), and EG353 cells at 0, 3, 6, 9, 15, and 21 h of growth without xylose. Bars = 2 µm.
To more precisely assess the physiological role of DipM, we generated an in-frame deletion of the dipM gene (Fig. 2B). We were able to obtain ΔdipM cells (EG590) indicating that it is not essential for viability. However, three independent ΔdipM isolates showed a growth defect in liquid culture (data not shown), a reduction in colony size, and an approximately 100-fold reduction in colony forming units/mL/OD as compared to wild type (Fig. 2C). Moreover, examination of cells lacking DipM by phase contrast microscopy revealed gross morphological defects (Fig. 2D). Notably, cells displayed varying degrees of filamentation, indicating that DipM has a critical role in supporting efficient cell division. Filamentous ΔdipM cells were heterogeneous in appearance: we observed long, apparently smooth filaments, as well as shorter, wide cells. Additionally, the surface of cells lacking DipM frequently appeared rough and uneven. When grown in minimal M2G media, the phenotype of cells lacking DipM was less severe than when grown in rich PYE media (Fig. S1). Nevertheless, we observed varying degress of filamentation and cell surface abnormalities under both grown conditions. All subsequent experiments were performed by growth in PYE.
Because of the severe phenotype and growth defects of ΔdipM cells, we generated two DipM depletion strains for ease of genetic manipulation and downstream experiments (Fig. 2B). In the first strain, we simultaneously truncated the dipM gene and integrated a xylose-inducible full-length version at the dipM locus (EG353). In the second strain, we introduced vanillate-inducible dipM at the vanA locus in the ΔdipM strain background (EG598). In each of the depletion strains, cells looked morphologically wild type in the presence of inducer, but became elongated and wider, and had frequent cell envelope roughening by 9 to 15 hours of depletion (Figs 2E, S2). Immunoblot analysis with anti-DipM antibodies confirmed that DipM was undetectable in the deletion strain and that it was depleted to undetectable levels between 9 and 15 hours without inducer in the depletion strains (Fig. 2F, data not shown). Complete rescue of the morphological defects associated with dipM deletion in the presence of vanillate in EG598 confirmed that they were directly attributable to loss of dipM (Fig. S2). We quantified the apparent cell widening effect of loss of DipM by manually measuring cell widths in phase contast images of the xylose-dependent DipM depletion strain at time 0 and at 24 hours without xylose inducer. In the presence of DipM, cells measured 668 ± 57 nm (mean ± standard deviation (S.D.), n = 171) in diameter, whereas cells lacking DipM had a diameter of 732 ± 78 nm (n = 171, P < 0.0001 for statistically significant difference of means). Thus, DipM plays an important role in regulating both the length and width of Caulobacter cells.
DipM requires FtsZ and its LysM domains for division-site localization
Our genetic and cell biological analyses indicated that DipM is critical for efficient cell division. Since the protein contains a number of functional domains, we sought to identify the determinants of DipM mid cell localization and function during growth and division. To identify the regions of the protein that are important for its localization, a series of truncation mutants of DipM fused to mCherry were created (Fig. 3A) and the localization of the resulting fusion proteins was determined. Specifically, we generated DipM truncations containing the N-terminal 123 amino acids (DipM-123), the N-terminus and first set of LysM domains (DipM-226), the N-terminus and all LysM domains (DipM-399), the first 123 amino acids fused to the LytM peptidase domain (DipM-123-pep), and the LytM domain (DipM-pep), alone. Both DipM-226-mCherry and DipM-399-mCherry, each of which contains at least one set of LysM domains, localized to the division site (Fig. 3B), while those truncations lacking the LysM domains failed to do so. The localization of DipM-399-mCherry was indistinguishable from that of full length DipM, whereas DipM-226-mCherry was recruited more weakly, indicating that the full complement of LysM domains is required to achieve the wild type localization pattern.
DipM-123-mCherry, DipM-123-pep-mCherry, and DipM-pep-mCherry were never observed at the division site or cell poles. To verify that the expected fusion proteins were produced, cell lysates from the above strains were probed with antibodies that recognize mCherry and DipM (Fig. S3). Fusion proteins of the expected molecular weights were detected using both antibodies for full-length DipM, DipM-123, DipM-226, and DipM-399 in similar abundance. However, only free mCherry was detected for DipM-123-pep and DipM-pep, indicating that it was completely cleaved from those truncation derivatives. DipM antibodies recognized a band corresponding to the expected size for DipM-123-pep that had been cleaved from mCherry, but no free DipM-pep was detected. This suggests that the LytM domain or some other feature of the C-terminus of DipM contains a protease cleavage site and that, when not fused to the N-terminus and/or exported to the periplasm, the LytM domain is highly unstable. We conclude that the LysM domains of DipM are the major mediators of its localization to the division site, presumably via direct interaction with peptidoglycan. However, since both DipM-123-pep and DipM-pep were cleaved from mCherry, we cannot rule out the possibility that the LytM domain contains localization determinants, as well.
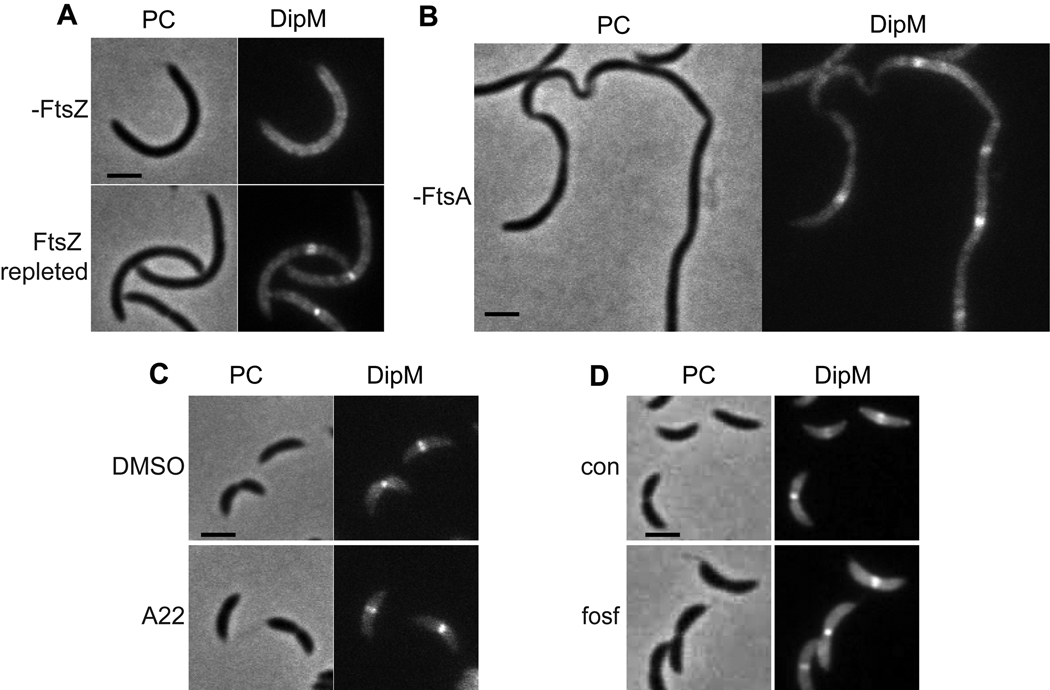
Every division-site localized protein investigated thus far requires FtsZ for its localization. To determine if this is true for DipM, a strain was made bearing vanillate inducible dipM-mCherry at the chromosomal vanA locus in a background where expression of the only functional copy of ftsZ is under the control of the xylose-inducible promoter. Cells grown in the absence of xylose were depleted of FtsZ and became filamentous, and DipM-mCherry was observed in a diffuse peripheral pattern in these cells (Fig 4A). Repletion of FtsZ restored the localization of DipM to mid cell bands. Thus, DipM requires FtsZ for its localization to the division site. It does not require FtsA, a protein anticipated to be shortly downstream of FtsZ in the divisome hierarchy (Goehring and Beckwith, 2005), however. DipM localized robustly to discrete mid cell bands in filamentous cells depleted of FtsA (Fig 4B).
Figure 4.
FtsZ is required for mid cell localization of DipM, but FtsA, active MreB, and active PG synthesis are dispensible. A. DipM-mCherry in cells (EG340) depleted of FtsZ for 3 h (-FtsZ) and with FtsZ depleted for 3 h then repleted for 1 h. Cells were grown in PYE with 0.2% glucose for 3h and 0.5 mM vanillate for 2 h prior to application on M2G agarose (-FtsZ) or M2X agarose (FtsZ repleted). B. Localization of DipM-mCherry in cells depleted of FtsA (EG406). Cells were grown in PYE with 0.2% glucose for 7 h prior to imaging. C. DipM-mCherry in cells (EG326) treated with 10 µg/mL A22 or DMSO control for 15 minutes prior to application on agarose pads containing A22. Cells were grown in PYE. D. DipM-mCherry in cells (EG327) treated (fosf) or not (con) with fosfomycin to inhibit PG synthesis. Cells were grown in PYE ± 100 µg/mL fosfomycin for 2.5 h and 0.5 mM vanillate for 1.5 h prior to imaging. Bars = 2 µm.
Given the importance of DipM’s peptidoglycan binding motifs for its localization, we also determined if MreB, which localizes to the division site early (Figge et al., 2004, Gitai et al., 2004) and is implicated in PG synthesis ((Pichoff and Lutkenhaus, 2007)), is required for its localization. Cells were treated with the MreB-perturbing drug A22 (Gitai et al., 2005) on a time-scale that is sufficient to completely delocalize MreB but not long enough to cause morphological defects (Fig. S4A), and DipM-mCherry was imaged. In the presence of A22, DipM-mCherry was still robustly recruited to the division site (Fig 4C), arguing that active MreB is not directly required for its localization. Interestingly, active PG synthesis was also not required for recruitment, as treatment with fosfomycin, which inhibits MurA and thus PG precursor synthesis, did not cause delocalization of DipM-mCherry (Fig 4D) on a time scale when it halted cell growth (Fig S4B). We propose that FtsZ recruits DipM to the division site via DipM’s LysM domains by establishing a PG subdomain that is distinct from that of the rest of the cell, yet is independent of active PG synthesis.
The LytM domain of DipM is sufficient to promote normal cell division and morphology
For many proteins, localization is intimately linked to function. Having established that the LysM domains are critical for localization of DipM to the division plane, we aimed to determine which domains mediate its role in cell division and growth. To this end, the vanillate-inducible mCherry fusions to the DipM truncations described above were introduced into the xylose-dependent DipM depletion strain. In the absence of xylose but the presence of vanillate, expression of dipM was turned off and production of the mCherry fusion was induced. Fusion of mCherry to full-length DipM did not appreciably perturb its function, as cells producing DipM-mCherry displayed wild type morphology in the absence of xylose and presence of vanillate (Fig. 3C). Strikingly, only one of the truncations, DipM-123-pep, containing the LytM domain targeted to the periplasm by fusion to the N-terminal 123 amino acids, was able to support near wild type cell morphology. The majority of cells producing DipM-123-pep as the only version of DipM were of wild type length and did not display cell envelope roughening. However a population of these cells were filamentous (22%, n = 639 cells), indicating that this domain alone does not completely fulfill the required functions of DipM. Moreover, cells producing only DipM-123-pep-mCherry were wider in diameter than those expressing full-length DipM-mCherry, with diameters of 734 ± 75.8 nm (n = 127) and 699.7 ± 64.7 nm (n = 165), respectively (P<0.0001). Neither DipM-226-mCherry nor DipM-399–mCherry was able to rescue the morphological defects of DipM depletion, although they localized to discrete bands in the filamentous cells. Production of DipM-123-mCherry also failed to rescue the depletion phenotype and did not localize to mid cell bands (Fig. 3C), however we noted that it often accumulated in bright foci that were associated with the sides of cells or free on the agarose pads (Fig 5B, discussed below). These data suggest that the LysM domains of DipM are important for its mid cell localization but are mostly dispensible for function, whereas production of the LytM domain targeted to the periplasm can minimize the filamentation and cell envelope roughening defects associated with DipM depletion.
Figure 5.
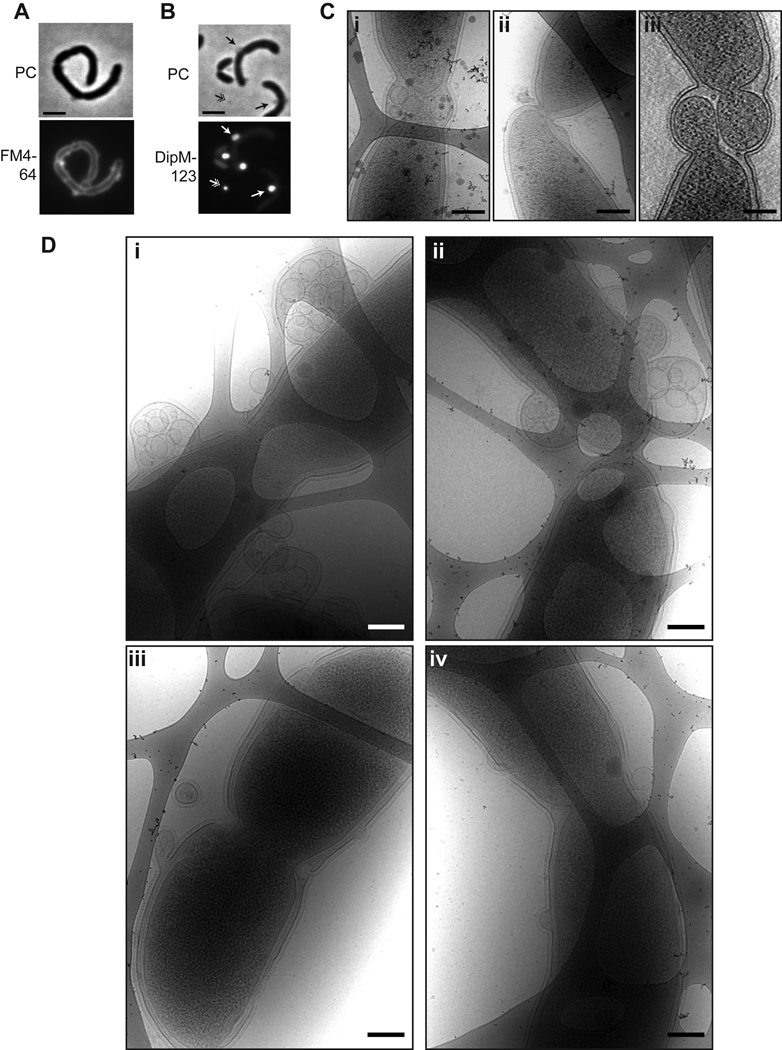
DipM is required for normal OM geometry. A. Membrane staining (FM4–64) in ΔdipM cells (EG590). Cells were grown in PYE. Bar = 2 µm. B. DipM-123-mCherry foci in cells (EG511) depleted of full-length DipM for 17 h. Foci appeared associated with the cell envelope (arrow) and free on the pads (hatched arrow). Cells were grown in PYE with 0.2% glucose and 0.5 mM vanillate for 17 h prior to imaging. Bar = 2 µm. C. Cryo electron micrographs (i and ii) and tomographic slice (iii) of cells (EG301) overproducing DipM. Cells were grown in PYE with 0.3% xylose for 17 h then prepared for cryoEM imaging. Bar = 200 nm. D. Cryo electron micrographs of cells (EG353) depleted of DipM. Cells were grown in PYE with 0.2% glucose for 11 h prior to cryo-plunging for cryo EM. Bar = 200 nm.
Overexpression or depletion of dipM causes defects in outer membrane geometry
Our phenotypic analysis of the ΔdipM and DipM depletion strains revealed frequent roughening and unevenness of the cell envelope. We reasoned that this might result from abnormalities in the cell membrane(s). Indeed, when imaged on agarose pads containing the lipophilic dye FM4–64, the majority of ΔdipM cells had membrane distortions or blebbing that were visible as uneven fluorescent foci or “bubbles” on the cell surface (Fig 5A). Interestingly, in cells depleted of full-length DipM, but expressing DipM-123-mCherry, we observed bright fluorescent foci associated with the surface of cells where apparent distortions of the cell membrane were located, as well as foci that were free on the agarose pads (Fig. 3C, Fig. 5B). Given the FM4–64 staining of dipM mutant cells, we interpret these foci as outer membrane blebs and free outer membrane vesicles that are filled with soluble DipM-123-mCherry. It appears, in fact, that DipM-123-mCherry is concentrated in the membrane blebs, as they are far brighter than the fluorescent signal in the rest of the periplasm. These bright foci were not present in images of any of the other truncation derivatives. We hypothesize that the LysM domain-containing truncations are prevented from filling the blebs by tight association with peptidoglycan.
To analyze the blebbing phenomenon in more detail, we turned to high-resolution cryo electron microscopy (cryoEM) and cryo electron tomographic (cryoET) imaging of mutant cells. When cells overproducing DipM were imaged at high resolution, lysed cells were apparent, as were cells with division defects, consistent with the phenotypes we observed by phase contrast microscopy (Figs 5C, S5A). Specifically, we noted aberrant invagination of all layers of the cell envelope in what appeared to be cell divisions that initiated but failed to complete (Fig 5C, ii and iii). Notably, cells overexpressing dipM also frequently accumulated vesicles contained within outer membrane blebs at presumptive division sites (Fig 5C, i) and exhibited waviness in their outer membranes near division sites (Fig 5C, ii and iii).
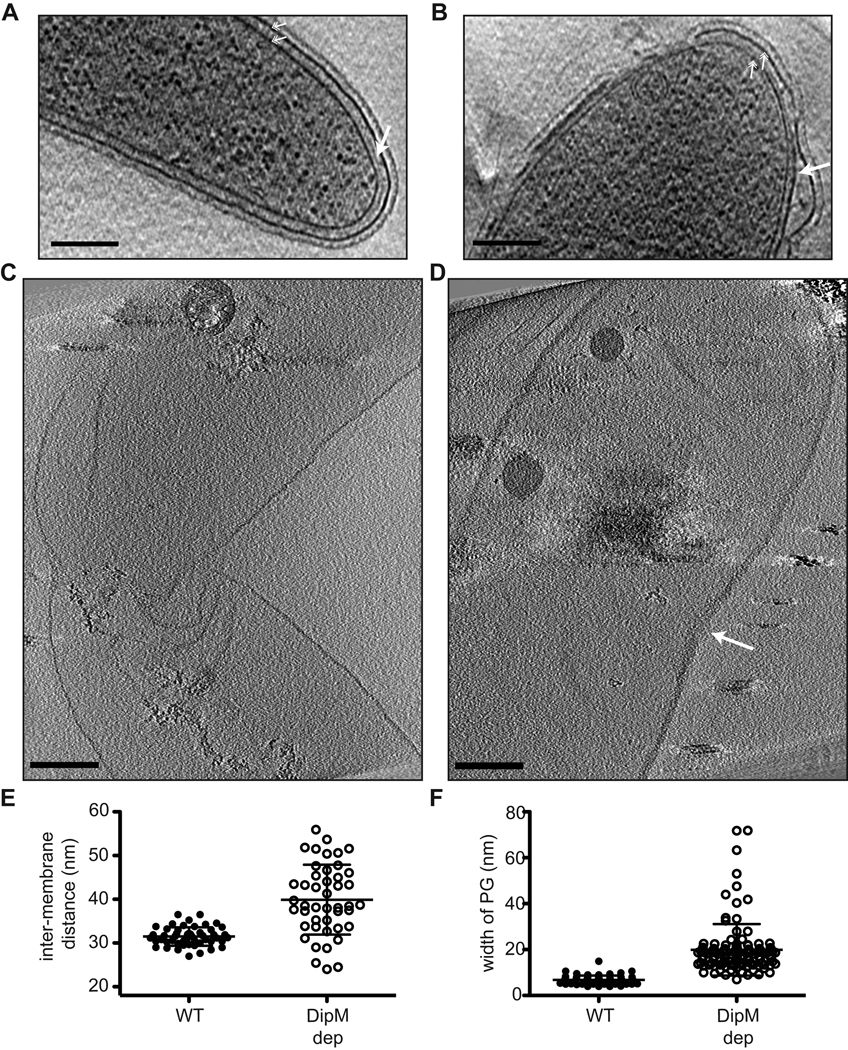
To assess the membrane morphology of cells lacking DipM, we imaged the xylose-dependent depletion strain EG353 by cryoEM and cryoET. Cultures were grown in the absence of xylose for 11 or 17 hours prior to cryo-plunging. A total of 29 mutant cells were imaged in 2D, and 3D cryoET reconstructions were generated of 6 mutant cells. As anticipated from FM4–64 staining, the majority of cells lacking DipM displayed varying degrees of OM distortions, vesicles, and blebbing of the outer membrane at both time points (Figs 5D, S5B). Blebs were found at division sites and cell poles, as might be predicted from DipM localization and the filamentation that occurs upon its depletion. However, OM distortions and blebs were detected along the cell sidewalls, as well, with no clear bias towards appearance at division sites and poles. Abundant vesicles were observed that were completely contained within outer membrane blebs (Fig. 5D). We hypothesize that these vesicles are derived from the outer membrane and contain periplasmic material, since they are located outside of the peptidoglycan and do not contain ribosomes (i.e. cytoplasm). However, although they may be derived from the OM, S-layer was not associated with the vesicles (Fig. 5D). This suggests that either S-layer is stripped from the membrane as the vesicles form or that they form from outer membrane material prior to assembly of S-layer on the membrane.
In addition to OM defects, cells depleted of DipM were frequently found to have only shallow invaginations at presumptive division sites (Fig. 5D, i and iii), and to have abnormal envelope organization at the division site (5D, ii–iv). Cells that had compartmentalized cytoplasms, but were connected via PG and/or OM linkages were observed (Fig. 5D, ii), as were cells with deeply invaginated IMs, but OMs that failed to invaginate (Fig. 5D, iv). Such envelope organization was not detected in wild type cells ((Judd et al., 2005) and data not shown). These results indicate that DipM activity is required for normal envelope invagination during cell division and to establish or maintain OM connections throughout the cell envelope.
DipM is required to maintain a sacculus of constant thickness
The finding that depletion of DipM, a factor presumed to have PG hydrolytic activity, causes disruption of OM geometry was surprising. A number of mutations that lead to OM blebbing and OM vesicle release have been identified in other Gram-negative bacteria, but these are generally mutations in factors that mediate IM-PG-OM connections such as Lpp and the Tol-Pal complex (Deatherage et al., 2009, McBroom and Kuehn, 2007, Bernadac et al., 1998, Sonntag et al., 1978). To gain insight into the mechanism behind this blebbing phenomenon, we explored the architecture of the cell envelope in cells lacking DipM in more detail. First, we asked whether the distance between the inner and outer membranes was altered in mutant cells throughout the cell envelope, or only in regions containing blebs. The inter-membrane distance was measured in both wild type and DipM depleted cells in regions where the IM and OM were parallel to each other (i.e. without blebbing). This analysis revealed an increase in average inter-membrane distance and in variability of inter-membrane distance in cells depleted of DipM as compared to wild type cells (Figs 6A,B,E). Wild type cells maintained fairly constant envelope geometry with an inter-membrane distance of 31.5 ± 2.1 nm (n = 50 measurements from 7 cells). Cells depleted of DipM, however, had an IM-OM distance of 39.9 ± 8.0 nm (n = 45 measurements from 6 cells) (P<0.0001 for statistically significant difference of mean IM/OM distances).
Figure 6.
DipM is required for maintenance of proper PG width. A and B. Tomographic slices of wild type (CB15N) (A) and DipM-depleted (EG353) (B) cells. Arrow indicates PG. Hatched arrows indicate inner and outer membranes. CB15N was grown in PYE, EG353 cells were grown as in 5D. Bar = 200 nm. C and D. Slices through 3D cryoET reconstructions of sacculi isolated from wild type (C) and DipM-depleted (EG353) cells. Arrow indicates division site in DipM depleted sacculus. Bar = 200 nm. E. Scatter dot plot of inter-membrane distances in wild type (WT) and DipM-depleted cells (calculated from images as in A and B). Mean and standard deviation are plotted. F. Scatter dot plot of PG width in sacculi from wild type and DipM-depleted cells (calculated from images as in C and D). Mean and standard deviation are plotted.
We reasoned that, given the putative PG peptidase activity of DipM, cells depleted of this protein might be defective in removing and/or inserting PG strands into the cell wall in coordination with PG biosynthetic machinery. This could lead to altered dimensions of the PG (e.g. PG thickening) that could, in turn, cause the IM and OM to be spaced further apart. To address this possibility, we attempted to directly visualize PG in slices from cryo electron tomograms of mutant and wild type cells. In tomogram slices from wild type cells, PG is observed as a faint line between the inner and outer membranes (Fig 6A, solid arrow). We found that, in general, PG was more readily visible in images of DipM-depleted cells than in wild type cells, indicating that it might be thicker, and we could identify instances in which the PG appeared to be remarkably thick in mutant cells (Fig. 6B). However, in the presence of the cell membranes, and without the ability to specifically label the PG, we were unable to measure the thickness of the PG in whole cell preparations with confidence.
To overcome these limitations in assessing the structure of PG in wild type and dipM mutant cells, we turned to high resolution imaging of isolated sacculi from these two cell populations. The procedure for sacculus isolation removes cellular membranes, DNA, RNA, and cytoplasmic contents, but leaves the PG intact, thereby eliminating sources of ambiguity when assessing its structure. This technique was recently used to image Caulobacter and E. coli PG by cryoET to address existing models for the organization of glycan strands within the sacculus (Gan et al., 2008). We isolated sacculi from wild type Caulobacter and from cells depleted of DipM for 16 h and subjected them to cryoET imaging. In tomogram slices, sacculi from wild type cells (n = 5 3D reconstructions) looked similar to those described previously (Gan et al., 2008), with the PG visible as a thin dotted line that retains the shape of a Caulobacter cell (Fig. 6C, Movie S1). Sacculi showed frequent wrinkling, but were not completely flattened, as they retained enough volume to contain polyhydroxybutyrate granules (Fig. 6C, Movie S1). Strikingly, the PG in sacculi isolated from DipM-depleted cells (n = 4 3D reconstructions) was markedly thicker than that of wild type sacculi (Fig. 6D, Movie S2). This difference was directly visible in tomogram slices throughout the sacculus (compare Fig. 6C and D), and in quantification of relative PG thickness (Fig. 6F). Whereas PG in wild type sacculi measured 6.8 ± 1.9 nm (mean ± S.D.; n = 67 measurements from 2 sacculi), PG in DipM-depleted cells was nearly three times as thick on average, at 19.9 ± 11.2 nm (n = 112 measurements from 4 sacculi) (P<0.0001). The increase in PG thickness is not attributed to the fact that cells depleted of DipM were grown in 0.2% glucose, as sacculi from wild type cells grown in PYE + 0.2% glucose had a mean PG thickness of 7.9 ± 1.5 (36 measurements from 2 sacculi, P>0.05 when compared to wild type sacculi grown in PYE lacking glucose, indicating that they are not significantly different in thickness). In sacculi from dipM mutant predivisional cells, PG was particularly thick at the division site (Figs 6D, S5, Movie S2), although PG was thicker on average throughout the sacculus of mutant cells. This might be reconciled with the concentrated localization of DipM at mid cell by the fact that the majority of PG synthesis in Caulobacter, both during elongation and division, occurs near mid cell in a FtsZ-dependent manner (Aaron et al., 2007).
Glycan strands in wild type Caulobacter sacculi were previously reported to lie in a monolayer within the plane of the sidewall, roughly perpendicular to the long axis of the cell (Gan et al., 2008). Thicker PG in cells depleted of DipM could arise from stacking of strands within the same plane to generate a multi-layered sacculus, or, alternatively, new strands could be inserted in different orientations, for example perpendicular to the plane of the sidewall. To determine the orientation of glycan strands in DipM depleted sacculi, we looked at close up views of the sidewalls of wild type and DipM depleted sacculi that were rotated such that the sidewall lay in the plane of the page (Fig. 7). In wild type sacculi, we observed tubes or fibers (interpreted as glycan strands (Gan et al., 2008)) that lay mostly within the plane of the sacculus, as described previously (Fig. 7). The glycan strands in sacculi from DipM depleted cells were generally in the same plane as in WT sacculi, but they were more abundant and displayed a wider range of orientations and an increase in apparent branching (Fig. 7). These data indicate that cells depleted of DipM generate thicker sacculi by addition of new PG strands that are parallel to the existing monolayer, but in a manner that leads to formation of a multi-layered, more disordered sacculus. We propose that, as a putative endopeptidase, DipM plays a critical role in the balance between PG synthesis and PG hydrolysis by cleaving peptide bonds in the sacculus that facilitate proper insertion and/or removal of glycan strands.
Figure 7.
Glycan strands in wild type and DipM depleted sacculi lie primarily in the same plane. A–C. (Top) Slices through 3D cryoET reconstructions of sacculi isolated from wild type (A) and DipM-depleted (B–C) cells. Bar = 100 nm. (Bottom) Boxed areas are rotated orthogonal to the planes in the top images and 90° counter-clockwise so that glycan strands run up-and-down in the plane of the page as illustrated in A (bottom).
Discussion
Coordinated invagination of the multiple layers of the Gram-negative cell envelope is central to successful cell division. We have shown here that a putative PG endopeptidase, DipM, plays a critical role in maintaining wild type PG dimensions that is surprisingly important for proper OM organization. Based on its early mid cell localization, peak of accumulation in pre-divisional cells, and functional importance during cytokinesis, DipM can be classified as an early recruit to the Caulobacter divisome that also plays a more general role in overall PG and OM architecture.
Physiological function of DipM and roles of its LytM and LysM domains
The domain layout of DipM led us to speculate that it is a periplasmic PG endopeptidase and thus might be a player in the balance between PG synthesis and PG hydrolysis during cellular growth and division. Indeed, our phenotypic analysis of cells lacking or overproducing DipM is consistent with the speculation that it possesses the endopeptidase activity attributed to LytM-domain containing proteins. Our finding that loss of DipM leads to PG thickening (Figs 6,7) further indicates that this activity is crucial for removal of glycan strands (i.e. PG turnover) and/or proper insertion of strands to form a monolayered sacculus during cellular growth and division. The precise substrate(s) of LytM-domain proteins in Gram-negative organisms, however, is currently unknown. The biochemically best studied members of the LytM family, lysostaphin and LytM, come from Gram-positive Staphylococci. These proteins cleave pentaglycine substrates (Browder et al., 1965, Ramadurai et al., 1999) that are not present in Gram-negative PG; therefore this result offers little insight into the substrate upon which DipM acts in the Caulobacter cell wall. Moreover, although the E. coli LytM family protein EnvC has been shown to have hydrolytic activity in an in-gel zymogram assay (Bernhardt and de Boer, 2004), peptidase activity has not been detected for the protein in solution, and analysis of the chemical composition of PG from E. coli cells lacking all LytM family proteins offered no information about potential substrates in vivo (Uehara et al., 2009). Finally, our cryoET images of DipM-depleted sacculi were not of sufficient resolution to suggest specifics of the altered chemistry of the PG as compared to wild type. Therefore, although all signs point to DipM having endopeptidase activity, the underlying biochemical mechanism of action for this enzyme and its LytM family relatives in Gram-negative organisms remains to be determined.
In addition to the LytM domain, DipM has four LysM motifs that we have shown are major determinants for localization of the protein to the division site. A large number of PG hydrolytic enzymes across bacterial species possess LysM domains, and the physical arrangement of these domains relative to the region of the protein with hydrolytic activity has been posited to play a role in substrate positioning (Buist et al., 2008). We were surprised, therefore, to find that the LysM domains of DipM are mostly dispensible for DipM function in vivo, as a truncation completely lacking the LysM domains almost entirely rescues the morphological defects associated with loss of DipM (Fig. 3C). This suggests that neither the ability of the LysM domains to localize DipM to mid cell, nor their putative role in positioning the peptidase domain such that it correctly interacts with its substrate are essential. The localization of DipM to mid cell may not be essential for its function if its activity were coupled to PG synthesis by other means, since the majority of PG synthesis in Caulobacter is concentrated at mid cell (Aaron et al., 2007). However, we cannot rule out the possibility that the periplasmic-targeted LytM domain was still able to localize to the division site independently of the LysM domains, since it was cleaved from the fluorescent reporter (Fig. S3). Furthermore, a significant fraction of cells expressing only the DipM truncation that lacks the LysM domains were filamentous, indicating that although the LysM domains are not essential for the physiological function of DipM, they may play a role in optimizing its activity.
Conservation of DipM and functional redundancy of LytM family proteins
DipM is an apparent ortholog of E. coli NlpD. However, the two proteins have substantially diverged, and the region of similarity between them resides only in the C-terminal half of the DipM, which is 29% identical and 50% similar to the C-terminal 273 amino acids of NlpD. Close homologs of DipM are found throughout α-proteobacteria, although it is absent from Rickettsia species. Moreover, proteins with similar domain organization (i.e. containing both LysM and LytM domains) are widely distributed among the eubacteria. Of the LytM domain containing proteins in Gram-negative organisms, however, only those in E. coli have been studied previously in any detail.
There are a number of notable differences in the phenotypes of Caulobacter cells lacking DipM and mutants in E. coli LytM family proteins. First, we were struck by the severity of the effects of dipM deletion on cellular growth and morphology under normal laboratory conditions as compared to the effects of deleting genes encoding E. coli LytM family proteins. Whereas deletion of dipM alone causes growth rate defects (Fig. 2C), cell filamentation (Fig. 2D), and gross distortions in outer membrane arrangement (Fig. 5), deletion of envC is the most severe of the E. coli LytM family mutants, but displays only mild cell chaining (Bernhardt and de Boer, 2004, Uehara et al., 2009). Surprisingly, deletion of the dipM ortholog nlpD causes no apparent cell division or outer membrane defect on its own. When all four LytM family proteins are absent in E. coli, cells form chains that are connected by PG septa at fairly regular intervals. Again, this differs from deletion of dipM: although we observe some cases where compartmentalized cells are connected by PG and/or OM linkages (though at irregular intervals), we predominantly see filamentation of cells that fail, partially or completely, to invaginate all layers of the envelope (Figs 5D, S5B). Moreover, OM defects are widespread in the dipM deletion and depletion mutants, but were not reported for E. coli LytM family mutants. These phenotypic differences could arise from any number of causes. Notably, the compositions of peptide crosslinks in the PG of E. coli and Caulobacter differ substantially: pentapeptides are reported to constitute close to 60% of the peptide crosslinks in Caulobacter PG (Markiewicz et al., 1983), but less than 1% of the peptides in E. coli PG (Glauner et al., 1988). Thus, the relative importance of various endopeptidases might differ in the two organisms, depending on their substrates. Understanding the molecular basis for these phenotypic differences between organisms will require more intimate knowledge of the enzymatic activities of the various LytM family proteins, as well as their modes of regulation.
E. coli appears to rely on functional redundancy among its PG hydrolytic enzymes: deleting the genes encoding multiple LytM family proteins, amidases, or other peptidases elicits much more dramatic cell separation defects than single deletions. Like E. coli, Caulobacter encodes multiple LytM family proteins: dipM, CC1872, CC2248, CC2297, CC3034, CC3301, and CC3434 are all annotated as encoding LytM/M23 peptidase family proteins. We explored the localization and function of a conserved subset of these (CC1872, CC2297, CC3034, and CC3434) and found that none showed a distinct localization pattern or readily apparent phenotype upon depletion (K.E.F., E.D.G., and L.S., unpublished). This argues that, of the putative LytM endopeptidases in Caulobacter, DipM may play the major role in PG hydrolysis during growth and division. However, the other LytM domain proteins may be able to compensate for lack of DipM under conditions of slower growth, e.g. growth in M2G, since ΔdipM cells grown in M2G display less severe morphological defects than those grown in PYE. Current studies are aimed at dissecting the functional redundancy of this family of proteins in Caulobacter by deleting multiple LytM proteins at once.
A role for regulated PG architecture in OM blebbing
The most surprising finding from our analysis of the dipM mutant was the widespread OM blebbing that occurred in the majority of cells grown in rich media. We were struck by the similarity between disorganization of the OM observed in cells depleted of DipM and disorganization of the OM observed in cells depleted of members of the Tol-Pal complex in Caulobacter (Y.-C.Yeh, L.S., and H. McAdams, unpublished). The Tol-Pal complex is thought to be a principal mediator of connection of the OM with the rest of the cell envelope, and is essential in Caulobacter, which lacks Lpp (Y.-C. Yeh, L.S., and H. McAdams, unpublished). The similarity in OM morphology between DipM depleted cells and Tol-Pal depleted cells leads us to hypothesize that these phenotypes have the same molecular basis, i.e. loss of connections between the IM and OM that are normally mediated by binding of TolA to Pal (Cascales et al., 2000). In the absence of this critical molecular contact, the OM would be free to bulge out from the rest of the envelope. In fact, in addition to the OM defects observed in mutants completely lacking Tol-Pal complex proteins (Bernadac et al., 1998, Deatherage et al., 2009, Gerding et al., 2007, McBroom and Kuehn, 2007), specific loss of the TolA-Pal interaction by deletion of the C-terminal 30 amino acids of Pal in Salmonella leads to increased OM vesicle release at division septa (Deatherage et al., 2009).
Our cryoEM and cryoET data provide a fairly simple explanation for how deletion of this single PG endopeptidase might lead to loss of TolA-Pal contacts. Specifically, the PG is nearly three times as thick in cells depleted of DipM than in wild type cells, leading to a ~9 nm increase in the distance between the inner and outer membranes (Fig. 6). This substantial increase in inter-membrane distance would reasonably be expected to put TolA (anchored in the IM) and Pal (bound to the OM) out of reach from one another, thereby mimicking loss of the Tol-Pal complex. Although this is our favored hypothesis, other explanations for the ΔdipM blebbing phenotype are certainly feasible, for example a more densely packed PG network might limit transport of a critical OM protein through the sacculus to the OM. Alternatively, the rate of increase in PG surface area may lag behind the rate of OM biosynthesis in the ΔdipM mutant, causing OM bulging and blebbing. Further work is required to clarify the molecular basis of this defect.
Increased OM permeability has been reported for chain forming mutants containing deletions of multiple PG hydrolases in E. coli (Heidrich et al., 2002, Korsak et al., 2005). However, to our knowledge this is the first report of a PG hydrolase mutant (or combination of mutants) causing OM blebbing and vesicle release. We speculate that this may be attributed to two key details: 1) differences in the molecules mediating connection of the OM to the PG and/or IM and 2) differences in the relative contributions of various PG-hydrolases to coordinating PG synthesis and PG hydrolysis/turnover among different Gram-negative organisms. Specifically, the effect of loss of Tol-Pal complex is especially profound in Caulobacter as compared to E. coli or Salmonella, since Caulobacter lacks the major OM anchoring lipoprotein, Lpp. Moreover, although chains of E. coli cells bearing LytM family gene deletions or amidase triple deletions are connected by PG septa, significant overall thickening of PG was not reported (Uehara et al., 2009, Heidrich et al., 2001). This may be owing to functional redundancies or differential contributions of various classes of hydrolases in maintaining the dimensions of the sacculus in E. coli versus Caulobacter. If PG is not thickened, one would not expect loss of TolA-Pal interactions in the E. coli hydrolase mutants.
It is notable that outer membrane blebbing and vesicle release are observed under normal conditions in a variety of Gram-negative organisms. It is thought that outer membrane vesicle (OMV) secretion plays important physiological roles in pathogenesis and intercellular communication, for example as a secretion mechanism for delivering key toxins or signaling molecules (Beveridge, 1999, Kuehn and Kesty, 2005). The regulation of OMV release is not well understood, however. Most of the factors implicated in OMV formation and release are proteins that mediate IM-PG-OM contacts (i.e. Lpp and Tol-Pal, discussed above) and other components of the OM (Beveridge, 1999, Deatherage et al., 2009, Kuehn and Kesty, 2005, McBroom and Kuehn, 2007). A mutation in the bifunctional transglycosylase transpeptidase PBP1B was found to promote OMV release in E. coli (McBroom and Kuehn, 2007). Together with our findings for DipM, this puts forth the intriguing possibility that a bacterial cell could exploit locally regulated modification of PG architecture to promote or limit OMV release as necessary. Future research aimed at a thorough understanding of the delicate balance between synthesis of PG and its dynamic interactions with cellular membranes will help illuminate the mechanisms behind this fascinating phenomenon.
Experimental procedures
Bacterial growth conditions and synchronization
All Caulobacter crescentus strains were derived from the synchronizable strain CB15N and grown at 28–30°C in PYE rich media (Poindexter, 1964) or M2 minimal media (Johnson and Ely, 1977) with 0.2% D-glucose (M2G). All experiments were performed in PYE with the exception of large scale synchronization of CB15N for determination of DipM protein levels over the course of the cell cycle, for which cells were grown in M2G. All experiments were performed with cells in log phase of growth. When appropriate, antibiotics were added to liquid cultures at the following concentrations: 1 µg/mL gentamycin, 5 µg/mL kanamycin, or 25 µg/mL spectinomycin. Sodium vanillate was used at 0.5 mM and D-xylose was used at 0.3% when indicated. Depletions were performed by washing the indicated strains in PYE lacking inducer three times and resuspension in media ± inducer. Large scale synchrony for preparation of cell lysates (Evinger and Agabian, 1977) and small scale synchrony for microscopy (Tsai and Alley, 2001) were performed as described previously. For assessing growth by spot dilution, cells at OD660 = 0.35 were serially diluted and 5 µL of the indicated dilution was spotted onto PYE agarose plates.
Bacterial strains, plasmids, and cloning
Strains and plasmids used are listed in Table S1 in Supporting Information. Standard protocols for electroporation and ϕCR30 transduction were used to generate Caulobacter strains. Cloning was performed in E. coli using standard procedures. Relevant plasmid and primer sequences are available upon request.
Phase contrast and fluorescence microscopy
Log phase cells were applied to thin pads of 1% agarose in M2G media prior to visualization. For membrane staining, FM4–64 (Invitrogen) was included in agarose pads at 2 µg/mL. All phase contrast and fluorescence imaging was performed on a Leica DM 6000 B microscope using a HCX PL APO 100x/1.40 Oil PH3 CS objective, Hamamatsu EM-CCD C9100 camera, and KAMS software (Michael Fero, Stanford University, unpublished). Images were processed with Adobe Photoshop and/or ImageJ. Measurements of cell diameter of phase contrast images was performed manually in ImageJ.
Cell fractionation
Fractionation of cells into periplasm and spheroplast was performed as described previously (Judd et al., 2005) except that 2 µg/mL lysozyme was used.
Protein purification and antibody production
His6-DipM-His6 was overproduced in Rosetta (DE3) pLysS E.coli from plasmid pEG411 by induction with 0.5 mM IPTG for 3 h at 37°C. Cell pellets were washed in phosphate buffered saline, frozen in liquid nitrogen and stored at −80°C. To purify the His-tagged protein, cells were thawed, resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole, 1 mM EDTA, 10% glycerol, 10 U/mL DNase I, and 2 mM PMSF), lysed by passage through a french pressure cell and cleared by centrifugation at 10,000 × g for 30 minutes. Initial purification was accomplished by Ni-affinity chromatography using Ni-NTA agarose (QIAGEN). DipM was further purified by cation exchange chromatography (HiTrap SP Sepharose FF, GE Healthcare) in S buffer (50 mM HEPES pH 7.2, 0.1 mM EDTA, 10% glycerol, 0.05% Triton X-100) using a 50 mM to 1 M KCl gradient over 40 column volumes. Peak fractions were pooled and concentrated. Protein was frozen in liquid nitrogen and stored at −80°C. HU2-His6 (CC2331) was overproduced from plasmid pMT166 in Tuner(DE3)/placI (Novagen) grown in LB/carb50 by induction with 0.5 mM IPTG for 3 h at 37°C. The protein was purified by affinity chromatography using Ni-NTA agarose (QIAGEN), adjusted to 20% glycerol, and frozen in liquid nitrogen. Purified His6-DipM-His6 and His6-HU2 proteins were used to immunize rabbits for antibody production (Josman, LLC). Anti-DipM antisera strongly recognize a major band that runs at ~72 kDa by SDS-PAGE and a second band at ~62 kDa, both of which are specifically present in wild type lysates but absent from ΔdipM cell lysates.
Immunoblotting
Cells were harvested in log phase of growth and lysed in SDS-PAGE loading buffer by boiling for 5 minutes. For a given experiment, equivalent OD units of cell lysate were loaded. SDS-PAGE and transfer of protein to PVDF membrane was performed using standard procedures. Samples were probed with primary antibodies at the following concentrations: α-DipM (1:10000 or 1:20000), α-FtsZ (1:10000) (Mohl et al., 2001), α-mRFP1 (recognizes mCherry) (1:10000) (Chen et al., 2005), α-HU2 (1:2000) and α-PodJ (1:10000) (Viollier et al., 2002). Secondary horse radish peroxidase-conjugated goat anti-rabbit antibody was used at a 1:10000 dilution and chemiluminescent substrate was applied prior to exposure to film.
Isolation of sacculi
CB15N or EG353 cells depleted of DipM for 16 hours were harvested in log phase of growth and sacculi were isolated essentially as described previously (Gan et al., 2008). 1 L of cells were cooled on ice for 10 min then harvested by centrifugation at 6000 × g for 15 min. Cells were resuspended in 12 mL ice cold water before drop-wise addition to 4% SDS pre-heated to 80–90°C. 12 mL 8% SDS pre-heated to 80–90°C was added and solution was heated for 1 h with constant stirring. Samples were pelleted at ~44000 × g for 30 min at 20°C and washed twice in water to remove SDS. Pellets were resuspended in 9 mL 10 mM sodium phosphate pH 7.8. Magnesium sulfate was added to 10 mM, DNase I was added to 10 µg/mL and the sample was incubated for 30 min at 30°C without shaking. RNase A was added to 20 µg/mL and incubated with shaking at 28°C for 30 min. Trypsin was added to 1 mg/mL and the sample was incubated with shaking at 28°C overnight (final volume ~10 mL). The sample was diluted by addition of 8 mL water and sacculi were pelleted at 55310 × g for 30 min at 25°C. Pellet was resuspended in 3 mL 4% SDS at 95°C. The suspension was added to 7 mL 4% SDS at 95°C and stirred for 30 min. 8 mL water was added and sample was centrifuged at 55310 × g for 30 min at 25°C. Pellet was resuspended in 18 mL water and 300 µL 0.5 M EDTA was added. Sacculi were pelleted as above, then washed twice in 18 mL water. Final sample was resuspended in 1 mL water and stored at 4°C for less than a week prior to application to grids for imaging.
CryoEM Specimen Preparation
For cryo electron microscopy, aliquots of 5 µl were taken directly from cell cultures or sacculus preparations and placed onto lacey carbon grids (Ted Pella 01881) that were pre-treated by glow-discharge. The Formvar support was not removed from the lacey carbon. The grids were manually blotted and plunged into liquid ethane by a compressed air piston, then stored in liquid nitrogen. For cryoET, fresh sample aliquots were deposited onto support grids pre-loaded with 10 nm colloidal gold particles.
CryoEM and cryoET Imaging
Samples were flash frozen in liquid ethane and stored in liquid nitrogen until they were loaded into the holder for data acquisition. Images were acquired on a JEOL–3100 electron microscope equipped a FEG electron source operating at 300 kV, an Omega energy filter, a Gatan 795 2Kx2K CCD camera, and cryo-transfer stage. The stage was cooled using liquid nitrogen to 80 K. Approximately half of the data was acquired using a Gatan 795 4Kx4K CCD camera mounted at the exit of an Electron Decelerator (Downing and Mooney, 2008). The decelerator was operated at 248 kV, resulting in images formed by a 52 kV electron beam at the CCD. The mutant samples presented a technical challenge owing to their increased length and width and their propensity to bleb, thus we were obliged to work with effectively thicker samples.
In order to have a statistically relevant survey of cell sizes and morphologies, 228 images were recorded on both cameras. 76 images were recorded on the 2Kx2K camera using magnifications of 48Kx and 25Kx at the CCD giving a pixel size of 0.68 nm or 1.2 nm at the specimen respectively. Underfocus values ranged between 6 µm ± 0.5 um to 14 µm ± 0.5 µm, and energy filter widths were typically around 22 eV ± 2 eV. 52 images were recorded on the Decelerator 4Kx4K camera using a magnification of 40Kx at the CCD giving a pixel size of 0.373 nm. A total of 100 images were acquired in the Search Diffraction Mode using both cameras, for large field of view and low resolution high contrast. A total of 17 ideal targets for 3D data acquisition were selected from these surveys.
Tomographic tilt series were acquired under low dose conditions, typically over an angular range between +62° and −62°, ± 2° with increments of 2°. 64 ± 2 images were recorded for each series. 8 tilt series were acquired on the 2Kx2K CCD, and 9 tilt series were acquired on the Decelerator-coupled 4Kx4K CCD, with the program Serial-EM (http://bio3d.colorado.edu/) adapted to JEOL microscopes. For tilt series data sets images were acquired using a magnification of 25Kx at the 2Kx2K CCD giving a pixel size of 1.2 nm at the specimen and 40kX at the 4Kx4K Decelerator-coupled CCD giving a pixel size of 0.373 nm at the specimen respectively. Most of the data acquired on the 4Kx4K CCD were subsequently binned. Underfocus values ranged between 6 µm ± 0.5 um to 12 µm ± 0.5 µm, depending on the goal of the data set, and energy filter widths ranged between 22 eV to 28 eV, also depending on the data set. For all data sets the maximum dose used per complete tilt series was approximately 150 e-/Å2, with typical values of approximately 100 e-/Å2.
CryoEM and CryoET image processing and analyses
All tomographic reconstructions were obtained with the program Imod (http://bio3d.colorado.edu/). The program ImageJ (NIH, http://rsb.info.nih.gov/ij/) was used for analysis of the two-dimensional image projections. Volume rendering and image analysis of tomographic reconstructions was done using the program Paraview (http://www.paraview.org). All movies were created with the package ffmpeg (www.ffmpeg.org). Measurements of PG thickness (15–39 measurements/sacculus) and IM-OM distance (6–11 measurments/cell) were taken manually using ImageJ at random intervals around the sacculus or cell. For measurements of IM-OM distance, regions of blebbing were avoided in DipM depleted cells.
Supplementary Material
Acknowledgments
We thank Martin Thanbichler and Christine Jacobs-Wagner for informing us that they are working on DipM and for collectively agreeing on a gene and protein name. We are especially grateful to members of the Shapiro and McAdams laboratories for helpful discussions and to Esteban Toro, Monica Schwartz, Natalie Dye, and Grant Bowman for comments on the manuscript. We thank Martin Thanbichler for HU2 antibody preparation, Michael Fero for developing KAMS imaging software, Lu Gan for the detailed sacculus isolation protocol, James Gober for α-FtsZ antibody, and John Werner and Zemer Gitai for the collaboration that led to identification of DipM. EDG is a Helen Hay Whitney Foundation postdoctoral fellow. This work was supported in part by the US Department of Energy under Contracts No. DE-FG02-05ER64136 (LS) and No. DE-AC02-05CH11231 (KDH), and by the NIH under grant R01 GM 32506 (LS).
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubes R. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol. 1998;180:4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol. 2003;48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol. 2004;52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman ME, Kannenberg K, et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol. 2006;61:675–690. doi: 10.1111/j.1365-2958.2006.05280.x. [DOI] [PubMed] [Google Scholar]
- Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browder HP, Zygmunt WA, Young JR, Tavormina PA. Lysostaphin: Enzymatic Mode of Action. Biochem Biophys Res Commun. 1965;19:383–389. doi: 10.1016/0006-291x(65)90473-0. [DOI] [PubMed] [Google Scholar]
- Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- Burdett ID, Higgins ML. Study of pole assembly in Bacillus subtilis by computer reconstruction of septal growth zones seen in central, longitudinal thin sections of cells. J Bacteriol. 1978;133:959–971. doi: 10.1128/jb.133.2.959-971.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett ID, Murray RG. Electron microscope study of septum formation in Escherichia coli strains B and B-r during synchronous growth. J Bacteriol. 1974a;119:1039–1056. doi: 10.1128/jb.119.3.1039-1056.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett ID, Murray RG. Septum formation in Escherichia coli: characterization of septal structure and the effects of antibiotics on cell division. J Bacteriol. 1974b;119:303–324. doi: 10.1128/jb.119.1.303-324.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Gavioli M, Sturgis JN, Lloubes R. Proton motive force drives the interaction of the inner membrane TolA and outer membrane pal proteins in Escherichia coli. Mol Microbiol. 2000;38:904–915. doi: 10.1046/j.1365-2958.2000.02190.x. [DOI] [PubMed] [Google Scholar]
- Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- Costa T, Priyadarshini R, Jacobs-Wagner C. Localization of PBP3 in Caulobacter crescentus is highly dynamic and largely relies on its functional transpeptidase domain. Mol Microbiol. 2008;70:634–651. doi: 10.1111/j.1365-2958.2008.06432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT. Biogenesis of bacterial membrane vesicles. Mol Microbiol. 2009;72:1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Blaauwen T, Aarsman ME, Vischer NO, Nanninga N. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol Microbiol. 2003;47:539–547. doi: 10.1046/j.1365-2958.2003.03316.x. [DOI] [PubMed] [Google Scholar]
- den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- Downing KH, Mooney PE. A charge coupled device camera with electron decelerator for intermediate voltage electron microscopy. Rev Sci Instrum. 2008;79 doi: 10.1063/1.2902853. 043702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci U S A. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci U S A. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Glauner B, Holtje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–R526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Hara H, Narita S, Karibian D, Park JT, Yamamoto Y, Nishimura Y. Identification and characterization of the Escherichia coli envC gene encoding a periplasmic coiled-coil protein with putative peptidase activity. FEMS Microbiol Lett. 2002;212:229–236. doi: 10.1111/j.1574-6968.2002.tb11271.x. [DOI] [PubMed] [Google Scholar]
- Harry E, Monahan L, Thompson L. Bacterial cell division: the mechanism and its precison. Int Rev Cytol. 2006;253:27–94. doi: 10.1016/S0074-7696(06)53002-5. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Ursinus A, Berger J, Schwarz H, Holtje JV. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol. 2002;184:6093–6099. doi: 10.1128/JB.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa JK, Li C, Fu J, Clarke S. A gene at 59 minutes on the Escherichia coli chromosome encodes a lipoprotein with unusual amino acid repeat sequences. J Bacteriol. 1994;176:1630–1638. doi: 10.1128/jb.176.6.1630-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T, Yamazoe M, Maeda M, Wada C, Hiraga S. Proteolytic activity of YibP protein in Escherichia coli. J Bacteriol. 2002;184:2595–2602. doi: 10.1128/JB.184.10.2595-2602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC, Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH. Distinct constrictive processes, separated in time and space, divide caulobacter inner and outer membranes. J Bacteriol. 2005;187:6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsak D, Liebscher S, Vollmer W. Susceptibility to antibiotics and beta-lactamase induction in murein hydrolase mutants of Escherichia coli. Antimicrob Agents Chemother. 2005;49:1404–1409. doi: 10.1128/AAC.49.4.1404-1409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- Lange R, Hengge-Aronis R. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol Microbiol. 1994;13:733–743. doi: 10.1111/j.1365-2958.1994.tb00466.x. [DOI] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- Markiewicz Z, Glauner B, Schwarz U. Murein structure and lack of DD- and LD-carboxypeptidase activities in Caulobacter crescentus. J Bacteriol. 1983;156:649–655. doi: 10.1128/jb.156.2.649-655.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, Tan MH, et al. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, den Blaauwen T. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol. 2007;65:1106–1121. doi: 10.1111/j.1365-2958.2007.05851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl DA, Easter J, Jr, Gober JW. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol Microbiol. 2001;42:741–755. doi: 10.1046/j.1365-2958.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Overview of cell shape: cytoskeletons shape bacterial cells. Curr Opin Microbiol. 2007;10:601–605. doi: 10.1016/j.mib.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JS. Biological Properties and Classification of the Caulobacter Group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, Popham DL, Young KD. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J Bacteriol. 2006;188:5345–5355. doi: 10.1128/JB.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus E, Din N, Brun YV. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc Natl Acad Sci U S A. 1996;93:6314–6319. doi: 10.1073/pnas.93.13.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadurai L, Lockwood KJ, Nadakavukaren MJ, Jayaswal RK. Characterization of a chromosomally encoded glycylglycine endopeptidase of Staphylococcus aureus. Microbiology. 1999;145(Pt 4):801–808. doi: 10.1099/13500872-145-4-801. [DOI] [PubMed] [Google Scholar]
- Sackett MJ, Kelly AJ, Brun YV. Ordered expression of ftsQA and ftsZ during the Caulobacter crescentus cell cycle. Mol Microbiol. 1998;28:421–434. doi: 10.1046/j.1365-2958.1998.00753.x. [DOI] [PubMed] [Google Scholar]
- Sonntag I, Schwarz H, Hirota Y, Henning U. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol. 1978;136:280–285. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt BG. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J Bacteriol. 1977;131:293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Alley MR. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats P, Shih YL, Rothfield L. Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol Microbiol. 2009;72:170–182. doi: 10.1111/j.1365-2958.2009.06632.x. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci U S A. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Wang L, Khattar MK, Donachie WD, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Pogliano K, Carson M, Guzman LM, Fraipont C, Nguyen-Disteche M, et al. Localization of the Escherichia coli cell division protein Ftsl (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci U S A. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.