Abstract
Insulin-like growth factor-binding protein (IGFBP)-3 is overexpressed frequently in esophageal squamous cell carcinoma. Yet, the role of IGFBP3 in esophageal tumor biology remains to be elucidated. We find that IGFBP3 facilitates transforming growth factor (TGF)-β1-mediated epithelial-to-mesenchymal transition (EMT) in transformed human esophageal epithelial cells, EPC2–hTERT–EGFR–p53R175H. In organotypic 3D culture, a form of human tissue engineering, laser-capture microdissection revealed concurrent upregulation of TGF-β target genes, IGFBP3 and EMT-related genes in the cells invading into the stromal compartment. IGFBP3 enhanced TGF-β1-mediated EMT as well as transcription factors essential in EMT by allowing persistent SMAD2 and SMAD3 phosphorylation. TGF-β1-mediated EMT and cell invasion were enhanced by ectopically expressed IGFBP3 and suppressed by RNA interference directed against IGFBP3. The IGFBP3 knockdown effect was rescued by IGFBP3I56G/L80G/L81G, a mutant IGFBP3 lacking an insulin-like growth factor (IGF)-binding capacity. Thus, IGFBP3 can regulate TGF-β1-mediated EMT and cell invasion in an IGF or insulin-like growth factor 1 receptor-independent manner. IGFBP3I56G/L80G/L81G also promoted EMT in vivo in a Ras-transformed human esophageal cell line T-TeRas upon xenograft transplantation in nude mice. In aggregate, IGFBP3 may have a novel IGF-binding independent biological function in regulation of TGF-β1-mediated EMT and cell invasion.
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the most aggressive squamous cell carcinomas due to locally advanced states and distant metastasis at the time of presentation, compromising therapy, resulting in poor prognosis (1). Common genetic changes associated with ESCC and its precursor lesions include epidermal growth factor receptor (EGFR) overexpression, tumor suppressor p53 mutations (2–5) and telomerase (TERT) activation (6). The presence of p53 mutations is positively correlated with EGFR overexpression (7). EGFR overexpression and p53 mutations contribute to malignant transformation of human esophageal epithelial cells (8).
Insulin-like growth factor-binding protein (IGFBP)-3 is a major regulator of insulin-like growth factor (IGF)-I or -II among six secretory glycoproteins of the IGFBP family (9,10). While biological activities of IGFBP3 are in part attributed to its ability to neutralize IGFs, preventing insulin-like growth factor 1 receptor (IGF1R) activation, IGF (or IGF1R)-independent IGFBP3 activities have also been documented in vitro and in vivo (9,10). Growth inhibitory or proapoptotic effects of IGFBP3 have been described extensively. However, IGFBP3 may stimulate cell proliferation in a context-dependent manner (9,10).
IGFBP3 is overexpressed frequently in ESCC with concurrent EGFR overexpression (11). While wild-type (WT) IGFBP3 induces G1 cell-cycle arrest by preventing IGF-I from activating IGF1R in non-transformed human esophageal cells, apoptosis is induced by neither WT IGFBP3 nor IGFBP3I56G/L80G/L81G (GGG), a mutant IGFBP3 with a greatly reduced affinity to IGFs (12). Thus, IGFBP3 overexpression alone is not sufficient to trigger apoptosis in culture and that the IGF neutralization effect by IGFBP3 can be compensated by medium components such as insulin that can stimulate IGF1R, but cannot be neutralized by IGFBP3. Interestingly, WT IGFBP3 and IGFBP3I56G/L80G/L81G display contrasting phenotypes in xenograft transplantation models using T-TeRas, an oncogenic Ras-transformed human esophageal cell line (13). While WT IGFBP3 prevented tumor formation by inducing massive apoptosis, indicating the crucial role of IGF1R signaling for tumor cell survival in vivo, IGFBP3I56G/L80G/L81G stimulated tumor growth in nude mice (14). These observations suggest that IGFBP3 may have an IGF/IGF1R-independent role in esophageal tumor progression in response to factors in the tumor microenvironment.
Epithelial-to-mesenchymal transition (EMT) occurs during cancer cell invasion and metastasis (15–17). Transforming growth factor (TGF)-β, a potent EMT inducer present in the tumor microenvironment (18), regulates genes essential in cell–cell adhesion, cell polarity and cell–extracellular matrix interactions through transcription factors such as SNAI1, zinc finger E-box-binding proteins (ZEB) (ZEB1 and ZEB2). In primary ESCC tumor tissues, EMT-related genes are upregulated at the invasive front (19–23). Since TGF-β1 is expressed by tumor as well as stromal cells in ESCC (24), it is conceivable that TGF-β contributes to EMT in ESCC. We have demonstrated recently that EGFR overexpression and p53 mutations lead to enrichment of a subpopulation of human esophageal cells expressing ZEB1 and ZEB2, which promotes EMT in response to TGF-β stimulation (25). Although IGFBP3 has been implicated in TGF-β-mediated cell growth stimulatory or inhibitory effects (9,10), its role in EMT is not known.
Herein, we investigated the role of IGFBP3 in esophageal tumor biology to find its novel IGF/IGF1R-independent functions in modulating TGF-β1-mediated EMT and cell motility.
Materials and methods
Cell lines and treatment
EPC2–hTERT–neo-puro, EPC2–hTERT–EGFR–p53R175H, T-TeRas and their derivatives were grown as described previously (8,13,14,25). Cells were treated with 5 ng/ml of recombinant human TGF-β1 (R&D Systems, Minneapolis, MN) reconstituted in 4 mM HCl containing 0.1% bovine serum albumin. Recombinant human insulin-like growth factor-binding protein-3 (rhIGFBP3) (Upstate Biotechnology) was reconstituted in 10 mM acetic acid and used at the final concentration of 1 μg/ml. Phase contrast images were acquired using a Nikon Eclipse TS100 microscope. Spindle-shaped cells were scored by counting at least 100 cells per high-power field (n = 6) under light microscopy as described previously (25).
Retrovirus and lentivirus-mediated gene transfer
Replication-incompetent retroviruses expressing WT and GGG-mutant IGFBP3 were produced with pBABE-bla vector and infected as described previously (8,12,14). Lentiviral pGIPZ vectors carrying short hairpin RNA (shRNA) directed against human IGFBP3 (V2LHS_111628 and 225584) and a non-silencing scramble control sequence (RHS4346) along with green fluorescent protein (Open Biosystems, Huntsville, AL) were used as described previously (25). Drug selection was done for 7 days with 5 μg/ml of Blasticidin S (Invitrogen, Carlsbad, CA) to establish stable cell lines expressing WT and GGG-IGFBP3. Cells transduced with shRNAs were selected by flow sorting for the GFP brightest cells (top 20%).
Organotypic 3D culture
Stratified squamous epithelia were reconstituted in organotypic 3D culture as described previously (8). In brief, 0.5 × 106 of EPC2–hTERT–EGFR–p53R175H and its derivatives were seeded on top of the collagen/Matrigel matrices containing FEF3 human fetal esophageal fibroblasts and grown in submerged conditions for 4 days. Cultures were then raised to the air–liquid interface for additional 4 days and harvested for hematoxylin and eosin staining or immunofluorescence as well as laser-capture microdissection (LCM). Each organotypic culture experiment was performed in triplicate. Representative images were obtained and quantified by using Adobe Photoshop CS4 extended (Adobe Systems, San Jose, CA).
Laser-capture microdissection
Frozen sections (8 μm) of organotypic cultures were fixed immediately with 75% ethanol, stained with hematoxylin and dehydrated. LCM was done using a Nikon Eclipse TE 2000-5 ultraviolet laser microscope (Molecular Machines and Industries, Glattbrugg, Switzerland). Captured cells were lysed in 50 μl of RNA extraction buffer (Arcturus, Mountain View, CA).
Migration and invasion assays
Cell migration and invasion assays were done using Boyden chambers with the BD Falcon™ FluoroBlok™ 24-Multiwell Insert System (BD Biosciences, Bedford, MA) as described previously (8). In brief, 3 × 104 cells were suspended in Keratinocyte Basal Medium devoid of growth factors (Lonza, Basel, Switzerland) and placed in each insert coated with (for invasion) or without (for migration) Matrigel matrix. Cells were allowed to move toward keratinocyte-serum free medium (Invitrogen) supplemented with 5 μg/ml insulin, 1 ng/ml epidermal growth factor and 40 μg/ml bovine pituitary extract filled in the bottom well for 24 h and labeled with 4 μg/ml Calcein AM dye (Invitrogen) for 1 h prior to detection of the fluorescence excited at 485 nm and emitted at 528 nm. All experiments were performed at least three times in triplicate.
Luciferase assays
Transient transfection was carried out using the Lipofectamine™ LTX and Plus reagents (Invitrogen) according to the manufacturer's instructions. Briefly, 2 × 105 cells were seeded per well in 24-well plates 24 h before transfection. Four hundred nanograms of 3TP-Lux (26) (gift of Dr Rebecca G. Wells) was transfected along with 10 ng of pRL-SV40 Vector (Promega, Madison, WI) to calibrate the variation of transfection efficiencies among wells. Cells were incubated for 48 h before cell lysis. Luciferase activities were determined using the Dual-Luciferase Reporter Assay system (Promega) and the ORION Microplate Luminometer (Berthold Detection Systems USA, Oak Ridge, TN). The mean of firefly luciferase activity was normalized with the co-transfected renilla luciferase activity. Transfection was carried out at least three times.
Xenograft transplantation experiments
Xenograft transplantation experiments were performed as described previously (14). Briefly, 3 × 106 cells of the T-TeRas cell derivatives were suspended in 50% Matrigel and implanted subcutaneously in quadruplicate fashion into the dorsal skin of athymic nu/nu mice (4–6 weeks old) (Charles River Laboratories International, Wilmington, MA). Mice were killed to analyze the tumors morphologically and biochemically. A part of a tumor was used for RNA extraction and the other part of the tumor was fixed in 4% paraformaldehyde and embedded in paraffin.
RNA isolation, complementary DNA synthesis and real-time reverse transcription–polymerase chain reaction
Total RNA from LCM was purified with the Arcturus PicoPure RNA isolation kit (Arcturus), determined by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and further subjected to linear amplification and complementary DNA synthesis using the WT-Ovation™ RNA amplification system (NuGEN Technologies, San Carlos CA). RNA was extracted from monolayer cultures and snap-frozen tissues to synthesize complementary DNA as described previously (11). Each complementary DNA was amplified in triplicate using the Taqman® Universal polymerase chain reaction Master Mix or Power SYBR® Green polymerase chain reaction Master Mix (Applied Biosystems, Foster City, CA) with the TaqMan® Gene Expression Assays (Applied Biosystems) or primers listed in supplementary Table 1 (available at Carcinogenesis Online) and detected in the ABI PRISM® 7000 Sequence Detection System (Applied Biosystems). Relative gene expression was determined based upon corresponding threshold cycle values and normalized to β-actin as an internal control as described previously (11).
Western blot analysis
Cells were lysed with 1× Cell Lysis Buffer (Cell Signaling, Danvers, MA) supplemented with 2 mM phenylmethylsulfonyl fluoride and Complete Mini ethylenediaminetetraacetic acid-free (Roche, Basel, Switzerland). Twenty micrograms of denatured protein was fractionated on a NuPAGE Bis–Tris 4–12% gel (Invitrogen). Following electrotransfer, Immobilon-P membranes (Millipore, Billerica, MA) were incubated with primary antibodies listed in supplementary Table 2 (available at Carcinogenesis Online) and then with the appropriate horseradish peroxidase-conjugated secondary antibody (GE Healthcare, Piscataway, NJ). β-Actin served as loading controls. Western blots were analyzed by densitometry using the Scion Images software (Scion, Frederick, MD).
Immunofluorescence
Deparaffinized sections were blocked with 1% bovine serum albumin for 30 min. Slides were incubated with mouse anti-pancytokeratin (CK) (1:3000; Sigma, St Louis, MO), anti-fibroblast-specific protein 1 (FSP1) (1:1500; DAKO Cytomation, Glostrup, Denmark), anti-SV40 T-antigen (T-Ag) (1:1500; Santa Cruz Biotechnology, Santa Cruz, CA) or anti-E-cadherin (1:200; BD Biosciences, San Jose, CA) overnight at 4°C and then with an appropriate Cy2- or Cy3-conjugated secondary antibody (1:400; Jackson ImmunoResearch, West Grove, PA) for 30 min at room temperature. Nuclei were counterstained by 4′,6-diamidino-2-phenylindole (1:10 000; Invitrogen). Stained objects were examined with a Nikon Microphot microscope and imaged with a digital camera.
Statistical analyses
Data from triplicate and hexaduplicate experiments in real-time reverse transcription–polymerase chain reaction, Boyden chamber assays, luciferase assays, densitometry and scoring of spindle-shaped cells in photomicrographs were presented as mean ± SE and were analyzed by two-tailed Student's t-test. P <0.05 was considered significant.
Results
IGFBP3 is induced as a TGF-β-signaling modulator during invasive growth of EMT-competent-transformed human esophageal cells
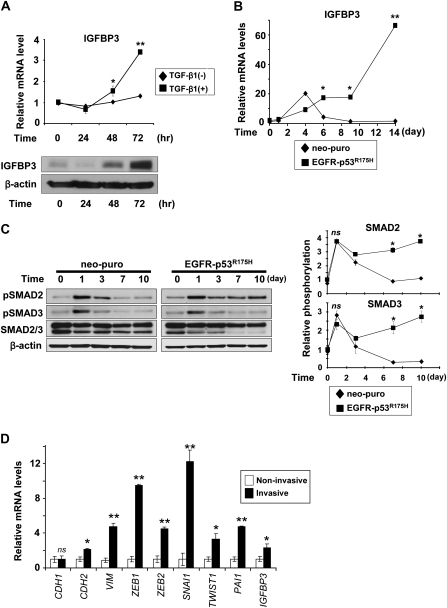
EGFR oncogene and mutant p53 co-operate to enrich a unique subset of transformed human esophageal epithelial cells designated EPC2–hTERT–EGFR–p53R175H, which are highly capable of undergoing EMT in response to TGF-β stimulation (25). As observed in a variety of cell types (27–30), TGF-β induced IGFBP3 in EPC2–hTERT–EGFR–p53R175H cells as a function of time (Figure 1A and B). Interestingly, IGFBP3 was induced only transiently in immortalized, yet non-transformed human esophageal cells EPC2–hTERT–neo-puro (Figure 1B), which underwent EMT only minimally (<10%) upon TGF-β treatment (25). IGFBP3 modulates TGF-β signaling (31,32). TGF-β receptor activation, indicated by SMAD2 and SMAD3 phosphorylation persisted in EPC2–hTERT–EGFR–p53R175H, but not control cells (Figure 1C). Interestingly, phosphorylation of both SMAD2 and SMAD3 was sustained longer in EPC2–hTERT–EGFR–p53R175H cells than control cells (Figure 1C). Moreover, rhIGFBP3 prolonged TGF-β-induced SMAD2/3, but not p38 phosphorylation (supplementary Figure 1 is available at Carcinogenesis Online), raising the possibility that IGFBP3 may contribute to EMT by enhancing TGF-β signaling.
Fig. 1.
TGF-β induction of IGFBP3 in EMT-competent-transformed human esophageal cells is associated with induction of EMT-related genes in the engineered tissue microenvironment in organotypic 3D culture. Cells were stimulated with TGF-β1 for indicated time periods (A–C). (A) IGFBP3 mRNA (upper panel) and protein (lower panel) were determined in EPC2–hTERT–EGFR–p53R175H cells by real-time reverse transcription–polymerase chain reaction and western blotting, respectively. In the upper panel, cells untreated with TGF-β1 served as a control. **P < 0.001 and *P < 0.05 versus TGF-β1 (−) at indicated time point (n = 3). (B) IGFBP3 mRNA levels were determined by real-time reverse transcription–polymerase chain reaction in either EMT-competent (EGFR-p53R175H) or EMT-incompetent (neo-puro) EPC2–hTERT cell derivative at indicated time point. **P < 0.001 and *P < 0.01 versus neo-puro (n = 3). (C) SMAD2/3 phosphorylation was determined in (B). pSmad2, phospho-Smad2 (Ser465/467) and pSmad3, phospho-Smad3 (Ser423/425). Diagrams (right panel) represent densitometry of the western blot (left panel). *P < 0.01 versus neo-puro; ns, not significant versus neo-puro (n = 3). (D) Relative mRNA for IGFBP3, TGF-β target and EMT-related genes was determined by real-time reverse transcription–polymerase chain reaction in the invasive EPC2–hTERT–EGFR–p53R175H cells comparing with the non-invasive EPC2–hTERT–EGFR–p53R175H cells isolated by LCM as shown in supplementary Figure 2B, available at Carcinogenesis Online. **P < 0.001; *P < 0.01; ns, not significant versus non-invasive (n = 3).
EPC2–hTERT–EGFR–p53R175H, but not control cells, exhibited invasive growth in organotypic 3D culture, a form of human tissue engineering (supplementary Figure 2A is available at Carcinogenesis Online) as described previously (8). We suspected that IGFBP3 may be involved in tumor cell invasion through regulation of TGF-β signaling. LCM and real-time reverse transcription–polymerase chain reaction revealed upregulation of Plasminogen activator inhibitor type 1, a TGF-β target gene, IGFBP3, EMT markers and transcription factors essential in EMT such as SNAI1, ZEB1 and ZEB2 in EPC2–hTERT–EGFR–p53R175H cells invading into the stromal compartment (Figure 1D and supplementary Figure 2B, available at Carcinogenesis Online). Although E-cadherin (CDH1) messenger RNA (mRNA) was not downregulated at the mRNA level (Figure 1D), E-cadherin protein was mislocalized from the cell membrane to the cytoplasm in the invasive cells (supplementary Figure 2C, available at Carcinogenesis Online), indicating that an E-cadherin protein recycling/degradation may occur prior to transcriptional repression at the onset of EMT as described previously (33). These observations prompted us to hypothesize that IGFBP3 may have a unique role in the TGF-β-mediated EMT program, allowing invasive growth of EPC2–hTERT–EGFR–p53R175H cells.
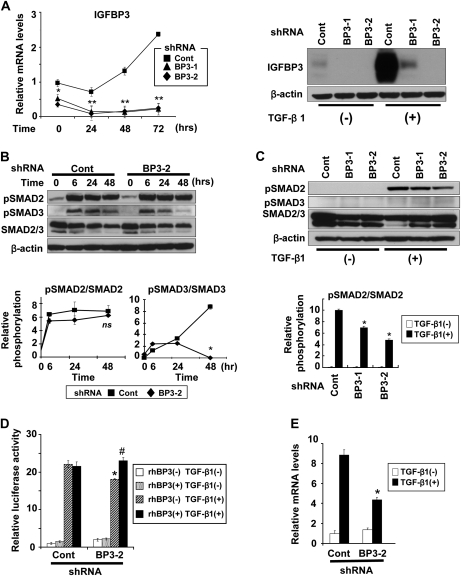
To address the roles of IGFBP3 in TGF-β-mediated EMT and invasive growth of EPC2–hTERT–EGFR–p53R175H cells, we conducted RNA interference (RNAi) experiments. Stable expression of two independent shRNA sequences directed against IGFBP3 prevented TGF-β from inducing IGFBP3 (Figure 2A). IGFBP3 knockdown did not affect acutely induced SMAD2 phosphorylation within the first 24–48 h during TGF-β stimulation (Figure 2B). After a long-term TGF-β stimulation for 14 days, however, IGFBP3 knockdown reduced the relative SMAD2 phosphorylation by 30–50% (Figure 2C). Interestingly, IGFBP3 knockdown suppressed SMAD3 phosphorylation within 48 h after TGF-β stimulation (Figure 2B). SMAD3 phosphorylation was barely detectable after a long-term TGF-β stimulation (Figure 2C). Moreover, total SMAD3 protein level was downregulated upon TGF-β stimulation and that was antagonized partially by IGFBP3 knockdown (Figure 2C and supplementary Figure 3, available at Carcinogenesis Online). Similar trend was observed for total SMAD2, but to a lesser extent (Figure 2C and supplementary Figure 3, available at Carcinogenesis Online). Such observations may be accounted for by the reduced SMAD activities upon IGFBP3 knockdown since TGF-β signaling is terminated by proteasomal degradation for SMAD proteins (34).
Fig. 2.
IGFBP3 knockdown suppresses TGF-β signaling in EPC2–hTERT–EGFR–p53R175H cells. Cells were transduced stably with shRNA directed against IGFBP3 (BP3-1 and BP3-2) or a scrambled control shRNA (Cont). Cells were stimulated with TGF-β1 for 14 days in (A) (right panel), (C) and (E) and for 48 h in (D), unless indicated otherwise. pSmad2, phospho-Smad2 and pSmad3, phospho-Smad3 in (B) and (C). (A) IGFBP3 mRNA (left) and protein (right) were determined by real-time reverse transcription–polymerase chain reaction upon IGFBP3 knockdown. **P < 0.01; *P < 0.05 versus Cont (n = 3). (B) SMAD2/3 phosphorylation in (A) was determined by western blotting. The relative phosphorylation levels for SMAD2 and SMAD3 (lower panel) represent densitometry of the western blots. *P < 0.01 versus Cont; ns, not significant versus Cont (n = 3). (C) Western blotting was done with or without TGF-β1 stimulation for 14 days to determine the long-term impact of IGFBP3 silencing upon SMAD2/3 expression and phosphorylation. Histograms represent the relative phosphorylation levels for SMAD2/3 (lower left panel) and total SMAD2/3 contents (lower right panel) determined by densitometry of the western blots. *P < 0.01 versus Cont with TGF-β1 (+) (n = 3). (D) TGF-β reporter activity was determined by luciferase assays with 3TP-Lux. Indicated reagents were added 5 h after transfection. rhBP3, rhIGFBP3; *P < 0.05 versus TGF-β1 (+) and rhBP3 (−) in Cont; #P < 0.05 versus TGF-β1 (+) and rhBP3 (−) in BP3-2 (n = 6). (E) plasminogen activator inhibitor type 1 mRNA was determined by real-time reverse transcription–polymerase chain reaction. *P < 0.01 versus TGF-β1 (+) in Cont (n = 3).
IGFBP3 knockdown effects were observed in the TGF-β-mediated transcriptional activity (Figure 2D) as well as plasminogen activator inhibitor type 1 mRNA induction (Figure 2E). Although IGFBP3 silencing appeared to prevent TGF-β reporter activation only to a limited extent, such an inhibitory effect of IGFBP3 shRNA was antagonized by recombinant IGFBP3 protein added into culture medium (Figure 2D), implying an IGFBP3-mediated effect upon TGF-β signaling. Thus, IGFBP3 appeared to modulate TGF-β signaling, if not to a full extent, in EPC2–hTERT–EGFR–p53R175H cells.
IGFBP3 promotes EMT and invasive growth of transformed human esophageal cells in an IGF-independent fashion
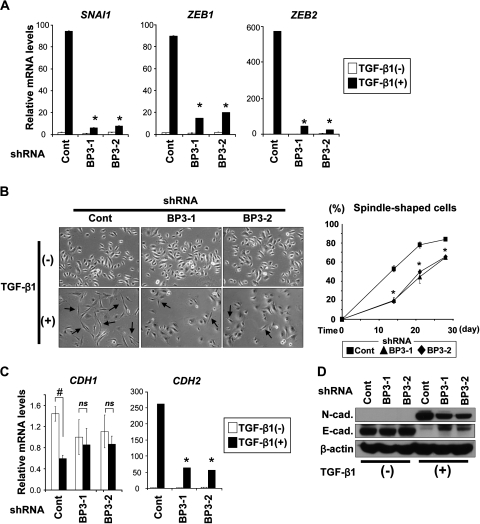
TGF-β induced SNAI1, ZEB1 and ZEB2 in EPC2–hTERT–EGFR–p53R175H cells, where a characteristic spindle-shaped cell morphology emerged, displaying loss of E-cadherin and gain of mesenchymal markers such as N-cadherin and vimentin (25). IGFBP3 knockdown delayed significantly TGF-β induction of SAI1, ZEB1 and ZEB2 (Figure 3A) as well as spindle-shaped cells (Figure 3B). Moreover, a class switch from E-cadherin-to-N-cadherin was suppressed partially in the IGFBP3 knockdown cells (Figure 3C and D). These findings indicate that IGFBP3 may promote EMT by stimulating TGF-β induction of the transcription factors essential in EMT.
Fig. 3.
IGFBP3 knockdown delays TGF-β-mediated EMT in EPC2–hTERT–EGFR–p53R175H cells. Cells expressing shRNA directed against IGFBP3 (BP3-1 and BP3-2) or a scrambled control shRNA (Cont) were stimulated with TGF-β1 for 14 days in (A), (B, left panel), (C) and (D), unless indicated otherwise. (A) mRNA for SNAI1, ZEB1 and ZEB2 was determined by real-time reverse transcription–polymerase chain reaction. *P < 0.01 versus Cont plus TGF-β1 (+) (n = 3). (B) Phase contrast photomicrograph was taken (left). Arrows indicate spindle-shaped cells suggesting EMT as described previously (25); magnification, ×200. Spindle-shaped cells were scored at the indicated time points (right). *P < 0.05 versus Cont (n = 6). (C) mRNA for E-cadherin (CDH1) and N-cadherin (CDH2) was determined by real-time reverse transcription–polymerase chain reaction. #P < 0.01 TGF-β1 (−); ns, not significant versus TGF-β1 (−); *P < 0.01 TGF-β1 (+) (n = 3). (D) E-cadherin (E-cad.) and N-cadherin (N-cad.) proteins were determined by western blotting.
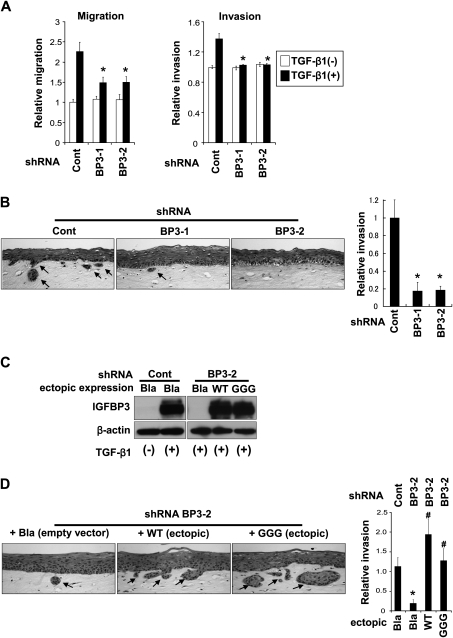
EGFR overexpression and p53 mutations confer co-operatively invasive characteristics to EPC2–hTERT cells as determined by the conventional Boyden chamber assays as well as organotypic 3D culture (8). Since EMT increases cellular motility, we first examined the effect of IGFBP3 knockdown upon cell migration and invasion by the Boyden chamber assays. Although a small subset of EPC2–hTERT–EGFR–p53R175H cells undergo EMT spontaneously, the vast majority of them does not without TGF-β stimulation (25). In agreement, RNAi affected neither migration nor invasion in the cells untreated with TGF-β (Figure 4A). TGF-β stimulated cell migration (∼2-fold) and invasion (∼1.3-fold) in scrambled shRNA-transduced control cells (Figure 4A) compared with those without TGF-β treatment. IGFBP3 knockdown reduced the TGF-β-stimulated cell migration by 30% (Figure 4A, left panel). In addition, TGF-β-mediated enhancement of invasion was cancelled upon IGFBP3 knockdown (Figure 4A, right panel). Moreover, IGFBP3 knockdown impaired invasion of EPC2–hTERT–EGFR–p53R175H cells in organotypic 3D culture (Figure 4B). These observations suggest that IGFBP3 may stimulate motility of transformed human esophageal cells by enhancing TGF-β-mediated EMT.
Fig. 4.
IGFBP3 knockdown impaired EMT program to prevent cellular motility. EPC2–hTERT–EGFR–p53R175H cells expressing shRNA directed against IGFBP3 (BP3-1 and BP3-2) or a scrambled control shRNA (Cont) were subjected to Boyden chamber assays (A) or organotypic 3D culture (B and D). (A) Cells were stimulated with or without TGF-β1 for 14 days prior to the Boyden chamber assays. *P < 0.01 versus Cont plus TGF-β1 (+) (n = 3). (B) Cells were grown in organotypic 3D culture in the absence of exogenous TGF-β. Hematoxylin and eosin staining visualized the reconstituted stratified squamous epithelia and cells displaying downward invasive growth into the stromal compartment. The invasive area, indicated by arrows, was measured and represented as a histogram; magnification, ×200. *P < 0.01 versus Cont (n = 3). (C) Western blotting validates that IGFBP3 expression was restored by ectopic WT or GGG-mutant IGFBP3 in the cells expressing IGFBP3 shRNA. Note that empty vector (Bla) failed to antagonize the IGFBP3 shRNA effect. (D) Cells in (C) were grown in organotypic 3D culture and analyzed as in (B); magnification, ×200. *P < 0.01 versus Cont plus Bla; #P < 0.01 versus BP3-2 plus Bla (n = 3).
In organotypic 3D culture, not every single basal keratinocyte invades into the stromal compartment (supplementary Figure 2, available at Carcinogenesis Online and Figure 4B), implying cellular heterogeneity in the invasive capability. IGFBP3 does not have either antiproliferative or growth stimulatory effect in non-transformed and transformed human esophageal cells in culture (12,14). However, there is the possibility that RNAi directed against IGFBP3 has eliminated a subset of invasive cells that depend upon IGFBP3 for survival, resulting in reduced tumor cell invasion (Figure 4A and B). We addressed this possibility by restoring IGFBP3 expression in EPC2–hTERT–EGFR–p53R175H cells expressing an shRNA sequence targeting endogenous IGFBP3 mRNA at the 3′ untranslated region. Such shRNA does not affect ectopically expressed IGFBP3 transduced by retrovirus carrying its open reading frame only. Cells were stably transduced with either WT or GGG-IGFBP3. GGG-mutant IGFBP3 is used to interrogate the IGF (or IGF1R)-independent biological activities of IGFBP3 (12). While endogenous IGFBP3 was silenced, we detected ectopic IGFBP3 at the mRNA and protein levels (supplementary Figure 4, available at Carcinogenesis Online, and Figure 4C). In organotypic 3D culture, both WT and GGG-mutant IGFBP3 restored the invasiveness of EPC2–hTERT–EGFR–p53R175H cells (Figure 4D), indicating a direct regulation of tumor cell invasiveness by IGFBP3 in 3D culture.
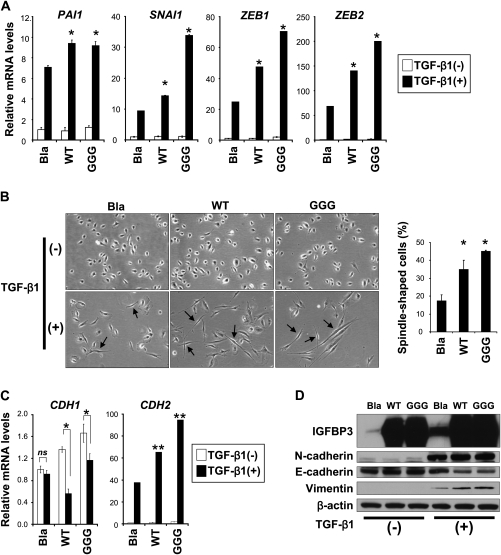
We next assessed how IGFBP3 overexpression may impact upon TGF-β-mediated EMT and invasive growth of EPC2–hTERT–EGFR–p53R175H cells. Ectopically expressed IGFBP3 boosted TGF-β-stimulated SMAD2/3 phosphorylation modestly (i.e. a 10% increase by densitometry) (data not shown). Moreover, both WT and GGG-mutant IGFBP3 accelerated TGF-β induction of plasminogen activator inhibitor type 1 as well as SANI1, ZEB1 and ZEB2 (Figure 5A) and spindle-cell shape morphology (Figure 5B), accompanied by cadherin class switch and vimentin induction (Figure 5C and D) within 7 days after TGF-β1 stimulation. Thus, ectopically expressed IGFBP3 facilitated TGF-β1-mediated EMT initially. However, such an effect became less overt by 14 days after TGF-β stimulation (data not shown) as endogenous IGFBP3 was induced robustly as a function of time (Figure 1B and data not shown). In organotypic 3D culture, both WT and GGG-mutant IGFBP3 enhanced invasion of EPC2–hTERT–EGFR–p53R175H cells (supplementary Figure 5, available at Carcinogenesis Online). These data suggest that IGFBP3 may promote TGF-β1-mediated EMT and invasion in an IGF (or IGF1R)-independent manner.
Fig. 5.
IGFBP3 overexpression stimulates TGF-β signaling to promote EMT and cell invasion. EPC2–hTERT–EGFR–p53R175H cells were stably transduced with WT or GGG-mutant IGFBP3 or an empty control vector (Bla). Cells were stimulated with or without TGF-β1 for 7 days in (A–D). (A) mRNA for indicated genes was determined by real-time reverse transcription–polymerase chain reaction. *P < 0.01 versus Bla plus TGF-β1 (+) (n = 3). (B) Phase contrast photomicrograph was taken. Arrows indicate spindle-shaped cells suggesting EMT; magnification, ×200. Spindle-shaped cells were scored. *P < 0.01 versus Bla (n = 6). (C) mRNAs for CDH1 and CDH2 was determined by real-time reverse transcription–polymerase chain reaction. ns, not significant versus Bla plus TGF-β1 (−); *P < 0.05 versus Bla plus TGF-β1 (−); **P < 0.01 versus Bla plus TGF-β1 (+) (n = 3). (D) Western blotting determines IGFBP3 and EMT marker proteins.
IGFBP3 promotes EMT in vivo in Ras-transformed esophageal cell tumors
Exploring the biological activities of IGFBP3 in vivo, we used T-TeRas cells in a xenograft transplantation model in athymic nude mice since we have investigated previously the functional consequence of IGFBP3 overexpression in T-TeRas cells in vitro and in vivo (14). WT IGFBP3 prevented tumor formation by triggering a massive apoptosis, indicating an IGF (or IGF1R)-dependent growth inhibitory effect of IGFBP3. In contrast, an empty vector (Bla) allowed tumor formation, overcoming modest apoptosis elicited by endogenous IGFBP3. Interestingly, GGG-IGFBP3 did not induce apoptosis, and instead stimulated tumor growth, implying IGF-independent biological activities (14). GGG-IGFBP3-expressing tumors appeared to consist of both spindle-shaped tumor cells and epithelioid cells (supplementary Figure 6, available at Carcinogenesis Online), prompting us to hypothesize that IGFBP3 may have a role promoting EMT in vivo.
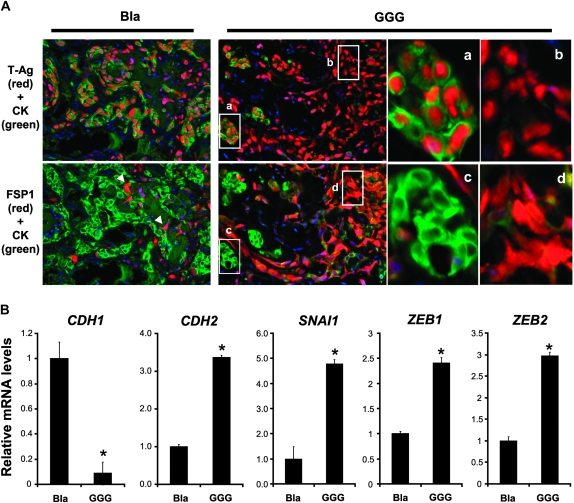
We performed immunofluorescence to further characterize the xenografted T-TeRas cell derivatives. FSP1 and CK were used as mesenchymal and epithelial markers, respectively. Injected tumor cells were confirmed by SV40 T-Ag, whereas mouse-derived stromal cells were double negative for T-Ag and CK (Figure 6A). In empty vector (Bla) control tumors, most of the cells were found to be T-Ag positive and CK bright, but negative for FSP1, corroborating their epithelial nature (Figure 6A). Consistent with hematoxylin and eosin staining results (supplementary Figure 6, available at Carcinogenesis Online), GGG-IGFBP3-expressing tumors were represented predominantly by T-Ag positive, FSP1 positive and CK dim spindle-shaped cells (60–70%) among all T-Ag-positive cells (Figure 6A). Moreover, GGG-IGFBP3-expressing T-TeRas tumors displayed downregulation of E-cadherin and upregulation of N-cadherin as well as SNAI1, ZEB1 and ZEB2 (Figure 6B). These results indicate that IGFBP3 may have an IGF-independent EMT-promoting effect upon Ras-transformed human esophageal cells in vivo.
Fig. 6.
IGFBP3 overexpression promotes EMT in xenografted esophageal tumors. T-TeRas tumors stably expressing GGG-mutant IGFBP3 or an empty control vector (Bla) were harvested at 3 weeks after subcutaneous injection into nude mice. Cells expressing WT IGFBP3, failing to form tumors due to massive apoptosis as described previously (14), was excluded from the analysis. (A) Tumor tissues were stained by immunofluorescence for CK (green) along with either T-Ag (red) or FSP1 (red). Note that the tumor tissues contain CK bright [demarcated area was enlarged in (a) and (c)] and CK dim or negative cells [arrow heads and enlarged in (b) and (d)]. Nuclear T-Ag was detected in both CK bright and dim or negative cells, but not in host-derived stromal cells stained blue only by 4′,6-diamidino-2-phenylindole. (B) mRNA for indicated genes was determined by real-time reverse transcription–polymerase chain reaction in the tumors. *P < 0.01 versus Bla (n = 3).
Collectively, our data indicate that IGFBP3 may have novel IGF-binding-independent biological functions to facilitate EMT and cell motility through modulation of TGF-β signaling in transformed human esophageal cells.
Discussion
This study reveals a novel unique modulatory role of IGFBP3 in TGF-β-mediated EMT and cell invasion in transformed human esophageal cells. IGFBP3 was induced by TGF-β, which was associated with persistent activation of SMAD2/3 in EMT-competent EPC2–hTERT–EGFR–p53R175H cells during TGF-β stimulation (Figure 1A–C). IGFBP3 was found also to be upregulated in the invasive cells expressing EMT-regulated genes and TGF-β target genes in reconstituted human esophageal tissues (Figure 1D). RNAi experiments documented modulation of TGF-β signaling by IGFBP3 (Figure 2), where IGFBP3 appeared to be essential in induction of transcription factors SNAI1, ZEB1 and ZEB2 and delayed EMT significantly (Figure 3). Moreover, lack of IGFBP3 induction impaired cell motility (Figure 4). In contrast, ectopically expressed IGFBP3 stimulated TGF-β target genes, facilitating TGF-β1-mediated EMT and invasion in organotypic 3D culture (Figure 5 and supplementary Figure 5, available at Carcinogenesis Online). Importantly, GGG-mutant IGFBP3 restored the IGFBP3 knockdown effects (Figure 4) as well as enhanced TGF-β signaling and its downstream events (Figure 5), implying IGF-independent biological activities of IGFBP3. Finally, such IGF-independent effects were observed in xenografted tumors of T-TeRas cells (Figure 6), providing an in vivo context.
IGFBP3 is often silenced epigenetically in many tumor types (10). In Ewing sarcoma, the EWS/FLI-1 fusion gene targets IGFBP3 for transcriptional repression (35). Thus, inactivation of the proapoptotic and antiproliferative functions of IGFBP3 is important in cancer biology. Nonetheless, IGFBP3 is overexpressed frequently in esophageal cancers (11,36). In addition to TGF-β, as implied in this study, the hypoxic tumor microenvironment is thought to play an essential role in IGFBP3 induction (13,14,36). T-TeRas tumors grow by overcoming the proapoptotic effects of endogenously induced IGFBP3, implicating possible adaptation to the microenvironment (14). Moreover, IGFBP3 has an IGF-independent growth stimulatory effect in T-TeRas tumors (14). Thus, IGFBP3 may display differential biological functions in the early and late stages of carcinogenesis (37,38). IGFBP3 has dichotomous effects in cell proliferation in T47D breast cancer cells, where cell proliferation was suppressed initially by IGFBP3, yet became refractory to its growth inhibition in culture, resulting in tumor formation in nude mice with enhanced EGFR signaling (39,40). However, little is known about the role of IGFBP3 in EMT and cell motility.
EMT as a functional target for IGFBP3 through regulation of TGF-β signaling is a novel finding based upon our studies. IGFBP3 stimulates TGF-β receptors directly in certain cell types (31,32). TGF-β is expressed by EPC2–hTERT–EGFR–p53R175H cells as enzyme-linked immunosorbent assay detected a low level (100–200 pg/ml) of TGF-β1 in the culture supernatant (data not shown). However, rhIGFBP3 alone failed to enhance TGF-β receptor signaling in EPC2–hTERT–EGFR–p53R175H cells (supplementary Figure 1, available at Carcinogenesis Online), whereas rhIGFBP3 or ectopically expressed IGFBP3-prolonged SMAD2/3 phosphorylation upon TGF-β stimulation (supplementary Figure 1, available at Carcinogenesis Online and data not shown). Moreover, ectopically expressed IGFBP3 did not stimulate EMT without simultaneous TGF-β treatment (Figure 5). The impact of RNAi upon SMAD2/3 phosphorylation and the TGF-β reporter activity was partial despite 80–90% IGFBP3 silencing efficiency (Figure 2). These observations suggest that IGFBP3 may not necessarily be indispensible in TGF-β signaling but serve as an essential modifier in esophageal cancer cells.
IGFBP3 shows an inhibitory effect upon cell motility in prostate and ovarian carcinoma as well as Ewing sarcoma cells, where IGFBP3 also inhibits cell proliferation (41–43). In Hs578T breast cancer cells, however, IGFBP3 exerts promigratory functions in an IGF-independent fashion (44). IGFBP3 appears to play an essential role in cutaneous melanoma cell motility and proliferation promoting metastasis (45). IGFBP3 also mediates endothelial progenitor cell migration during capillary tube formation in angiogenesis (46). Thus, IGFBP3 can stimulate cell motility in several cell types other than transformed human esophageal cells. Our data indicate also that IGFBP3 requires factors (e.g. TGF-β) in the tumor microenvironment to exert its biological activity stimulating cell motility as implicated in organotypic 3D culture, where TGF-β target genes were induced along with IGFBP3 in invasive EPC2–hTERT–EGFR–p53R175H cells (Figure 1D). In fact, IGFBP3 silencing alone did not affect cell invasion, whereas TGF-β-mediated effect was impaired in Boyden chamber assays (Figure 4A). IGFBP3 interacts physically with numerous extracellular matrix components including type I α-collagen, fibronectin, vitronectin and latent TGF-β-binding protein-1, all pertinent to cellular motility (9,47–50), suggesting that molecular interactions between IGFBP3 and these factors may influence TGF-β signaling. For example, fibronectin regulates TGF-β via latent TGF-β-binding protein-1 interactions (51).
In conclusion, our unique model systems demonstrate that IGFBP3 plays a critical role in esophageal tumor progression by facilitating EMT and invasive growth of transformed human esophageal cells in concert with the TGF-β pathway in the context of the tumor microenvironment.
Supplementary material
Supplementary Tables 1 and 2 and Figures 1–6 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (NIH) to MN, SO, HN (R01DK077005), HN, MN, SO, GSW, DB, AJPK, MH, JAD (P01-CA-098101), HN (K01-DK-066205), GSW (NIH/NCI T32-CA115299), RK, AA (P30-DK050306); NIH/National Institute of Diabetes and Digestive and Kidney Diseases Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306).
Supplementary Material
Acknowledgments
We thank Dr Charles H.Pletcher (Flow Cytometry & Cell Sorting Facility), Dr Gary P.Swain (Morphology Core), Dr Gary D.Wu and Dr Sue Keilbaugh (Molecular Biology/Gene Expression Core) and Dr Richard Carroll (Cell Culture Core). We thank Ben Rhoades for technical assistance, members of the Nakagawa and Rustgi labs and Claudia D.Andl (Vanderbilt University) for helpful discussions as well as Dr Rebecca G. Wells for reagents. Finally, we are grateful to Anil K.Rustgi for his support and advice and critical review of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- EGFR
epidermal growth factor receptor
- EMT
epithelial-to-mesenchymal transition
- ESCC
esophageal squamous cell carcinoma
- FSP1
fibroblast-specific protein 1
- IGF
insulin-like growth factor
- IGFBP
insulin-like growth factor-binding protein
- IGF1R
insulin-like growth factor 1 receptor
- LCM
laser-capture microdissection
- mRNA
messenger RNA
- CK
pancytokeratin
- rhIGFBP3
recombinant human insulin-like growth factor-binding protein-3
- RNAi
RNA interference
- shRNA
short hairpin RNA
- T-Ag
T-antigen
- TGF
transforming growth factor
- WT
wild-type
- ZEB
zinc finger E-box-binding proteins
References
- 1.Enzinger PC, et al. Esophageal cancer. N. Engl. J. Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Nakagawa H, et al. Biology of esophageal cancer. In: Rustgi AK, editor. Gastrointestinal Cancers. London: Elsevier; 2003. pp. 241–251. [Google Scholar]
- 3.Itakura Y, et al. Epidermal growth factor receptor overexpression in esophageal carcinoma. An immunohistochemical study correlated with clinicopathologic findings and DNA amplification. Cancer. 1994;74:795–804. doi: 10.1002/1097-0142(19940801)74:3<795::aid-cncr2820740303>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 4.Parenti AR, et al. p53 overexpression in the multistep process of esophageal carcinogenesis. Am. J. Surg. Pathol. 1995;19:1418–1422. doi: 10.1097/00000478-199512000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Volant A, et al. p53 protein accumulation in oesophageal squamous cell carcinomas and precancerous lesions. J. Clin. Pathol. 1995;48:531–534. doi: 10.1136/jcp.48.6.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiyama T, et al. Overexpression of human telomerase RNA is an early event in oesophageal carcinogenesis. Virchows Arch. 1999;434:483–487. doi: 10.1007/s004280050372. [DOI] [PubMed] [Google Scholar]
- 7.Esteve A, et al. Correlation of p53 mutations with epidermal growth factor receptor overexpression and absence of mdm2 amplification in human esophageal carcinomas. Mol. Carcinog. 1993;8:306–311. doi: 10.1002/mc.2940080414. [DOI] [PubMed] [Google Scholar]
- 8.Okawa T, et al. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21:2788–2803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firth SM, et al. Cellular actions of the insulin-like growth factor binding proteins. Endocr. Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 10.Jogie-Brahim S, et al. Unraveling insulin-like growth factor binding protein-3 actions in human disease. Endocr. Rev. 2009;30:417–437. doi: 10.1210/er.2008-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takaoka M, et al. Epidermal growth factor receptor regulates aberrant expression of insulin-like growth factor-binding protein 3. Cancer Res. 2004;64:7711–7723. doi: 10.1158/0008-5472.CAN-04-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takaoka M, et al. EGF-mediated regulation of IGFBP-3 determines esophageal epithelial cellular response to IGF-I. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G404–G416. doi: 10.1152/ajpgi.00344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S-H, et al. Tumorigenic conversion of primary human esophageal epithelial cells using oncogene combinations in the absence of exogenous Ras. Cancer Res. 2006;66:10415–10424. doi: 10.1158/0008-5472.CAN-06-2104. [DOI] [PubMed] [Google Scholar]
- 14.Takaoka M, et al. IGFBP-3 regulates esophageal tumor growth through IGF-dependent and independent mechanisms. Cancer Biol. Ther. 2007;6:534–540. doi: 10.4161/cbt.6.4.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalluri R, et al. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peinado H, et al. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 17.Thiery JP, et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Zavadil J, et al. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 19.Yuen HF, et al. Upregulation of Twist in oesophageal squamous cell carcinoma is associated with neoplastic transformation and distant metastasis. J. Clin. Pathol. 2007;60:510–514. doi: 10.1136/jcp.2006.039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Usami Y, et al. Snail-associated epithelial-mesenchymal transition promotes oesophageal squamous cell carcinoma motility and progression. J. Pathol. 2008;215:330–339. doi: 10.1002/path.2365. [DOI] [PubMed] [Google Scholar]
- 21.Uchikado Y, et al. Slug expression in the E-cadherin preserved tumors is related to prognosis in patients with esophageal squamous cell carcinoma. Clin. Cancer Res. 2005;11:1174–1180. [PubMed] [Google Scholar]
- 22.Natsugoe S, et al. Snail plays a key role in E-cadherin-preserved esophageal squamous cell carcinoma. Oncol. Rep. 2007;17:517–523. [PubMed] [Google Scholar]
- 23.Ansieau S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Natsugoe S, et al. Smad4 and transforming growth factor beta1 expression in patients with squamous cell carcinoma of the esophagus. Clin. Cancer Res. 2002;8:1838–1842. [PubMed] [Google Scholar]
- 25.Ohashi S, et al. EGFR and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70:4174–4184. doi: 10.1158/0008-5472.CAN-09-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrana JL, et al. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 27.Gucev ZS, et al. Insulin-like growth factor binding protein 3 mediates retinoic acid- and transforming growth factor beta2-induced growth inhibition in human breast cancer cells. Cancer Res. 1996;56:1545–1550. [PubMed] [Google Scholar]
- 28.Huang SS, et al. TGF-beta control of cell proliferation. J. Cell. Biochem. 2005;96:447–462. doi: 10.1002/jcb.20558. [DOI] [PubMed] [Google Scholar]
- 29.Martin JL, et al. Transforming growth factor-beta stimulates production of insulin-like growth factor-binding protein-3 by human skin fibroblasts. Endocrinology. 1991;128:1425–1433. doi: 10.1210/endo-128-3-1425. [DOI] [PubMed] [Google Scholar]
- 30.Rajah R, et al. Insulin-like growth factor (IGF)-binding protein-3 induces apoptosis and mediates the effects of transforming growth factor-beta1 on programmed cell death through a p53- and IGF-independent mechanism. J. Biol. Chem. 1997;272:12181–12188. doi: 10.1074/jbc.272.18.12181. [DOI] [PubMed] [Google Scholar]
- 31.Fanayan S, et al. Growth inhibition by insulin-like growth factor-binding protein-3 in T47D breast cancer cells requires transforming growth factor-beta (TGF-beta) and the type II TGF-beta receptor. J. Biol. Chem. 2000;275:39146–39151. doi: 10.1074/jbc.M006964200. [DOI] [PubMed] [Google Scholar]
- 32.Fanayan S, et al. Signaling through the Smad pathway by insulin-like growth factor-binding protein-3 in breast cancer cells. Relationship to transforming growth factor-beta 1 signaling. J. Biol. Chem. 2002;277:7255–7261. doi: 10.1074/jbc.M108038200. [DOI] [PubMed] [Google Scholar]
- 33.Janda E, et al. Raf plus TGFbeta-dependent EMT is initiated by endocytosis and lysosomal degradation of E-cadherin. Oncogene. 2006;25:7117–7130. doi: 10.1038/sj.onc.1209701. [DOI] [PubMed] [Google Scholar]
- 34.Heldin CH, et al. SMAD destruction turns off signalling. Nat. Cell Biol. 1999;1:E195–E197. doi: 10.1038/70223. [DOI] [PubMed] [Google Scholar]
- 35.Prieur A, et al. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol. Cell. Biol. 2004;24:7275–7283. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JJ, et al. Hypoxia activates the cyclooxygenase-2-prostaglandin E synthase axis. Carcinogenesis. 2010;31:427–434. doi: 10.1093/carcin/bgp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silha JV, et al. Insulin-like growth factor (IGF) binding protein-3 attenuates prostate tumor growth by IGF-dependent and IGF-independent mechanisms. Endocrinology. 2006;147:2112–2121. doi: 10.1210/en.2005-1270. [DOI] [PubMed] [Google Scholar]
- 38.Cohen P. Insulin-like growth factor binding protein-3: insulin-like growth factor independence comes of age. Endocrinology. 2006;147:2109–2111. doi: 10.1210/en.2006-0195. [DOI] [PubMed] [Google Scholar]
- 39.Firth SM, et al. Development of resistance to insulin-like growth factor binding protein-3 in transfected T47D breast cancer cells. Biochem. Biophys. Res. Commun. 1998;246:325–329. doi: 10.1006/bbrc.1998.8615. [DOI] [PubMed] [Google Scholar]
- 40.Butt AJ, et al. Insulin-like growth factor binding protein-3 expression is associated with growth stimulation of T47D human breast cancer cells: the role of altered epidermal growth factor signaling. J. Clin. Endocrinol. Metab. 2004;89:1950–1956. doi: 10.1210/jc.2003-030914. [DOI] [PubMed] [Google Scholar]
- 41.Torng PL, et al. Insulin-like growth factor binding protein-3 (IGFBP-3) acts as an invasion-metastasis suppressor in ovarian endometrioid carcinoma. Oncogene. 2008;27:2137–2147. doi: 10.1038/sj.onc.1210864. [DOI] [PubMed] [Google Scholar]
- 42.Massoner P, et al. Novel mechanism of IGF-binding protein-3 action on prostate cancer cells: inhibition of proliferation, adhesion, and motility. Endocr. Relat. Cancer. 2009;16:795–808. doi: 10.1677/ERC-08-0175. [DOI] [PubMed] [Google Scholar]
- 43.Benini S, et al. Insulin-like growth factor binding protein 3 as an anticancer molecule in Ewing's sarcoma. Int. J. Cancer. 2006;119:1039–1046. doi: 10.1002/ijc.21929. [DOI] [PubMed] [Google Scholar]
- 44.O'Han MK, et al. Effects of endogenous insulin-like growth factor binding protein-3 on cell cycle regulation in breast cancer cells. Growth Factors. 2009;27:394–408. doi: 10.3109/08977190903185032. [DOI] [PubMed] [Google Scholar]
- 45.Xi Y, et al. Association of insulin-like growth factor binding protein-3 expression with melanoma progression. Mol. Cancer Ther. 2006;5:3078–3084. doi: 10.1158/1535-7163.MCT-06-0424. [DOI] [PubMed] [Google Scholar]
- 46.Chang KH, et al. IGF binding protein-3 regulates hematopoietic stem cell and endothelial precursor cell function during vascular development. Proc. Natl Acad. Sci. USA. 2007;104:10595–10600. doi: 10.1073/pnas.0702072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gui Y, et al. Insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3) binds to fibronectin (FN): demonstration of IGF-I/IGFBP-3/fn ternary complexes in human plasma. J. Clin. Endocrinol. Metab. 2001;86:2104–2110. doi: 10.1210/jcem.86.5.7472. [DOI] [PubMed] [Google Scholar]
- 48.Kricker JA, et al. Structural and functional evidence for the interaction of insulin-like growth factors (IGFs) and IGF binding proteins with vitronectin. Endocrinology. 2003;144:2807–2815. doi: 10.1210/en.2002-221086. [DOI] [PubMed] [Google Scholar]
- 49.Liu B, et al. Type Ialpha collagen is an IGFBP-3 binding protein. Growth Horm. IGF Res. 2003;13:89–97. doi: 10.1016/s1096-6374(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 50.Gui Y, et al. Interaction of insulin-like growth factor binding protein-3 with latent transforming growth factor-beta binding protein-1. Mol. Cell Biochem. 2003;250:189–195. doi: 10.1023/a:1024990409102. [DOI] [PubMed] [Google Scholar]
- 51.Dallas SL, et al. Fibronectin regulates latent transforming growth factor-beta (TGF beta) by controlling matrix assembly of latent TGF beta-binding protein-1. J. Biol. Chem. 2005;280:18871–18880. doi: 10.1074/jbc.M410762200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.