Abstract
The phosphoinositide-3 kinase (PI3K)–AKT– mammalian target of rapamycin (mTOR) pathway is an important cellular pathway controlling cell growth, tumorigenesis, cell invasion and drug response. We hypothesized that genetic variations in the PI3K–AKT–mTOR pathway may affect the survival in muscle invasive and metastatic bladder cancer (MiM-BC) patients. We conducted a follow-up study of 319 MiM-BC patients to systematically evaluate 289 single-nucleotide polymorphisms (SNPs) of 20 genes in the PI3K–AKT–mTOR pathway as predicators of survival. In multivariate Cox regression, AKT2 rs3730050, PIK3R1 rs10515074 and RAPTOR rs9906827 were significantly associated with survival. In combined analysis, we found a cumulative effect of these three SNPs on survival. With the increasing number of unfavorable genotypes, there was a significant trend of higher risk of death in multivariate Cox regression (P for trend <0.001) and shorter median survival time in Kaplan–Meier estimates (P log rank <0.001). This is the first study to evaluate the role of germ line genetic variations in the PI3K–AKT–mTOR pathway genes as predictors of MiM-BC clinical outcomes. These findings warrant further replication in independent populations and may provide information on disease management and development of target therapies.
Background
Bladder cancer is the second most common genitourinary malignancy in the USA with an increasing incidence in recent years (1). Over 90% of bladder cancers are transitional cell carcinomas (TCCs) with 70% of TCC being non-muscle-invasive tumors (stages Ta, Tis and T1). The remaining TCCs are either muscle-invasive tumors (stages T2 and T3; 25%) or metastatic tumors (stage T4; 5%) (2). Radical cystectomy with bilateral pelvic lymph node dissection remains the gold standard for the treatment of muscle-invasive bladder cancer (3,4). Radiotherapy and chemotherapy may also be applied to these patients (3,4). For metastatic bladder cancer, systemic chemotherapy is the only treatment option (3,4). Despite these treatments, the overall median survival time (MST) for metastatic bladder cancer is ∼14 months (5). The majority of muscle invasive and metastatic bladder cancer (MiM-BC) patients will die of metastasis within 3 years (6). Therefore, identification of prognostic biomarkers is important in the achievement of molecular-based targeted therapy therefore prolonged survival of MiM-BC patients.
The phosphoinositide-3 kinase (PI3K)–AKT–mammalian target of rapamycin (mTOR) pathway is an important cellular pathway involved in cell growth, tumorigenesis, cell invasion and drug response (7–9). This pathway is frequently activated in many cancers and uncontrolled PI3K–AKT–mTOR signaling may also result in poor clinical outcome in lung, cervical, ovarian and esophageal cancers (7,8,10–12). In bladder cancer, it was reported that this pathway may regulate tumor invasion (13). Therefore, we hypothesize that genetic variations in this pathway may affect bladder cancer survival. There have been no studies to assess the association between germ line variations in the PI3K–AKT–mTOR pathway and clinical outcomes of bladder cancer. Herein, we applied a pathway-based approach to systematically evaluate single-nucleotide polymorphisms (SNPs) in PI3K–AKT–mTOR pathway genes as predictors of MiM-BC prognosis.
Materials and methods
Study subjects
Bladder cancer patients were recruited from the University of Texas M. D. Anderson Cancer Center and Baylor College of Medicine through a daily review of computerized appointment schedules as a part of an ongoing bladder cancer case–control study since 1995. As described previously (14), patients were all newly diagnosed within 1 year prior to recruitment, histologically confirmed and previously untreated with chemotherapy and radiotherapy. Patients with a long history (>1 year) of non-muscle-invasive disease before referral to M. D. Anderson or with a prior medical history of recurrence were excluded. There were no age, gender and stage restrictions on case recruitment. In our population, because >99% were pure TCCs, and 90.6% were Caucasians, all analyses were limited to TCC and Caucasians. In this paper, we restricted the study subjects to the muscle invasive and metastatic bladder cancer patients (stages T2–T4) accounting for ∼40% of the total bladder cancer patient of our program.
Epidemiological and clinical data collection
Epidemiological data were collected by M.D. Anderson interviewers during a 45 min interview for demographics, family history and smoking status. Immediately after each interview, a 40 ml blood sample was collected into heparinized tubes for lymphocyte isolation and DNA extraction. The participation rate was 92%. The clinical data was collected from our clinical station system by trained chart reviewers as described previously (15). Standard forms and tables were used for abstracting clinicopathological variables including tumor grade, size, stage, as well as treatment variables such as chemotherapy, radiotherapy, surgery, toxicity data were also abstracted using standard forms and tables. The end point of study was overall survival, which was calculated from the date of diagnosis to death or the date of last follow-up whichever came first. All of the human participation procedures were approved by the University of Texas M. D. Anderson Cancer Center and Baylor College of Medicine institutional review boards. Written consent forms were obtained from patients before interview.
Genotyping
Genomic DNA was isolated from peripheral blood using the QIAamp DNA blood Maxi Kit (QIAGEN, Valencia, CA) according to the manufacturer’s protocol. We combined literature exploration and database mining to select candidate genes in the PI3K–AKT–mTOR pathway following a procedure as described previously (16). A total of 289 SNPs, including haplotype tagging SNPs and potential functional SNPs from 20 PI3K–AKT–mTOR pathway genes were selected for genotyping. Among them, 185 SNPs were genotyped previously as part of our genome-wide association study of bladder cancer risk using Illumina’s Human-Hap610 BeadChips. The genotyping of the remaining SNPs was done using Illumina’s iSelect custom SNP array platform together with other cancer-related pathway SNPs according to the manufacturer’s Infinium II assay protocol (Illumina, San Diego, CA). All of the patients’ genotypes were called and exported using BeadStudio software (Illumina). The average call rate for the SNP array was 99.7%.
Statistical analysis
Most statistical analyses were performed using the Intercooled Stata 10 statistical software package (Stata Co. College Station, TX). Pearson’s χ2 test or Fisher’s exact tests was used to compare the difference in distribution of categorical variables, and Wilcoxon rank-sum test or the Student’s t-test was used for continuous variables where appropriate. For the main effect of individual SNP on overall survival, hazard ratios (HRs) and 95% confidence intervals(CIs) were estimated by multivariate Cox proportional hazard regression, adjusting for age, gender, smoking status, tumor grade, tumor stage and treatments. The patients who lost to follow-up were censored. Since many SNPs and tests were performed in the analysis, the Q value, a measure of significance in terms of the false discovery rate, was used to adjust the significance level for individual SNPs (17–19). We calculated Q value by the Q value package implemented in the R software. We applied a bootstrap resampling method to internally validate the results. We generated 100 bootstrapped samples. Each time, a bootstrap sample was drawn from the original dataset and the P value was obtained for each SNP among the dominant, recessive and additive models. All statistical analyses were two sided. For the cumulative effect of multiple variants, unfavorable genotypes were collapsed together. Taking the low-risk group carrying none of these unfavorable genotypes as the reference group, the HRs and 95% CIs were calculated for the other groups using multivariate Cox regression. Kaplan–Meier curve were plotted by each genotypes, and log rank tests were applied to compare the difference between the survival time of each genotype or group.
Results
Subject characteristics
There were 319 MiM-BC patients included in this study. The median follow-up time was 49.7 months. During the follow-up period, 134 patients had died and 185 were alive. We compared the demographic and clinical variables between groups of patients who had died and those surviving in Table I. The mean age of the patients who were dead was 69.1 (SD: 11.1) years compared with 64 (SD: 9.7) years in surviving patients. Clinical variables significantly associated with survival status included tumor stage (P < 0.001), grade (P = 0.05) and treatments (P = 0.001).
Table I.
Demographic and clinical variables for MiM-BC patients
| Variables | Death N (%) | Alive N (%) | P* |
| Yes (N = 134) | No (N = 185) | ||
| Sex | |||
| Male | 102 (76.1) | 147 (79.5) | 0.48 |
| Female | 32 (23.9) | 38 (20.5) | |
| Age (years) | |||
| Mean (SD) | 69.1 (11.1) | 64.0 (9.7) | <0.001 |
| Smoking statusa | |||
| Never | 27 (20.2) | 49 (26.5) | 0.42 |
| Former | 71 (53.0) | 89 (48.1) | |
| Current | 36 (26.8) | 47 (25.4) | |
| Stage | |||
| T2 | 71 (54.6) | 147 (81.2) | <0.001 |
| T3 | 30 (23.1) | 23 (12.7) | |
| T4 | 29 (22.3) | 11 (6.1) | |
| Grade | |||
| G1 | 0 | 0 | 0.05 |
| G2 | 1 (0.8) | 9 (4.9) | |
| G3 | 129 (99.2) | 174 (95.1) | |
| Treatmentsb | |||
| 1 | 35 (26.1) | 67 (36.2) | 0.001 |
| 2 | 27 (20.1) | 11 (5.9) | |
| 3 | 33 (24.6) | 63 (34.1) | |
| 4 | 14 (10.5) | 14 (7.6) | |
| 5 | 25 (18.7) | 30 (16.2) | |
Smoking status: individuals who had smoked >100 cigarettes in their lifetime were defined as ever-smokers; others were never-smokers. Smokers included current smokers and former smokers. Individuals who had quit smoking at least 1 year before diagnosis were categorized as former smokers.
Treatments: 1, Transurethral resection (TUR) + cystectomy; 2, TUR + chemotherapy; 3, TUR + cystectomy + chemotherapy; 4, TUR only; 5, others.
P values were derived from Pearson’s χ2 test or Fisher’s exact test for categorical variables and Student’s t-test for continuous variables. The significant P values (less than 0.05) were in bold.
Single SNP effect on survival
We assessed the association of each individual SNP with overall survival using multivariate Cox model, adjusting by age, sex, smoking status, stage, grade and treatments. We found that four SNPs, AKT2 rs3730050, PIK3R1 rs10515074, RAPTOR rs9906827 and RAPTOR rs7208502, were significantly associated with survival (Table II) and remained significant after correction for multiple comparison testing. Among them, RAPTOR rs9906827 and rs7208502 were in strong linkage disequilibrium (r2 = 0.99, D′ = 1.00), thus we only included RAPTOR rs9906827 in the subsequent joint analysis. Compared with the homozygous wild-type, the AKT2 rs3730050 heterozygous and homozygous variant showed 1.51-fold (95% CI: 1.02–2.23) and 2.99-fold (95% CI: 1.65–5.42) increased risk of death, respectively (P for trend <0.001). There was a significant trend of shorter MST as the number of variant allele increased (P log rank = 0.006, data not shown). For PIK3R1 rs10515074, carriers of at least one variant allele showed a 1.83-fold (95% CI: 1.24–2.69) increased risk of death. RAPTOR rs9906827 variants were observed to confer a protective effect on death with a HR of 0.55 (95% CI: 0.37–0.81) and longer MST (81.9 months, data not shown) compared with the wild-type genotype (MST, 32.3 months, P log rank = 0.013, data not shown). To internally validate the associations, we performed bootstrap sampling. The overall odds ratios and 95% CIs generated by bootstrapping were consistent with our initial results. Table II lists the number of times that the bootstrap-generated P value was 0.05, 0.001 or 0.0001 for each SNP. In 100 bootstrap samplings, each of the SNPs reached significance at the P 0.05 level in >80% of the samplings. This indicates that the results for these SNPs are unlikely to be due to chance alone.
Table II.
SNPs associated with survival for MiM-BC patients
| SNP’s genes | Genotypes | Death/alive | HRa (95% CI) | Pb | Bootstrapc |
||
| 134/185 | P < 0.0001 | P < 0.001 | P < 0.05 | ||||
| rs3730050 AKT2 (G>A) | ww | 52/97 | Ref. | ||||
| wv | 57/69 | 1.51 (1.02–2.23) | 0.05 | ||||
| vv | 16/11 | 2.99 (1.65–5.42) | 2 × 10−4 | ||||
| Dominant | 1.68 (1.16–2.44) | 6 × 10−3 | |||||
| Additive | P for the trend | 4 × 10−4 | 34 | 54 | 94 | ||
| rs10515074 PIK3R1 (A>G) | ww | 65/108 | Ref. | ||||
| wv | 56/61 | 1.88 (1.27–2.78) | 2 × 10−3 | ||||
| vv | 4/6 | 1.37 (0.48–3.94) | 0.56 | ||||
| Dominant | 1.83 (1.24–2.69) | 2 × 10−3 | 24 | 42 | 85 | ||
| Additive | P for the trend | 7 × 10−3 | |||||
| rs9906827 RAPTOR (G>A) | ww | 46/45 | Ref. | ||||
| wv | 50/83 | 0.55 (0.36–0.84) | 6 × 10−3 | ||||
| vv | 29/46 | 0.54 (0.34–0.88) | 0.01 | ||||
| Dominant | 0.55 (0.37–0.81) | 2 × 10−3 | 19 | 43 | 85 | ||
| Additive | 0.72 (0.56–0.92) | 0.01 | |||||
| rs7208502 RAPTOR (G>A) | ww | 46/45 | Ref. | ||||
| wv | 50/84 | 0.55 (0.36–0.84) | 6 × 10−3 | ||||
| vv | 29/46 | 0.54 (0.33–0.87) | 0.01 | ||||
| Dominant | 0.54 (0.37–0.80) | 2 × 10−3 | 15 | 37 | 82 | ||
| Additive | 0.72 (0.56–0.92) | 9 × 10−3 | |||||
The results of best genetic models are in bold.
Adjusted by age, sex, smoking status, stage, grade and treatments using Cox proportional hazard regression.
Remain significant after multiple comparison adjustment by Q test with false discovery rate at 10% level.
We did internal validation of the results choosing from the best genetic model using bootstrap 100 times.
Cumulative effect on survival
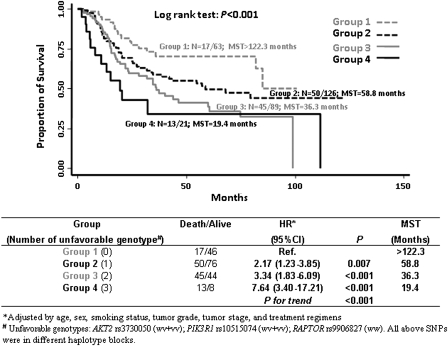
We defined genotypes with increased risks of death as unfavorable genotypes, which included RAPTOR rs9906827 wild-type, AKT2 rs3730050 variant and PIK3R1 rs10515074 variant genotypes, to perform the cumulative analysis. When we combined these unfavorable genotypes, there was a significant trend for an increasing risk of death with an increasing number of unfavorable genotypes. In Figure 1, compared with the reference (group 1 without any unfavorable genotypes), the HRs for individuals with 1 (group 2), 2 (group 3) and 3 (group 4) unfavorable genotypes were 2.17 (95% CI: 1.23–3.85), 3.34 (95% CI: 1.83–6.09) and 7.64 (95% CI: 3.40–17.21), respectively. In Kaplan–Meier estimates, there was a significant trend for shorter MST with the increasing number of unfavorable genotypes (log rank P value <0.001). The MSTs for groups 1–4 were >122.3, 58.8, 36.3 and 19.4 months, respectively.
Fig. 1.
Cumulative effect of unfavorable genotypes on survival of MiM-BC patients.
Discussion
This study evaluated effects of genetic variations of the PI3K–AKT–mTOR pathway on survival in MiM-BC patients. SNPs AKT2 rs3730050, PIK3R1 rs10515074 and RAPTOR rs9906827 were associated with overall survival. Moreover, a gene-dosage effect was observed, with the increasing number of unfavorable SNPs associated with an increased risk of death and shorter MST.
MiM-BC patients are a disease with poor clinical outcome. Currently, only clinicopathological variables, such as grade, stage, vascular and lymphatic extension and treatments, are used to predict prognosis and guide clinical management (20). Previous laboratory and population studies (5,15,21–24) have suggested that molecular markers in several pathways including DNA repair, apoptosis and inflammation may provide independent prognostic values for bladder cancer. Some putative molecular markers have been incorporated in the contemporary nomograms for predicting bladder cancer outcomes and are undergoing validation in large population (25). However, the predictive ability for prognosis and drug response is not optimal and many of the suggested markers needs further validation. To improve the prediction accuracy, there is still a need to identify molecular markers for MiM-BC clinical outcomes.
The PI3K–AKT–mTOR pathway is a candidate pathway for many human cancers. Inhibition of the PI3K–AKT signaling abrogated bladder cancer cell line invasiveness (13) indicating that this pathway may affect the natural history and prognosis of bladder cancer. Therefore, we evaluated germ line genetic variations in this pathway as predictors of prognosis in MiM-BC patients. The most interesting finding in this study is that the variant A allele of AKT2 rs3730050 was significantly associated with increased risk of death and shorter survival time. AKT, the cellular homologue of the retroviral oncogene v-AKT, is a serine/threonine kinase, which has three homologues, AKT1, AKT2 and AKT3 in mammals (26). Once, cells are stimulated by extracellular signals, such as hormones, growth factors and extracellular matrix components, PI3K kinase phosphorylates a second messenger, phosphatidylinositol (3–5)-trisphosphate, which directly anchors AKT to the cell member to be activated by other kinases. Phosphatase and tensin homolog is a phosphatase that can inhibit this process (26). AKT is involved in a number of cancer cell-acquired phenotypes as follows: for survival, AKT can interact with BCL2-associated agonist of cell death, nuclear factor kappaB and P53 and exert anti-apoptosis functions; for proliferation, AKT can regulate cell cycle machinery and for cell growth, AKT can activate mTOR-mediated biosynthesis. Constitutive amplification of AKT2 has been found in ovarian, pancreatic, breast cancers (27,28) and in ∼55% primary bladder cancers (13). Particularly, AKT2 activation was associated with high grade and more aggressive phenotypes (29). In addition, AKT amplification was reported to mediate cisplatin resistance in lung, cervical and ovarian cancer (7–10). A recent study on esophageal cancer also reported that SNPs in AKT2 were associated with poor prognosis (30). Therefore, it is biologically plausible that variations of AKT2 modulate survival of MiM-BC patients. The function of AKT2 rs3730050 remains to be elucidated. This SNP is an intronic SNP that may regulate gene expression. Alternatively, it may represent a tagging SNP in linkage with the real causal SNP from the same haplotype block.
Phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) is the regulatory subunit of PI3K, which is the upstream kinase of AKT. It directly associates with tyrosine kinases or through indirect interactions with tyrosine receptor kinases by insulin receptor substrate. PIK3R1 constitutively binds and inhibits the release of catalytic subunit p110 of PI3K. Mutation of PIK3R1 has been observed in ovarian and colon cancer (31), and higher kinase activity was detected in breast cancer (32). Compared with normal adjacent epithelium, primary bladder tumors of all stages had PI3K overexpression and 5- to 20-fold increased kinase activity (33). rs10515074 is located in intron 1 of PIK3R1. It may be a functional SNP or a surrogate marker for the causative SNP.
The function of RAPTOR is not fully understood. Raptor is a regulatory protein of mTOR, which is downstream of PI3K–AKT pathway. It may modify the mTOR activity or effectors of biosynthesis. rs9906827 is located in intron 3 of RAPTOR. Future studies are warranted to validate these three significant SNPs in independent population and to perform fine mapping in the vicinity of these gene regions to identify potential causal variants.
Recently, there has been an increasing application of the pathway-based approach in cancer genetic and association studies (34,35). In colorectal cancer, mutations of single genes in the PI3K–AKT pathway were only observed in 4% of the tumors. In contrast, mutations of multiple genes in the same pathway were detected in 40% of the patients (34). Studies from mouse models showed that single gene dysfunction in the PI3K–AKT pathway is insufficient to cause cancer (36). Evidences from epidemiological studies demonstrate that combining multiple variants would result in greater predictive power of disease risk and clinical outcomes (37–39). When we combined the three significant SNPs in different genes identified in this study, we found a 7.64-fold increased risk of death for patients with three unfavorable genotypes compared with those with no unfavorable genotypes, providing further support for the application of pathway-based approach in association studies.
This is the first study to evaluate the association between germ line genetic variations in the PI3K–AKT–mTOR pathway and survival in MiM-BC patients. We found that three significant SNPs (AKT2 rs3730050, PIK3R1 rs10515074, and RAPTOR rs9906827) and a cumulative effect of these three SNPs were associated with survival in MiM-BC patients. Although we adjusted for multiple testing and performed bootstrap sampling to internally validate these associations, we could not rule out the possibility of false-positive findings. Independent external patient cohorts are needed to validate our findings. If validated, these SNPs may be valuable biomarkers to complement clinicopathological variables in predicting prognosis of MiM-BC and to facilitate physicians in making individualized treatment decisions.
Funding
National Institutes of Health (CA74880 and CA91846).
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence interval
- HR
hazard ratio
- MST
median survival time
- MiM-BC
muscle invasive and metastatic bladder cancer
- PI3K–AKT–mTOR
phosphoinositide-3 kinase (PI3K)–AKT–mammalian target of rapamycin (mTOR)
- SNP
single-nucleotide polymorphism
- TCC
transitional cell carcinoma
References
- 1.Jemal A, et al. Cancer statistics. CA Cancer J. Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Parekh DJ, et al. Superficial and muscle-invasive bladder cancer: principles of management for outcomes assessments. J. Clin. Oncol. 2006;24:5519–5527. doi: 10.1200/JCO.2006.08.5431. [DOI] [PubMed] [Google Scholar]
- 3.Stenzl A, et al. The updated EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur. Urol. 2009;55:815–825. doi: 10.1016/j.eururo.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Oosterlinck W, et al. Guidelines on bladder cancer. Eur. Urol. 2002;41:105–112. doi: 10.1016/s0302-2838(01)00026-4. [DOI] [PubMed] [Google Scholar]
- 5.Abraham R, et al. Chromosomal deletions in bladder cancer: shutting down pathways. Front. Biosci. 2007;12:826–838. doi: 10.2741/2105. [DOI] [PubMed] [Google Scholar]
- 6.Ruggeri EM, et al. Adjuvant chemotherapy in muscle-invasive bladder carcinoma: a pooled analysis from phase III studies. Cancer. 2006;106:783–788. doi: 10.1002/cncr.21676. [DOI] [PubMed] [Google Scholar]
- 7.Liu LZ, et al. AKT1 amplification regulates cisplatin resistance in human lung cancer cells through the mammalian target of rapamycin/p70S6K1 pathway. Cancer Res. 2007;67:6325–6332. doi: 10.1158/0008-5472.CAN-06-4261. [DOI] [PubMed] [Google Scholar]
- 8.Faried LS, et al. Expression of an activated mammalian target of rapamycin in adenocarcinoma of the cervix: a potential biomarker and molecular target therapy. Mol. Carcinog. 2008;47:446–457. doi: 10.1002/mc.20402. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, et al. Akt-mediated cisplatin resistance in ovarian cancer: modulation of p53 action on caspase-dependent mitochondrial death pathway. Cancer Res. 2006;66:3126–3136. doi: 10.1158/0008-5472.CAN-05-0425. [DOI] [PubMed] [Google Scholar]
- 10.Faried LS, et al. Predictive and prognostic role of activated mammalian target of rapamycin in cervical cancer treated with cisplatin-based neoadjuvant chemotherapy. Oncol. Rep. 2006;16:57–63. [PubMed] [Google Scholar]
- 11.Hou G, et al. An activated mTOR/p70S6K signaling pathway in esophageal squamous cell carcinoma cell lines and inhibition of the pathway by rapamycin and siRNA against mTOR. Cancer Lett. 2007;253:236–248. doi: 10.1016/j.canlet.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, et al. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol. Oncol. 2005;97:26–34. doi: 10.1016/j.ygyno.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, et al. The phosphatidylinositol-3 kinase pathway regulates bladder cancer cell invasion. BJU Int. 2004;93:143–150. doi: 10.1111/j.1464-410x.2004.04574.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen M, et al. Genetic variations in PI3K-AKT-mTOR pathway and bladder cancer risk. Carcinogenesis. 2009;30:2047–2052. doi: 10.1093/carcin/bgp258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leibovici D, et al. Polymorphisms in inflammation genes and bladder cancer: from initiation to recurrence, progression, and survival. J. Clin. Oncol. 2005;23:5746–5756. doi: 10.1200/JCO.2005.01.598. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, et al. Profiling of genetic variations in inflammation pathway genes in relation to bladder cancer predisposition. Clin. Cancer Res. 2008;14:2236–2244. doi: 10.1158/1078-0432.CCR-07-1670. [DOI] [PubMed] [Google Scholar]
- 17.Storey JD, et al. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J. R. Stat. Soc. Series B Stat. Methodol. 2004;66:187–205. [Google Scholar]
- 18.Storey JD, et al. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey JD. A direct approach to false discovery rates. J. R. Stat. Soc. Series B Stat. Methodol. 2002;64:479–498. [Google Scholar]
- 20.Habuchi T, et al. Prognostic markers for bladder cancer: International Consensus Panel on bladder tumor markers. Urology. 2005;66:64–74. doi: 10.1016/j.urology.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 21.Mitra AP, et al. Molecular pathways in invasive bladder cancer: new insights into mechanisms, progression, and target identification. J. Clin. Oncol. 2006;24:5552–5564. doi: 10.1200/JCO.2006.08.2073. [DOI] [PubMed] [Google Scholar]
- 22.Gu J, et al. Nucleotide excision repair gene polymorphisms and recurrence after treatment for superficial bladder cancer. Clin. Cancer Res. 2005;11:1408–1415. doi: 10.1158/1078-0432.CCR-04-1101. [DOI] [PubMed] [Google Scholar]
- 23.Andrew AS, et al. Bladder cancer SNP panel predicts susceptibility and survival. Hum. Genet. 2009;125:527–539. doi: 10.1007/s00439-009-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karam JA, et al. Use of combined apoptosis biomarkers for prediction of bladder cancer recurrence and mortality after radical cystectomy. Lancet Oncol. 2007;8:128–136. doi: 10.1016/S1470-2045(07)70002-5. [DOI] [PubMed] [Google Scholar]
- 25.Shariat SF, et al. Nomograms for bladder cancer. Eur. Urol. 2008;54:41–53. doi: 10.1016/j.eururo.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson KM, et al. The protein kinase B/Akt signalling pathway in human malignancy. Cell. Signal. 2002;14:381–395. doi: 10.1016/s0898-6568(01)00271-6. [DOI] [PubMed] [Google Scholar]
- 27.Bellacosa A, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int. J. Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 28.Cheng JQ, et al. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc. Natl Acad. Sci. USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson FH, et al. Amplification of 19q13.1-q13.2 sequences in ovarian cancer. G-band, FISH, and molecular studies. Cancer Genet. Cytogenet. 1996;87:55–62. doi: 10.1016/0165-4608(95)00248-0. [DOI] [PubMed] [Google Scholar]
- 30.Hildebrandt MA, et al. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J. Clin. Oncol. 2009;27:857–871. doi: 10.1200/JCO.2008.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philp AJ, et al. The phosphatidylinositol 3'-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61:7426–7429. [PubMed] [Google Scholar]
- 32.Sun M, et al. Phosphatidylinositol-3-OH Kinase (PI3K)/AKT2, activated in breast cancer, regulates and is induced by estrogen receptor alpha (ERalpha) via interaction between ERalpha and PI3K. Cancer Res. 2001;61:5985–5991. [PubMed] [Google Scholar]
- 33.Benistant C, et al. A specific function for phosphatidylinositol 3-kinase alpha (p85alpha-p110alpha) in cell survival and for phosphatidylinositol 3-kinase beta (p85alpha-p110beta) in de novo DNA synthesis of human colon carcinoma cells. Oncogene. 2000;19:5083–5090. doi: 10.1038/sj.onc.1203871. [DOI] [PubMed] [Google Scholar]
- 34.Parsons DW, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vivanco I, et al. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 37.Sconce EA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 38.Wu X, et al. Germline genetic variations in drug action pathways predict clinical outcomes in advanced lung cancer treated with platinum-based chemotherapy. Pharmacogenet. Genomics. 2008;18:955–965. doi: 10.1097/FPC.0b013e32830efdd4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng SL, et al. Cumulative association of five genetic variants with prostate cancer. N. Engl. J. Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]