Abstract
Homodimerization of RON (MST1R), a receptor tyrosine kinase, usually occurs in cells stimulated by a ligand and leads to the downstream activation of signaling pathways. Here we report that bladder cancer cells, in response to physiological stress, use an alternative mechanism for signaling activation. Time-course studies indicated that RON migrated directly from the membrane to the nucleus of bladder cancer cells in response to serum starvation. Biochemical and genetic studies implied that this nuclear internalization was complexed with epidermal growth factor receptor (EGFR) and required the docking of importins. In vivo analysis confirmed that nuclear RON was present in 38.4% (28/73) of primary bladder tumors. Chromatin immunoprecipitation (ChIP) on microarray analysis further revealed that this internalized complex bound to at least 134 target genes known to participate in three stress-responsive networks: p53, stress-activated protein kinase/c-jun N-terminal kinase and phosphatidylinositol 3-kinase/Akt. These findings suggest that RON, in a complex with EGFR, acts as a transcriptional regulator in response to acute disturbances (e.g. serum starvation) imposed on cancer cells. In an attempt to re-establish homeostasis, these cells bypass regular mechanisms required by ligand stimulation and trigger the RON-directed transcriptional response, which confers a survival advantage.
Introduction
Receptor tyrosine kinases (RTKs) are transmembrane proteins that receive signaling from ligands (e.g. growth factors) produced by neighboring cells (1). Two RTK monomers usually dimerize upon ligand binding; partners can be formed from the same protein molecule (homodimerization), whereas in some cases, different molecules from the same RTK family are tethered together (hetero-dimerization) on the cell surface (2). After dimerization, phosphorylation occurs in their cytoplasmic tyrosine residues thereby providing docking sites for other proteins (e.g. mitogen-activated protein kinases) to bind and activate their respective downstream signaling cascades. This RTK-mediated signaling is a key mechanism by which most extracellular information is conveyed to the nucleus (3,4).
RON (MST1R) is a transmembrane protein of the RTK family related to c-Met (5). On the cell membrane, the precursor RON (185 kDa) is cleaved into two chains, α and β, of a mature 150 kDa monomer (5). The α chain is completely extracellular, whereas the β chain, which contains the tyrosine kinase regulatory element, traverses the cell membrane (5). When stimulated by the macrophage-stimulating protein (MSP) (6), two monomers of RON are combined to form a homodimer on the cell membrane. After the dimerization, phosphorylation occurs in cytoplasmic tyrosine residues of the β chain that provide docking sites for other proteins to bind and activate signaling cascades, including Ras, mitogen-activated protein kinase, PI3 kinase, nuclear factor kappaB and focal adhesion kinase pathways (7,8). Studies have shown that aberrantly expressed or mutated RON was observed in the carcinoma of the bladder, breast, colon, lung, ovary, pancreas and prostate (9–18). Increased levels of RON have also been found in aggressive tumors associated with poor patient survival (9,14). Further evidence has shown that RON can promote c-Src activities that mediate cell-cycle progression, angiogenesis and survival of tumor cells (14,19). Therefore, RON warrants further study on its role in tumorigenesis.
Toward this end, we studied the functions of RON in bladder cancer cells and unexpectedly found that this membrane protein was translocated to the cell nucleus without the need of MSP stimulation and homodimerization. Instead, RON formed a complex with another receptor tyrosine kinase, epidermal growth factor receptor (EGFR), in serum-starved bladder cancer cells. This complex was internalized into the nucleus through the importin transport machinery and then activated stress-responsive transcription. The finding highlights a previously unidentified mechanism of RON in bladder cancer cells in response to physiological stress.
Materials and methods
Cell culture, clinical specimens and immunohistochemical staining
Cells grown on glass coverslips were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 and blocked with 5% bovine serum albumin (BSA) to reduce non-specific binding. After thorough washing, primary antibody was incubated overnight at 4°C, followed by incubation with Rhodamine or fluorescein isothiocyanate-labeled secondary antibody (Chemicon International, Temecula, CA). 4′,6-Diamidino-2-phenylindole (Molecular Probes, Eugene, OR) was used for nuclear staining. Finally, cells were mounted and analyzed using a FV1000 confocal microscope (Olympus, Tokyo, Japan) and photographed with 1800-fold magnification. The number of cells expressing RON at subcellular locations was calculated from 10 optical fields under 400-fold magnification.
Immunofluorescence analysis
Cells grown on glass coverslips were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 and blocked with 5% BSA to reduce non-specific binding. After thorough washing, primary antibody was incubated overnight at 4°C, followed by incubation with Rhodamine or fluorescein isothiocyanate-labeled secondary antibody (Chemicon). 4′,6-Diamidino-2-phenylindole (Molecular Probes) was used for nuclear staining. Finally, cells were mounted and analyzed using a FV1000 confocal microscope (Olympus) and photographed with 1800-fold magnification. The number of cells expressing RON at subcellular locations was calculated from ten optical fields under 400-fold magnification.
Subcellular fractionation, co-immunoprecipitation and western blotting analysis
Cells were washed with phosphate-buffered saline, resuspended in a lysis buffer (20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.0, 10 mM KCl, 2 mM MgCl2, 0.5% NP40, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride and 0.15 U/ml aprotinin) and homogenized by 30 strokes in a tightly fitting Dounce homogenizer. The homogenate was centrifuged at 1500g for 5 min to collect the nuclear fraction of cells. The non-nuclear fraction was taken from the supernatant following second centrifugation at 15 000g for 5 min. The nuclear fraction for in vitro experiments was the supernatant of nuclear extract after centrifugation at 15 000g for 10 min. To exclude possible contaminations, each subcellular fraction was examined with both proliferating cell nuclear antigen (nuclear marker) and mitochondrial prohibitin (non-nuclear marker). Immunoblotting and co-immunoprecipitation were performed with antibodies against RON (C-20), p-Tyr (PY-20), importin α1 (C-20), importin β1 (H-300), Ran (H-96) or proliferating cell nuclear antigen (FL-261) (Santa Cruz Biotechnology, Santa Cruz, CA) and EGFR (clone 528) or prohibitin (II-14-10) (NeoMarkers, Lab Vision, CA).
RNA interference
Cells (2 × 105 cells) were seeded in a 6-well plate and transfected the next day with pre-designed small interfering RNAs (siRNAs) against RON (Santa Cruz Biotechnology), EGFR (QIAGEN, Valencia, CA) or importin α1 (obtained from National RNAi core facility, Academia Sinica, Taiwan) using the Fugene 6.0 transfection reagent (Roche Applied Science, Indianapolis, IN) according to manufacturer's protocol. After 48 h, the cells were starved for 24 h for immunoblotting or image analyses. Non-specific siRNAs (Santa Cruz Biotechnology) were used as a negative control.
Cell growth assay
Growth curves were determined by cell counting with trypan blue staining. Briefly, cells (103) seeded on a 10 cm dish was trypsinized and stained with trypan blue at indicated time periods (0, 2, 4, 6 and 8 days). The number of cells from a total of 10 fields per experiment was calculated using a counting chamber within ×40 magnification. In the anti-apoptosis experiment, cells were pretreated with methotrexate (100 μg/ml) for 72 h, irrespective of MSP treatment (100 ng/ml) and then cell viability was measured by the methylthiazoletetrazolium assay as described previously (20). Results are shown in the percentage of cell viability relative to absorbance of the wild-type RON transfectants treatment. Error bars represent the standard derivation of three separate determinations.
ChIP coupled with microarray (ChIP-chip)
ChIP was performed with anti-RON antibody using a ChIP assay kit (Upstate Biotechnology, New York, NY) as described previously (21). Incorporation of aminoallyl-deoxyuridine triphosphate (Sigma, St Louis, MO) into 2 μg ChIP–DNA or total input DNA was conducted with BioPrime DNA labeling system (Invitrogen, Carlsbad, CA). Cy5 and Cy3 fluorescent dyes (Amersham Biosciences Corp, Piscataway, NJ) were coupled to ChIP/total input DNA, respectively, and were co-hybridized to a human CpG island 185K microarray (∼185 000 45–60mers; Agilent Technologies, Santa Clara, CA). Microarray hybridization and post-hybridization washes have been described previously (22). The washed slides were scanned by a GenePix 4000B scanner (Axon, Sunnyvale, CA), and the acquired microarray images were analyzed with GenePix Pro 6.0 software. Two independent experiments were performed for each time point treatment and antibody. After excluding the spots flagged for bad quality, signal ratios were log2 transformed and normalized by using intensity-dependent normalization (23). Lowess-normalized data of duplicated microarray experiments were used to identify significant methylated loci (P < 0.001) using a peak detection algorithm in the ChIP Analytics 1.3 software (Agilent Technologies).
Quantitative ChIP–polymerase chain reaction
To confirm candidate RON target genes identified by ChIP-chip, polymerase chain reaction (PCR) primers targeting the enriched region were designed to measure the amount of the sequence in anti-RON-and anti-EGFR-immunoprecipitated samples by quantitative PCR with SYBR Green-based detection. Experimental quantitative ChIP–PCR values were normalized against values obtained by a standard curve (0.08–50 ng, 5-fold dilution, R2 > 0.99), constructed by input DNA with the same primer set.
Quantitative real-time–PCR
Total RNA (2 μg), isolated by the Trizol reagent (Invitrogen), was treated with DNase I to remove potential DNA contamination and then was reverse transcribed with the SuperScript III reverse transcriptase (Invitrogen). Quantitative real-time–PCR was performed by using SYBR Green as a marker for DNA amplification on a 7500 Real-Time PCR System apparatus (Applied Biosystems, Foster City, CA). Relative messenger RNA level of a given locus was calculated by relative quantitation of gene expression with β-actin messenger RNA as an internal control. Specific primers for amplification are available on request.
Motif analysis and functional classification of target genes
Chromosome sequences of 22 nuclear RON-binding regions identified by ChIP-chip and confirmed by ChIP–PCR were retrieved from human May 2004 (hg17) assembly (UCSC Genome Browser). Interspersed repeats and low complexity DNA sequences were removed by the Repeat Masker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) and the DUST program (Tatusoy,R.L. and Lipman, D.J., unpublished NCBI/Toolkit, downloaded from http://blast.wustl.edu/pub/dust). Then, a dataset within the above 22 chromosome sequences was established for the following motif analysis by the free software, the web version of Motif Discovery scan tool (MDscan; http://ai.stanford.edu/∼xsliu/MDscan). The logo of the consensus sequence to nuclear RON-targeting genes was drawn on http://weblogo.berkeley.edu. Gene ontology of the putative RON target genes was assessed by GOTree Machine (Gene Ontology Tree Machine; http://bioinfo.vanderbilt.edu/gotm). The biological function of genes were categorized into five groups and then plotted as a pie chart. The Ingenuity Pathway Analysis version 6.5 (Ingenuity Systems; http://www.ingenuity.com) was used to analyze nuclear RON targets. Filter was performed to remove genes with no annotation in Ingenuity Pathway Analysis, resulting in a list of 106 network eligible genes. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base.
Luciferase reporter assay
Luciferase assay were carried out using human embryonic kidney 293 (HEK293) cells grown in Dulbecco's modified Eagle's medium media supplemented with 10% fetal bovine serum and antibiotics. HEK293 cells (1 × 105) were plated in triplicate in 12-well culture plates on the day prior to transfection. About 60–70% confluent cells were transfected with pRLTK reporter (Renilla luciferase as internal control), pGL3 reporter (firefly luciferase), target gene plasmids and pLP-TRE2-RON (RON expression plasmid) by Lipofectamine 2000™ following the manufacturer's instructions (Invitrogen). The target gene promoter–reporter plasmids included pGL3-c-Jun (c-Jun/Luc), pGL3-BNIP3 and pGL3-P53. The cells were harvested at desired time points after transfection, respectively, and lysed in 100 ml of passive lysis buffer (Promega, Madison, WI). The cell lysate was assayed for both firefly and Renilla luciferase activities according to manufacturer's instructions. The relative luciferase units (RLU) were measured by a luminometer (EG&G Berthold, Bad Wildbad, Germany). Luciferase activity of the firefly luciferase was normalized for equal transfection efficiency using the Renilla luciferase activity in each lysate as the control. The RLUs were presented as the mean with standard deviation of three independent experiments performed in triplicate.
Results
Membranous RON is translocated into the nucleus as the dephosphorylated intact molecule in serum-starved bladder cancer cells
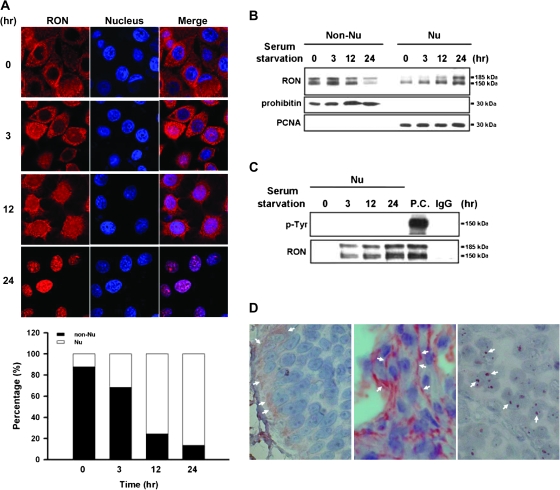
Our previous study reveals that RON overexpression is positively correlated to tumorigenicity of bladder carcinoma cells (13,20). Unexpected immunohistochemical results of RON staining in primary tumors indicate that RON is not only expressed on membrane, but it can be detected in the nucleus, suggestive of the potential transcriptional function of RON. To clarify the expression pattern of RON, immunofluorescence analysis was performed in a bladder cancer cell line, TSGH8301, with overexpression of RON (Figure 1A). In a time-course study, >80% of RON (red staining) were found to progressively enter into the nucleus (blue staining) of TSGH8301 cells within 24 h of serum starvation. Immunoblotting of non-nuclear and nuclear protein fractions confirmed this time-dependent nuclear entrance (Figure 1B). Furthermore, the detected two bands of RON, 185 kDa (precursor form of RON) and 150 kDa (mature form of RON) in immunoblotting results indicated that RON was expressed as the intact molecule in the nucleus. To exclude possible subcellular contaminations, these fractions were positively confirmed with a nuclear marker (proliferating cell nuclear antigen) and a cytosolic marker (mitochondrial prohibitin), respectively. This nuclear internalization was also found in other types of cancer cells undergoing serum starvation (supplementary Figure 1 is available at Carcinogenesis Online).
Fig. 1.
Kinetic analysis of nuclear localization of RON. TSGH8301 cells were serum starved for different time periods from 0, 3, 12 to 24 h. (A) Immunofluorescence analysis was conducted to monitor the time-dependent nuclear localization of RON. Treated cells were incubated with anti-RON (red) and then a Rhodamine-conjugated secondary antibody (upper panel). Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). The purple signals (merged) highlight the localization of RON in the nucleus. The results were quantified and expressed as a histogram (bottom panel). (B) Western blotting with anti-RON antibody on subcellular fractions of treated cells was exhibited to determine time-dependent nuclear import of RON. The prohibitin and proliferating cell nuclear antigen (PCNA) were chosen as markers for non-nucleus and nucleus fractions, respectively. (C) Nuclear fractions from treated cells were subjected to co-immunoprecipitation analysis with anti-RON antibody and then probed with anti-tyrosine (p-Tyr) antibody. Total cell lysate with MSP treatment was used as positive control (P.C.) for phospho-RON staining, and IgG was used as negative control of immunoprecipitation assay. (D) Immunohistochemical analysis was performed to detect RON expression on human bladder tissues from bladder cancer patients (n = 73). In this representative example, white arrows indicate positive staining areas of RON. Both ‘right’ and ‘middle panels’ are malignant bladder tissues; the ‘left panel’ is normal bladder tissue.
We then determined whether phosphorylated RON was present in the nucleus since this posttranslational event usually takes place upon stimulation by a ligand (24,25). Immunoprecipitation with anti-RON antibody was carried out, and the protein products were probed with an anti-phospho-tyrosine antibody. Phosphorylated RON was not found in these serum-starved cells (Figure 1C, lanes 2–4) compared with the total lysate of cells with ligand stimulation (lane 5). These in vitro findings suggest that the translocation of RON into the nucleus of TSGF8301 cells occurs in response to physiological stress and that this redistribution does not require phosphorylation of the protein.
The observation was confirmed in vivo, showing positive immunostaining of nuclear RON in 38.4% (28/73) of bladder tumor sections collected between 1999 and 2002 in our hospital (see a representative example in Figure 1D, right panel). Overexpressed RON was also found in the cell membrane (24.3%) of tumor sections (middle panel) compared with weaker staining on basal membrane areas of normal sections (left panel).
Nuclear import of RON is mediated by the importin family proteins
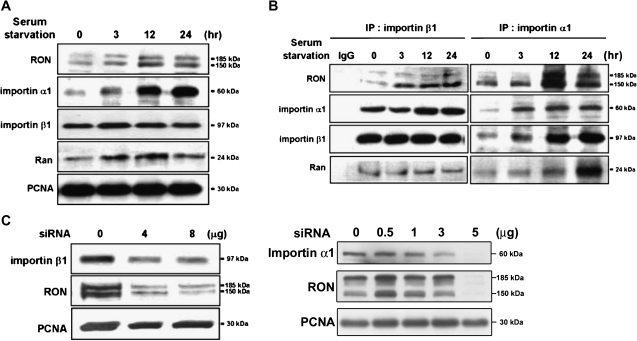
Studies have implicated that importins mediate the transport of macromolecules from the cytosol compartment into the nucleus (26). We therefore investigated whether these transport proteins were involved in the nuclear import of RON using immunoblotting assays. Both importin α1 and its associated protein, Ran, were accumulated in the nucleus in a time-dependent manner after serum starvation of TSGH8301 cells, whereas the increase of importin β1 in the nucleus was not remarkable (Figure 2A). Co-immunoprecipitation assays, with either importin β1 (Figure 2B, left panel) or α1 (right panel) antibody, of the TSGH8301 nuclear extract revealed the formation of a protein complex containing RON, importin α1, importin β1 and Ran (Figure 2B). The siRNA experiments showed that knockdown of importin α1 or β1 could effectively suppress the nuclear import of RON (Figure 2C), suggesting the involvement of these importin family members in this event.
Fig. 2.
Importins/NLS-mediated nuclear import of RON. TSGH8301 cells were starved for different time periods as indicated and then nuclear extracts were collected for the following western blotting and immunoprecipitation assays with the indicated antibodies. (A) Upon serum starvation, increasing expression of RON, importin α1, importin β1 and Ran in the nucleus were detected by immunoblotting analysis. Proliferating cell nuclear antigen (PCNA) was used as a nuclear marker. (B) The association of RON with nuclear importin proteins, including importin α1, importin β1 and Ran, was determined by co-immunoprecipitation assays. Nuclear extracts were pulled down by either anti-importin β1 (left) or anti-importin α1 (right) antibodies and then probed with the indicated antibodies. (C) Inhibition of nuclear RON expression by importin α1 or β1 siRNA knockdown. TSGH8301 cells were transfected with different dosage of importin α1 or β1 siRNA for 48 h and then nuclear extracts were collected for sodium dodecyl sulfate–polyacrylamide gel electrophoresis resolution. PCNA was as a nuclear marker and loading control. A dose-dependent suppressor effect was more apparent in knock down of importin β1.
Genetic studies were further conducted to determine whether the nuclear localization signal (NLS) sequence of RON is essential for this nuclear transport. NLS contains a stretch of 3–5 basic residues known to interact with importin β1 (26). Two possible NLS regions, N-terminus (aa 305-KRRRR-309) and C-terminus (aa 1389-RRPR-1391), of RON were identified by the PredNLS software program (supplementary Figure 2 is available at Carcinogenesis Online). Site-directed mutagenesis is being carried out to verify the putative NLS regions of RON. Taken together, importins may involve in nuclear transportation of RON in bladder cancer cells.
RON forms a nuclear complex with EGFR in serum-starved bladder cancer cells
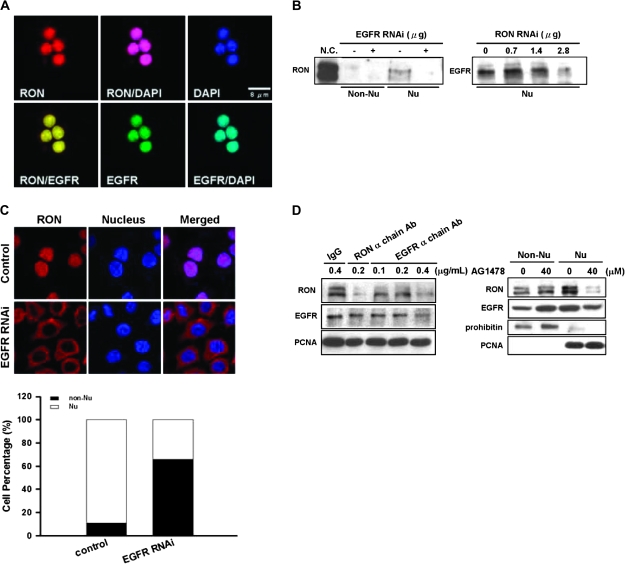
Our previous study showed that EGFR could activate RON in a ligand-independent manner (20). We therefore hypothesized that EGFR might involve in the nuclear import of RON. First, we found that EGFR was colocalized with RON in the nucleus of TSGF8301 cells with 24 h of serum starvation (Figure 3A). Attenuated EGFR by siRNA knockdown could suppress the nuclear translocation of RON (Figure 3B, left panel) and arrested the protein on the cell membrane of >70% cells (Figure 3C). A reciprocal experiment indicated that siRNA knockdown of RON could reduce the expression of nuclear EGFR (Figure 3B, right panel). Furthermore, nuclear import of RON was greatly restrained when cells were treated with a RON or EGFR neutralization antibody or AG1478, an EGFR-kinase inhibitor (Figure 3D). These results suggest that collaboration occurs between RON and EGFR in the nucleus of serum-starved cells. The complex may bind to DNA and trigger transcription of potential target genes.
Fig. 3.
Interactions between EGFR and nuclear RON in bladder cancer cells. (A) Immunofluorescence analysis showed that, upon 24 h of serum starvation, RON (red) was colocalized with EGFR (green) in the nucleus of TSFG8301 cells. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue) and images acquired in an Olympus Confocal Microscope. (B) Nuclear RON expression was reduced by EGFR siRNA in TSGH8301 cells. After 48 h of either EGFR (left) or RON (right) siRNA transfection, nuclear extracts were collected for immunoblotting analysis. (C) RON (red) location was redistributed upon attenuating EGFR expression by EGFR siRNA. After 48 h transfection of EGFR siRNA, TSGH8301 cells were subjected to immunofluorescence analysis. Nuclei were counterstained with DAPI (blue). Staining intensities were quantified and expressed as histogram (bottom panel). (D) TSGH8301 cells were treated with indicated doses of EGFR neutralization antibody (left panel; lanes 3–5) and nuclear extracts were collected for western blot analysis. Treatment of RON α chain antibody (lane 3) was utilized as a positive control, whereas normal IgG antibody (lane 1) was used as a negative control to the neutralization antibodies. In ‘right panel’, cells were pretreated with optimized doses of AG1478, an EGFR-specific inhibitor, and then serum-starved for 24 h. Subcellular fractions were collected for immunoblotting with antibodies as indicated.
Nuclear RON binds to DNA and activates stress-responsive transcription
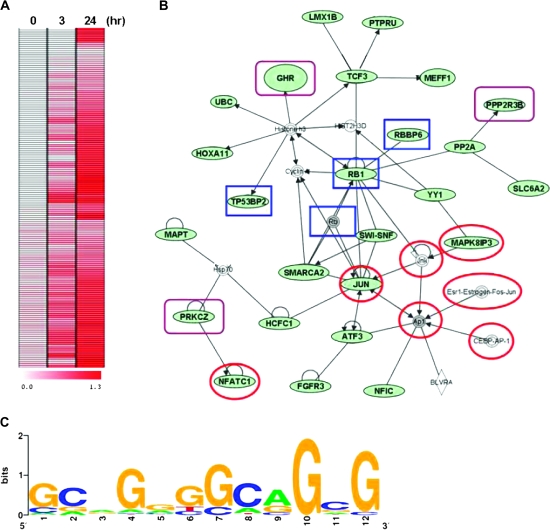
To determine whether nuclear RON functions as a transcriptional regulator, we used ChIP coupled with microarray (ChIP-chip) to identify its promoter targets. Chromatin was immunoprecipitated with anti-RON antibody in TSGH8301 cells harvested at 0, 3 and 24 h after serum starvation. Pull-down DNAs were fluorescently labeled and hybridized to a microarray panel containing ∼17 000 annotated promoters (22). Positive loci were identified with greater fluorescent intensities (>1.8-fold cut off) of hybridization signals relative to the control DNAs (i.e. total input) in two biological replicates. Increased binding of RON was observed in 134 targets 24 h after serum starvation of TSGH8301 cells (Figure 4A). Ontological analysis of these putative targets (listed in supplementary Table 1, available at Carcinogenesis Online) categorized their biological functions into five groups: 23.9% of these loci involved in transcriptional regulation (as transcription factors or cofactors), 21.6% in development (e.g. skeletal-and anatomical structure-related functions), 24.6% in cell metabolism, 11.2% in cell growth and 18.7% in physiological processes (e.g. cell migration) (supplementary Figure 3 is available at Carcinogenesis Online). Twenty-six of these target genes were known to participate in three starvation-responsive networks, p53, stress-activated protein kinase/c-jun N-terminal kinase and phosphatidylinositol 3-kinase (P13K)/Akt (Figure 4B). The predicted genes, RBBP6, RB1, TP53BP2 and JUN of p53 pathway were associated with pro-apoptotic activities in response to DNA damage (27–30). Activation of SMAPK/JNK (e.g. JUN, MAPK8IP3, NFATC1 and TRADD) and phosphatidylinositol 3-kinase/Akt (e.g. GHR, PPP2R3B and PRKCZ) pathways were related to the stress-induced survival of cancer cells (16,30–32). Motif scanning of these promoters identified a consensus sequence, GCA(G)GGGGCA GCG, to which nuclear RON binds its targets upon serum starvation of TSGH8301 cells (Figure 4C).
Fig. 4.
Identification of RON-targeting genes. (A) ChIP assay coupled with microarray (ChIP-chip) was conducted to identify the potential targets of RON, which is summarized into a heat map of 134 genes. Starved TSGH8301 cells at the indicated time periods (0, 3 and 24 h) were harvested for ChIP assay with anti-RON antibody. The dye-coupled-immunoprecipitated DNA was hybridized onto the 185K CGI microarrays. (B) Nuclear RON targets form a network of interactions by Ingenuity Pathway Analysis software. The input was all 134 significant genes from ChIP-chip assay. Genes depicted in green are the RON-targeting genes. Targets in red circles are associated with the stress-activated protein kinase/JNK signaling pathway, targets in blue rectangles are related to the p53 signaling pathways and targets in purple rectangles are associated with the phosphatidylinositol 3-kinase/Akt pathway. (C) The motif logo of consensus sequence to RON-targeting genes. The potential nuclear RON-targeting genes with ChIP–PCR validation were collected to establish a dataset for the motif analysis. The putative sequence was determined by MDscan (Motif Discovery Scan; http://ai.stanford.edu/∼xsliu/MDscan). The motif log was generated on http:weblogo.berkeley.edu.
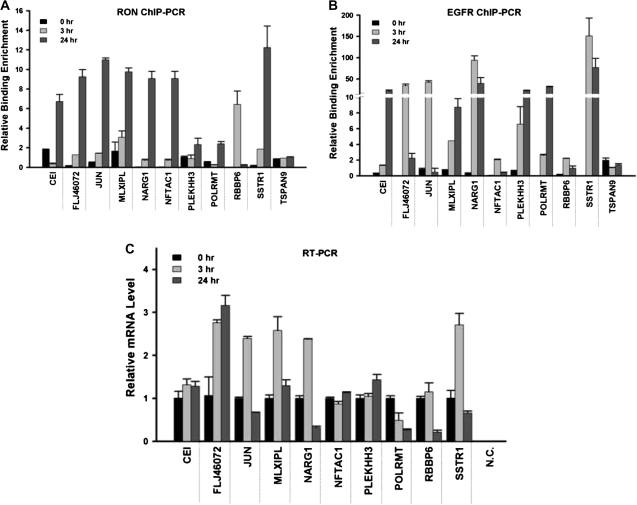
Quantitative ChIP–PCR assays were further developed to validate the binding of RON to, or near, this consensus sequence in 10 randomly selected promoter targets (CEI, FLJ46072, JUN, MLXIPL, NARG1, NFATC1, PLEKHH3, POLRMT, RBBP6 and SSTR1). These results confirmed nine targets (except for RBBP6) that showed 2- to 12-fold enrichment of RON binding, relative to the control immunoglobulin G at the 24 h time point of serum starvation (Figure 5A). The binding of RON to RBBP6 was maximized at 3 h but reduced to the basal level at 24 h after serum starvation of TSGH8301 cells. We additionally conducted ChIP–PCR assays on these targets using an anti-EGFR antibody (Figure 5B). Supporting the findings of the co-immunoprecipitation assays, we found that EGFR could bind to eight of these targets (except for NFATC1 and RBBP6) in the serum-starved condition. This collaborative binding between RON and EGFR resulted in transcriptional activation or repression of some of these targets as determined by quantitative real-time–PCR; five genes, FLJ46072, JUN, MLXIPL, NARG1 and SSTR1, were found to be upregulated, whereas the transcripts of RBBP6 and POLRMT were reduced after serum starvation (Figure 5C). The expression levels of three other target genes, CEI, NFATC1 and PLEKHH3, were not changed. It is possible that the RON or EGFR binding may be insufficient to activate or repress these transcripts; an additional recruitment of cofactors might be needed to alter their transcription status. Consistent with the aforementioned biochemical and genetic studies, these ChIP-based results suggest that RON aberrantly functions as a DNA-binding protein that activates or deactivates gene transcription necessary for a stress response in bladder cancer cells.
Fig. 5.
Quantitative PCR analyses of candidate target genes. Quantitative ChIP–PCR analyses were conducted to validate the potential target genes of nuclear RON. After serum starvation for the indicated time periods, TSGH8301 cells were harvested and ChIP assays were performed with antibodies directed against RON (A) or EGFR (B). The immunoprecipitation DNA corresponding to the interested targets was measured by quantitative PCR. Quantitation of specific RON targeting was determined as a percent of input DNA and each error bar represents standard deviation calculated from triplicates. (C) Quantitative real-time–PCR analysis was performed to investigate the expression level of candidate target genes. Each error bar represents standard deviation calculated from triplicates. Amplified samples without reverse transcription were used as negative controls.
Transcriptional activation of the target genes of nuclear RON
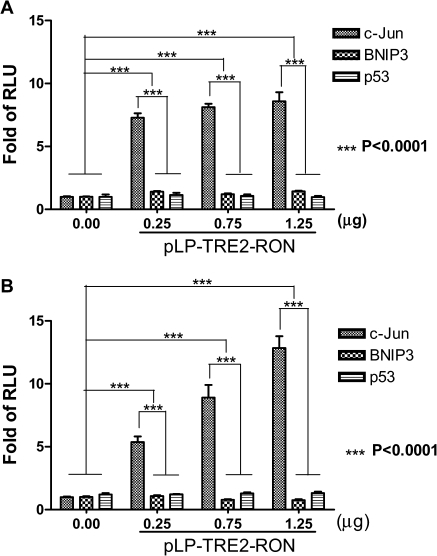
To confirm the transcriptional activation of the promoter targets by nuclear RON, luciferase reporter gene assay was performed using HEK293 cells. The luciferase activity in HEK293 cells was determined after transfection. The means ± SDs (error bars) were derived from three independent experiments. The JUN was selected for confirmation because of our ChIP–PCR and reverse transcription–PCR results and a recent report in vivo showing that c-jun is one of the RON-regulated genes in pancreatic cancer (16). Both BNIP3 and p53 were chosen as negative controls. We observed a time-dependent induction of JUN promoter activity by RON, with dose-dependent induction most noticeable at 3 and 24 h, respectively (P < 0.0001, by two-way analysis of variance test) (Figure 6A and B). The promoter activity reached plateau at 48 h (data not shown). As expected, luciferase activity of p53 or BNIP3 promoter target was not obviously induced by RON. The results support that nuclear translocation of RON is mediating activation of the JUN gene promoter in response to serum starvation.
Fig. 6.
Transcription activation of putative target genes of RON. The promoter activity in HEK293 cells co-transfected with the indicated target gene reporter plasmids together with RON plasmid at various dosages was determined. The means ± SDs (error bars) were derived from three independent experiments. The promoter activity represented by luciferase activity was analyzed by the Dual-Glo™ Luciferase Assay System at 0, 3, 24 and 48 h after transfection, respectively. The results at 3 and 24 h were shown in 6A and 6B, respectively (P < 0.0001, by two-way analysis of variance test). The promoter activity was shown as fold of RLU (relative to the level of control vector reporter gene after normalization with co-transfected Renilla luciferase activity). This experiment was conducted in triplicate. The promoter activity after EGF treatment was used as a positive control. The empty vector pRLTK was used as an internal control.
Discussion
Since RON gene was first cloned from human keratinocytes in 1993 (5), it was found to be an important proto-oncogene in many cancer types (9–18). When wild-type or mutant form of RON is overexpressed in cancer cells, its tumorigenic effect is thought to be initiated through MSP stimulation by neighboring macrophages (7). Based on the present finding, we suggest an alternative mechanism for RON, which acts as a transcriptional regulator in bladder cancer cells responding to environmental stress (e.g. serum starvation). At the cellular level, this environmental stress may be regarded as an acute disturbance to the homeostasis of cancer cells. In an attempt to re-establish homeostasis, these cancer cells bypass regular mechanisms and rapidly activate stress-responsive signaling without ligand stimulation in order to confer a survival advantage.
Here we propose that in response to serum starvation, RON directly migrates from the membrane to the nucleus of bladder cancer cells. Our previous study (13,20) indicates that in addition to homodimerization, RON may tether with receptor tyrosine kinases to form heterologous partner. This partner could be EGFR (20) that is shown to facilitate nuclear translocation of RON. The internalized RON is in a dephosphorylated state, which is unusual as other tyrosine kinases are frequently phosphorylated in the nucleus (24,25). One possible explanation is that dephosphorylated RON may prevent proteosome-mediated degradation and prolongs its nuclear actions. An example is β-catenin in which its dephosphorylated form can enhance the nuclear targeting (31). On the contrary, phosphorylated β-catenin in the cytoplasm can lead to rapid ubiquitination and subsequent degradation in the proteosome (32).
The translocation requires a docking of importins on the NLS sites of RON. Once the complex is in the nucleus, RON can act as a transcription factor to regulate the expression of its target genes. Further analysis indicates that RON contains three IPTs (Ig-like, Plexins, Transcription factors; see in supplementary Figure 4, which is available at Carcinogenesis Online) that bind a common sequence in target promoters. While the functions of the 134 targets identified by ChIP-chip remain to be explored, at least 26 genes are potential stress responders. Two of these genes, JUN and RBBP6, are discussed here in detail. JUN is an important early cellular responder to stresses, leading to upregulation of protective genes involved in cellular proliferation, apoptosis and antioxidant defense (33). Its activation is usually mediated by redox modulation of mitogen-activated protein kinases in the cytoplasm (30). However, the present finding provides new evidence that this activation can also be mediated by the direct binding of RON to the JUN promoter. The second gene is RBBP6, which encodes a negative regulator of p53 (27). Upregulation of RBBP6 may heighten the sensitivity of the p53 network toward apoptotic responses. Knockdown of RBBP6 is shown to induce both apoptosis and cell growth retardation in a p53-dependent manner (27). These two RON-regulated genes, as well as other apoptosis-related loci, are our future focuses on bladder cancer cells in response to stress.
It is interesting to note that RON can be activated through ligand-dependent or -independent mechanisms, which lead to responses important for tumorigenesis and metastasis, including cell scattering, proliferation, motility and survival (12–15,20,34). RON can also regulate the production of angiogenic chemokines that promote tumor growth and angiogenesis (16,18). In addition to serum starvation, oxidative stress may play a certain role in stimulation of RON for migration to the nucleus of cancer cells. Nuclear translocation of RON was abrogated by pretreatment with diphenyleneiodonium, a 5,10-methylenetetrahydrofolate reductase oxidase inhibitor, in a dose-dependent manner (supplementary Figure 5 is available at Carcinogenesis Online). In terms of transcriptional targets, recent experiment in vivo has identified novel RON-regulated genes, including Bcl-2 and members of the activator protein-1 transcription factor complex, c-jun, c-fos and activating transcription factor 3 (17). Because silencing of RON signaling sensitizes cancer cells to gemcitabine therapy (17), RON-targeted agents may ultimately form part of an effective multidrug approach to human cancer treatment.
In conclusion, our study suggests a dual role of RON in bladder tumorigenesis. The previous paradigm indicates that upon MSP stimulation, phosphorylated RON can induce the phosphatidylinositol 3-kinase/Akt signaling cascade that leads to cellular transformation (35). However, based on the present finding, nuclear transport of dephosphorylated RON may occur in transformed cells in response to stress. This non-ligand mechanism, achieved through interaction between RON and EGFR, may directly activate transcriptional machinery necessary for cancer cells to survive in a harsh environment. The finding therefore emphasizes the importance of co-targeting strategy in therapeutic designs for patients with overexpressing tyrosine kinases.
Supplementary material
Supplementary Figures 1–5 and Table 1 can be found at at http://carcin.oxfordjournals.org/.
Funding
National Science Council, Taiwan (NSC96-2320-B-006-025, 95-2320-B006-058-MY3); Ministry of Education Program for Promoting Academic Excellence in Universities, Taiwan (91-B-FA09-1-4); Department of Health, Executive Yuan, Taiwan (DOH-TD-B-111-004); National Health Research Institutes, Taiwan (NHRI-EX99-9930BI); National Institutes of Health, USA (R01CA 69065, U54CA11300).
Supplementary Material
Acknowledgments
We are grateful to Professor Tim H.-M.Huang and his laboratory for their excellent assistance in doing ChIP coupled with microarray and CHIP-PCR experiments.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ChIP
chromatin immunoprecipitation
- EGFR
epidermal growth factor receptor
- MSP
macrophage-stimulating protein
- NLS
nuclear localization signal
- PCR
polymerase chain reaction
- RTK
receptor tyrosine kinase
- siRNA
small interfering RNA
References
- 1.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 2.Li E, et al. Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry. 2006;23:6241–6251. doi: 10.1021/bi060609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorkin A, et al. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 4.Gschwind A, et al. Cell communication networks: epidermal growth factor receptor transactivation as the paradigm for inter-receptor signal transmission. Oncogene. 2001;20:1594–1600. doi: 10.1038/sj.onc.1204192. [DOI] [PubMed] [Google Scholar]
- 5.Ronsin C, et al. A novel putative receptor protein tyrosine kinase of the met family. Oncogene. 1993;8:1195–1202. [PubMed] [Google Scholar]
- 6.Skeel A, et al. Macrophage stimulating protein: purification, partial amino acid sequence, and cellular activity. J. Exp. Med. 1991;173:1227–1234. doi: 10.1084/jem.173.5.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang MH, et al. Oncogenic and invasive potentials of human macrophage-stimulating protein receptor, the RON receptor tyrosine kinase. Carcinogenesis. 2003;24:1291–1300. doi: 10.1093/carcin/bgg089. [DOI] [PubMed] [Google Scholar]
- 8.Danilkovitch-Miagkova A. Oncogenic signaling pathways activated by RON receptor tyrosine kinase. Curr. Cancer Drug Targets. 2003;3:31–40. doi: 10.2174/1568009033333745. [DOI] [PubMed] [Google Scholar]
- 9.Maggiora P, et al. Overexpression of the RON gene is human breast carcinoma. Oncogene. 1998;16:2927–2933. doi: 10.1038/sj.onc.1201812. [DOI] [PubMed] [Google Scholar]
- 10.Chen YQ, et al. Targeted expression of the receptor tyrosine kinase RON in distal lung epithelial cells results in multiple tumor formation: oncogenic potential of RON in vivo. Oncogene. 2002;21:6382–6386. doi: 10.1038/sj.onc.1205783. [DOI] [PubMed] [Google Scholar]
- 11.Zhou YQ, et al. Altered expression of the RON receptor tyrosine kinase in primary human colorectal adenocarcinomas: generation of different splicing variants and their oncogenic potential. Oncogene. 2003;22:186–197. doi: 10.1038/sj.onc.1206075. [DOI] [PubMed] [Google Scholar]
- 12.Maggiora P, et al. The RON and MET oncogenes are co-expression in human ovarian carcinomas and cooperate in activating invasiveness. Exp. Cell Res. 2003;288:382–389. doi: 10.1016/s0014-4827(03)00250-7. [DOI] [PubMed] [Google Scholar]
- 13.Cheng HL, et al. Co-expression of RON and MET is a prognostic indicator for patients with transitional-cell carcinoma of the bladder. Br. J. Cancer. 2005;92:1906–1914. doi: 10.1038/sj.bjc.6602593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feres KJ, et al. The RON receptor tyrosine kinase promotes MSP-independent cell spreading and survival in breast epithelial cells. Oncogene. 2009;28:279–288. doi: 10.1038/onc.2008.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thangasamy A, et al. Recepteur d'origine nantais tyrosine kinase is a direct target of hypoxia-inducible factor-1alpha-mediated invasion of breast carcinoma cells. J. Biol. Chem. 2009;284:14001–14010. doi: 10.1074/jbc.M809320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thobe MN, et al. The Ron receptor tyrosine kinase positively regulates angiogenic chemokine production in prostate cancer cells. Oncogene. 2010;29:214–226. doi: 10.1038/onc.2009.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan-Collins J, et al. Silencing of RON receptor signaling promotes apoptosis and gemcitabine sensitivity in pancreatic cancers. Cancer Res. 2010;70:1130–1140. doi: 10.1158/0008-5472.CAN-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas RM, et al. The RON tyrosine kinase receptor regulates vascular endothelial growth factor production in pancreatic cancer cells. Pancreas. 2010;39:301–307. doi: 10.1097/mpa.0b013e3181bb9f73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danilkovitch-Miagkova A, et al. Integrin-mediated RON growth factor receptor phosphorylation requires tyrosine kinase activity of both the receptor and c-Src. J. Biol. Chem. 2000;275:14783–15147. doi: 10.1074/jbc.C000028200. [DOI] [PubMed] [Google Scholar]
- 20.Hsu PY, et al. Collaboration of RON and epidermal growth factor receptor in human bladder carcinogenesis. J. Urol. 2006;176:2262–2267. doi: 10.1016/j.juro.2006.07.048. [DOI] [PubMed] [Google Scholar]
- 21.Leu YW, et al. Loss of estrogen receptor signaling triggers epigenetic silencing of down-stream targets in breast cancer. Cancer Res. 2004;64:8184–8192. doi: 10.1158/0008-5472.CAN-04-2045. [DOI] [PubMed] [Google Scholar]
- 22.Yan PS, et al. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- 23.Yang YH, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin SY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 25.Offterdinger M, et al. c-erbB-3: a nuclear protein in mammary epithelial cells. J. Cell Biol. 2002;157:929–939. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 27.Li L, et al. PACT is a negative regulator of p53 and essential for cell growth and embryonic development. Proc. Natl Acad. Sci. USA. 2007;104:7951–7956. doi: 10.1073/pnas.0701916104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallucci M, et al. Status of the p53, p16, RB1, and HER-2 genes and chromosomes 3, 7, 9, and 17 in advanced bladder cancer: correlation with adjacent mucosa and pathological parameters. J. Clin. Pathol. 2005;58:367–371. doi: 10.1136/jcp.2004.021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tidow H, et al. Effects of oncogenic mutations and DNA response elements on the binding of p53 to p53-binding protein 2 (p53BP2) J. Biol. Chem. 2006;281:32526–32533. doi: 10.1074/jbc.M604725200. [DOI] [PubMed] [Google Scholar]
- 30.Filosto M, et al. Transcription factors c-Jun/activator protein-1 and nuclear factor-kappa B in oxidative stress response in mitochondrial diseases. Neuropathol. Appl. Neurobiol. 2003;29:52–59. doi: 10.1046/j.1365-2990.2003.00411.x. [DOI] [PubMed] [Google Scholar]
- 31.Chan TA, et al. Targeted inactivation of CTNNB1 reveals unexpected effects of β-catenin mutation. Proc. Natl Acad. Sci. USA. 2002;99:8265–8270. doi: 10.1073/pnas.082240999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staal FJ, et al. Wnt signals are transmitted through N-terminally dephosphorylated β-catenin. EMBO Rep. 2002;3:63–68. doi: 10.1093/embo-reports/kvf002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eferl R, et al. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 34.Wang MH, et al. Macrophage- stimulating protein induces proliferation and migration of murine keratinocytes. Exp. Cell Res. 1996;226:39–46. doi: 10.1006/excr.1996.0200. [DOI] [PubMed] [Google Scholar]
- 35.Camp ER, et al. RON, a tyrosine kinase receptor involved in tumor progression and metastasis. Ann. Surg. Oncol. 2005;12:273–281. doi: 10.1245/ASO.2005.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.