Abstract
Background: It is well established that estrogen increases endometrial cancer risk, whereas progesterone opposes the estrogen effects. The PROGINS allele of the progesterone receptor (PGR) gene reduces the function of PGR and has been associated with increased risk of the endometrioid type ovarian cancer. We investigated whether genetic variation in PGR is also associated with endometrial cancer risk using a haplotype-based approach. Methods: We pooled data from two endometrial cancer case–control studies that were nested within two prospective cohorts, the Multiethnic Cohort Study and the California Teachers Study. Seventeen haplotype-tagging single nucleotide polymorphisms (SNPs) across four linkage disequilibrium (LD) blocks spanning the PGR locus were genotyped in 583 incident cases and 1936 control women. Odds ratios (ORs) and 95% confidence intervals (CIs) associated with each haplotype were estimated using conditional logistic regression, stratified by age and ethnicity. Results: Genetic variation in LD block 3 of the PGR locus was associated with endometrial cancer risk (Pglobal test = 0.002), with haplotypes 3C, 3D and 3F associated with 31–34% increased risk. Among whites (383 cases/840 controls), genetic variation in all four blocks was associated with increased endometrial cancer risk (Pglobal test = 0.010, 0.013, 0.005 and 0.020). Haplotypes containing the PROGINS allele and several haplotypes in blocks 1, 3 and 4 were associated with 34–77% increased risk among whites. SNP analyses for whites suggested that rs608995, partially linked to the PROGINS allele (r2 = 0.6), was associated with increased risk (OR = 1.30, 95% CI = 1.06–1.59). Conclusions: Our results suggest that genetic variation in the PGR region is associated with endometrial cancer risk.
Introduction
Endometrial cancer is the fourth most common cancer in USA women (1). Both endogenous and exogenous sources of estrogen increase endometrial cell proliferation and endometrial cancer risk, whereas progesterone opposes these effects (2,3). In ovulating premenopausal women, the endometrium proliferates during the follicular phase of the menstrual cycle in response to the preovulatory increase in estrogen, whereas the endometrium stops proliferating during the luteal phase when progesterone concentration is high (4). Among postmenopausal women with an intact uterus, estrogens given alone as hormone therapy (HT) increase the risk of endometrial cancer, but when a progestin is added for at least 10 days per month, risk is not elevated (5,6). Progesterone must bind to the progesterone receptor (PGR) in order to exert its effect. Therefore, we hypothesized that polymorphisms at the PGR gene locus may be associated with endometrial cancer risk.
The PROGINS allele is an Alu insertion in intron 7 of the PGR gene, which is in complete linkage disequilibrium (LD) with rs1042838 (V660L in exon 4) and rs1042839 (H770H in exon 5) (7). In vitro studies have shown that the PROGINS allele may reduce PGR transcription or signaling in several ovarian cancer cell lines (8). The PROGINS allele has been associated with increased ovarian cancer risk in a number of studies (9–13), although such associations have not been observed in some studies (14–16). Recently, a pooled analysis within the Ovarian Cancer Association Consortium showed that the PROGINS allele is associated with increased risk of endometrioid type ovarian cancer (17). Few studies have examined the role of the PROGINS allele on endometrial cancer risk. Consistent with the functional studies and association studies of the PROGINS allele in ovarian cancer, one study found a positive association between the PROGINS allele and endometrial cancer recurrence (18), whereas the results from two studies on the risk of incident endometrial cancer were inconclusive (19,20). Another potentially functional single nucleotide polymorphism (SNP) in the PGR (+331 G/A, rs10895068) has been shown to increase translation of PGR isoform B (19), which may be associated with progesterone-dependent proliferation of the endometrium (21,22). However, the results from association studies examining the effect of this SNP on risk of endometrial cancer have been mixed (19,23,24).
We have investigated the association between genetic variation spanning the PGR gene region and endometrial cancer risk using a haplotype-based approach. We also evaluated the role of potential functional SNPs of PGR on endometrial cancer risk.
Materials and methods
Subjects
This study is based on two case–control studies, nested within two prospective cohort studies.
Multiethnic cohort.
The multiethnic cohort (MEC) included >118 000 African-American, Native Hawaiian, Japanese American, Latino and white women, who were aged 45–75 years old and resided in Hawaii or Los Angeles when the cohort was formed (between 1993 and 1996) (25). MEC study participants provided information on demographic, anthropometric and reproductive factors as well as hormone use through a self-administered mail questionnaire. The endometrial cancer case–control study nested within the MEC included 299 incident invasive endometrial cancer cases diagnosed after women joined the cohort and before 1 January 2004 and 1533 control women (26). Case patients were identified through linkage with the Surveillance, Epidemiology and End Results cancer registries covering California and Hawaii. Of the 299 case women, we excluded 18 women who were diagnosed with either uterine sarcomas, mesodermal mixed tumors or mullerian mixed tumors (ICD-O-3 codes: 8930-8933, 8950, 8951 and 8980). Control participants were a subsample of women randomly selected to serve as controls for a breast cancer case-control study nested within the MEC. For this study, we also required that they had an intact uterus at baseline and remained free of breast and uterine cancer through 31 December 2003 (27). Over 65% of cases and controls provided a blood sample (26). For the current study, we excluded 4 cases and 84 controls who were aged <50 years to be consistent with the age criteria for the California Teachers Study (CTS). The majority (82%) of the participants in the current study were postmenopausal. The study was approved by the Institutional Review Boards of the University of Hawaii and University of Southern California. All participants provided written informed consent.
CTS.
The CTS was established in 1995–96, with the recruitment of 133 479 current and former female teachers and other public school professionals who were enrolled in the California State Teachers Retirement System (28). All participants joined the cohort by returning a self-administered questionnaire on lifestyle and other factors. Women were eligible to participate in the nested case–control study of endometrial cancer if, at diagnosis (cases) or ‘reference date’ (controls), they were aged ≥50 years, had no history of endometrial cancer, had an intact uterus and resided in California (29). Incident cancers of the corpus uteri were identified among CTS participants through 31 December 2004 by linking the cohort file with the California Cancer Registry, which comprised three Surveillance, Epidemiology and End Results registries. For each case, two controls were randomly selected from among eligible women in the cohort. Controls were frequency matched to cases on age (within 5 year age groups), race/ethnicity (white, African-American, Latina, Asian/Pacific Islander, Native American and other/mixed) and broad geographic region within California (corresponding to the 10 regional cancer registry regions). Control selection was based on eligibility at predetermined quarterly reference dates starting with 31 March 1996. Of 675 eligible cases, we interviewed 401 case women. Of these, we excluded seven women with in situ carcinomas and nine women with uterine sarcomas, mesodermal mixed tumors or mullerian mixed tumors (ICD-O-3 codes: 8930-8933, 8950, 8951 and 8980) and 37 women with previous history of breast cancer. Of the 1329 potentially eligible controls, 682 women were interviewed, among whom 59 women with previous history of breast cancer were excluded. Collection of blood or buccal samples from cases and controls was initiated in 2002. Over 96% of the interviewed cases and controls (99% for cases and 96% for controls) provided a sample for DNA. Because the majority of CTS participants are non-Latino white (86%) and the number of non-white women in the CTS who carried minor alleles of each polymorphic locus was too small, we restricted the present analyses to white women (320 endometrial cancer cases and 540 controls) from the CTS. The majority of women (93.7%) included in the current analyses were postmenopausal. The nested case–control study was approved by Institutional Review Boards at the Northern California Cancer Center and the University of Southern California. All participants provided written informed consent.
Haplotype-tagging SNP selection and genotyping
Detailed procedures to select haplotype-tagging SNPs (htSNPs) have been previously published (12). Briefly, 74 SNPs spanning the PGR locus, including putative regulatory regions, were identified from the public and Celera databases and genotyped in a multiethnic panel (whites, African-Americans, Native Hawaiians, Latinas and Japanese) from the MEC. Exclusion of 20 SNPs that failed to genotype with high reliability (8 SNPs) or that were not polymorphic in the screened panel (12 SNPs) resulted in 54 remaining SNPs (23.0 kb upstream of exon 1 to 7.6 kb downstream of the 3′ untranslated region) to be used for determination of haplotype block structure. Four haplotype blocks were identified following the method of Gabriel et al. (30), and haplotype frequency was estimated within each ethnic group using the expectation–maximization algorithm of Excoffier and Slatkin (31). The htSNPs in each haplotype block were selected using the TagSNP program (32) to ensure the minimum of Rh2 (i.e. the squared correlation between the true haplotypes and the estimated haplotypes) was >0.80 for all ethnic groups. This criterion resulted in 21 htSNPs including four African-American-specific htSNPs. However, as the MEC case–control study included only a small number of African-Americans (52 cases and 289 controls) and therefore was underpowered to specifically study haplotype effects in African-Americans, we limited our investigation to the common haplotypes (and tag SNPs) of whites. With the 17 htSNPs, the minimum Rh2 was >0.90 in each LD block for whites (the majority of this study population) and >0.80 in the all other ethnic groups except African-Americans. Exclusion of African-Americans (from the MEC) for ethnicity-combined analyses did not change the results, and therefore we kept African-Americans in the analyses.
After extracting DNA using the Qiagen 96 DNA Blood Kit, we genotyped the selected htSNPs and one additional potentially functional SNP in PGR (+331 C/T) (19) using the TaqMan assay (Applied Biosystems, Foster City, CA) in 597 incident invasive endometrial cancer cases and 1989 controls. We then excluded 67 women (6 cases and 38 controls including 4 African-American cases and 9 African-American controls from the MEC; 8 cases and 15 controls form the CTS) for whom the sample call rate was <70%. We included 89 random duplicates to assess reproducibility of genotyping. The concordance rate was 99.6%.
Statistical analyses
The final dataset includes 583 incident cases and 1936 control women. We tested for deviation from Hardy–Weinberg equilibrium (HWE) among controls within each ethnic/study group (MEC: whites, African-Americans, Latinos, Japanese and Native Hawaiian; CTS: whites). We defined HWE deviation as P < 0.01 in two or more ethnic groups, and all SNPs were in HWE. However, we noted that one SNP in haplotype block 1 (rs499590) was not in HWE in Native Hawaiians (P = 0.0003; supplementary Table 1 is available at Carcinogenesis Online) and evaluated whether exclusion of Native Hawaiians changed the results of the block 1 analysis. Since the results were similar, we present the analysis including Native Hawaiians.
Within each haplotype block, haplotype frequencies and expected haplotype counts were estimated from the genotype data using the TagSNP program (32) utilizing the expectation–maximization algorithm of Excoffier and Slatkin (31). Given that we used the htSNPs that were described by Pearce et al. (12), we labeled the estimated haplotypes with the same notation as in that study. Because the PGR haplotypes within a block were correlated with PGR haplotypes in adjacent blocks (12), we also estimated long-range haplotypes (i.e. throughout the PGR locus, not limiting to within-block estimation) using the TagSNP program. We performed both haplotype-based analyses and single SNP-based analyses using conditional logistic regression models to estimate odds ratios (ORs) and 95% confidence intervals (CIs). For the haplotype-based analyses, we first examined whether the haplotypes in each LD block were associated with endometrial cancer risk by examining the likelihood ratio test of a ‘global model’ for each LD block. In this model, the most common haplotype is used as the reference group, and the ORs associated with each of the other haplotypes are estimated. To correct for multiple testing, we used Bonferroni adjustments on the P-values of the global models. Secondly, we constructed ‘single-haplotype’ models, estimating the OR per copy of each haplotype compared with all other haplotypes combined (i.e. versus having one or more copies of any other haplotype).
All models were stratified by ethnic/study group and age at diagnosis/blood draw (50 to <60, 60 to <70 and 70–90). When we additionally adjusted for other factors known to influence endometrial cancer risk including menopausal status, HT use, oral contraceptive use, body mass index (BMI) and parity, or when we used different age categorization (quartiles or continuous), the results remained essentially identical and thus are not presented. We tested the heterogeneity of the genetic associations across ethnicity, BMI (<30 or ≥30 kg/m2), and postmenopausal HT use, using likelihood ratio tests to compare models with and without cross product terms for each covariate and haplotype, based on a single-haplotype model.
Considering that only Type 1, but not Type 2, endometrial cancer is considered to be hormone related (33), we repeated all analyses limiting the cases to the 530 Type 1 endometrial cancer cases (247 from MEC and 283 from CTS). Since the results were similar, we present all cases combined. Exclusion of premenopausal women did not change the results, and therefore we present the results based on all women. All statistical analyses were performed with SAS 9.2 (SAS Institute, Cary, NC).
Results
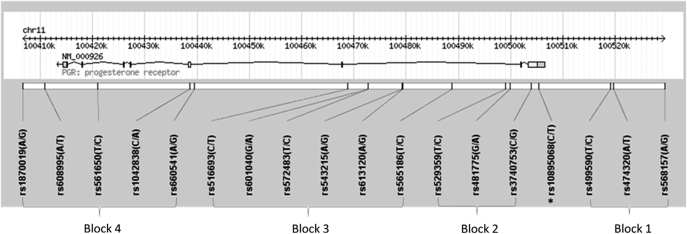
The structure of the PGR locus and the location of each htSNP are presented in Figure 1. The PROGINS allele (rs1042838) was more frequent in whites (minor allele frequency, MAF = 0.15) and Latinos (MAF = 0.15) than in other ethnic groups (MAF = 0.06 for Hawaiians; MAF < 0.05 for African-Americans and Japanese; supplementary Table 2 is available at Carcinogenesis Online). In every ethnic group, the PROGINS allele (rs1042838; in block 4) was in tight linkage (r2 > 0.8) with rs474320 in block 1, rs3740753 in block 2 (except among Japanese) and rs565186 in block 3. The PROGINS allele was located on only one haplotype in block 4 (4D) and was linked to one haplotype in each of the other blocks (1D, 2C and 3C; data not shown). When evaluating the long-range haplotypes across the PGR locus, only nine distinct haplotypes were observed with a haplotype frequency ≥ 0.5 in any ethnic group (Table I). Haplotype 4E was linked to a long-range haplotype of 1C-2D-3D-4E.
Fig. 1.
Location of SNPs along the PGR gene locus. Picture was generated using Haploview 4.1 and modified (34). Asterisk represents a potentially functional SNP (+331 C/T).
Table I.
Distribution of PGR gene haplotypes in cases and controls (%)
| Haplotype | MEC |
CTS |
|||||
| African-Americans | Native Hawaiians | Japanese | Latinos | Whites | Whites | ||
| 44 cases/280 controls | 15 cases/207 controls | 73 cases/330 controls | 68 cases/279 controls | 71 cases/315 controls | 312 cases/525 controls | ||
| Block 1 | |||||||
| 1A | AAC | 36.6/29.8 | 23.3/9.9 | 1.4/2.5 | 27.9/18.7 | 28.2/36.6 | 27.8/31.8 |
| 1Ba | GAT | 23.9/29.2 | 8.8/11.4 | 0/0.8 | 20.1/25.6 | 28.3/28.5 | 31.6/33.7 |
| 1C | AAT | 33.8/38.3 | 63.3/72.5 | 97.2/93.9 | 37.9/40.2 | 25.6/19.2 | 21.8/19.4 |
| 1Db | GTT | 5.7/2.7 | 4.5/6.3 | 1.4/2.4 | 14.1/15.4 | 15.6/15.6 | 18.7/15.0 |
| Block 2 | |||||||
| 2A | CGC | 56.8/44.5 | 71.8/71.9 | 82.8/81.6 | 50.0/43.2 | 34.3/40.5 | 33.4/35.6 |
| 2B | CAT | 9.0/9.5 | 10.7/10.4 | 0/0.8 | 17.6/24.3 | 31.9/32.5 | 32.3/36.7 |
| 2Cb | GGT | 3.8/2.4 | 6.7/6.6 | 1.3/1.4 | 14.7/15.2 | 14.1/15.5 | 18.8/14.2 |
| 2D | CGT | 26.3/43.3 | 10.8/11.2 | 15.8/16.1 | 17.7/17.3 | 17.5/11.1 | 15.5/13.4 |
| Block 3 | |||||||
| 3A | TAATGC | 9.1/10.4 | 8.8/10 | 0/0.8 | 17.7/23.9 | 32.7/32.9 | 32.4/36.6 |
| 3B | TGACGC | 5.7/3.1 | 9.2/2.4 | 0/0.2 | 16.9/11.6 | 15.2/20.6 | 15.3/19.0 |
| 3Cb | CGATGC | 4.5/2.5 | 6.7/6.7 | 1.4/1.4 | 16.2/15.2 | 12.8/15.9 | 19.0/14.5 |
| 3Da | TGACGT | 23.8/28.3 | 10.0/11.0 | 15.7/15.5 | 16.9/15.7 | 19.5/9.8 | 14.2/12.4 |
| 3E | TAGTAC | 31.8/24.8 | 8.9/2.3 | 1.4/2.1 | 9.0/6.1 | 7.0/7.7 | 6.6/7.4 |
| 3F | TAGTGC | 14.6/11.4 | 54.4/64.6 | 81.5/80.0 | 22.6/24.8 | 12.0/11.8 | 11.1/8.1 |
| 3G | TGATGC | 8.0/18.2 | 0/0.5 | 0/0 | 0/2.6 | 0.7/0.9 | 0.8/1.4 |
| Block 4 | |||||||
| 4A | GCTAA | 49.5/42.5 | 16.4/6.1 | 2.1/2.4 | 25.5/19.2 | 23.7/27.7 | 20.0/25.8 |
| 4B | ACTAG | 10.6/8.6 | 6.4/8.2 | 0/0.6 | 14.9/20.9 | 28.1/28.7 | 28.7/29.9 |
| 4C | ACTAA | 8.7/14.8 | 53.6/66.5 | 74.6/75.6 | 31.4/31.1 | 21.2/20.4 | 21.9/20.2 |
| 4Db | GATTA | 4.6/2.7 | 6.7/5.5 | 1.4/1.4 | 13.8/14.4 | 11.9/15.4 | 18.1/13.7 |
| 4E | GCCTA | 11.5/7.4 | 10.0/6.9 | 17.8/15.5 | 8.8/10.2 | 9.8/5.7 | 7.4/5.7 |
| 4F | GCTTA | 15.1/23.7 | 3.3/3.6 | 4.1/4.2 | 2.2/2.5 | 1.4/0.6 | 1.2/2.0 |
| Long-range | |||||||
| 1A-2A-3B-4A | AACCGCTGACGCGCTAA | 4.5/3.3 | 10.0/2.8 | 0/0.2 | 15.2/10.9 | 14.6/19.5 | 14.5/18.1 |
| 1C-2D-3D-4E | AATCGTTGACGTGCCTA | 5.0/6.7 | 10.0/6.7 | 15.8/15.0 | 6.6/9.8 | 9.1/4.5 | 6.3/4.6 |
| 1C-2A-3F-4C | AATCGCTAGTGCACTAA | 5.2/7.2 | 51.5/59.2 | 74.7/73.4 | 22.0/23.8 | 9.8/10.1 | 8.4/6.7 |
| 1A-2A-3E-4A | AACCGCTAGTACGCTAA | 17.4/18.5 | 8.5/2.1 | 1.4/1.5 | 6.9/4.9 | 5.8/7.1 | 4.5/5.9 |
| 1A-2B-3A-4B | AACCATTAATGCACTAG | 4.3/2.2 | 3.3/1.4 | 0/0 | 2.2/1.9 | 5.6/6.1 | 3.9/4.8 |
| 1B-2B-3A-4B | GATCATTAATGCACTAG | 2.3/5.3 | 5.2/5.7 | 0/0.6 | 10.3/16.9 | 18.9/18.7 | 21.7/22.3 |
| 1D-2C-3C-4Db | GTTGGTCGATGCGATTA | 4.5/2.3 | 6.7/5.8 | 1.4/1.4 | 12.4/13.4 | 11.3/14.4 | 16.4/12.9 |
| 1B-2D-3G-4A | GATCGTTGATGCGCTAA | 4.5/7.0 | 0/0 | 0/0 | 0/0.4 | 0/0 | 0/0 |
| 1B-2D-3D-4F | GATCGTTGACGTGCTTA | 1.5/7.3 | 0/0.4 | 0/0 | 0.6/0.5 | 0/0 | 0.2/0.4 |
African-American specific haplotypes (1E and 3H) in Pearce et al. (12) could not be distinguished from haplotypes 1B and 3D, respectively, in this study.
Haplotypes where PROGINS allele is located.
Results from the global model suggested that haplotypes in block 3 were statistically significantly associated with endometrial cancer risk, even after Bonferroni adjustment (P = 0.002, supplementary Table 3 is available at Carcinogenesis Online). Compared with the most common haplotype in block 3, haplotype 3C (a haplotype containing the PROGINS allele) was associated with increased risk of endometrial cancer (OR = 1.34; 95% CI = 1.07–1.70; P = 0.013). In addition, haplotypes 3D and 3F were associated with increased endometrial cancer risk, whereas haplotype 3G was associated with decreased risk. Although haplotypes in block 2 were also associated with risk (P = 0.008), the association was attenuated (P = 0.077) when dropping the term for rare (<5%) haplotypes categorized together. However, significant heterogeneity was observed across ethnic groups for the effects of haplotypes 1A, 2D, 3B and 4A [P for interaction (5 degrees of freedom) = 0.004, 0.008, 0.021 and 0.014, respectively]. Results from ethnic/study group-specific analyses are summarized in supplementary Table 3 (available at Carcinogenesis Online).
When we restricted the analyses to whites, all haplotype blocks showed a significant association with endometrial cancer risk (Table II). The associations for haplotype blocks 1 through 3 remained statistically significant after Bonferroni adjustments, but the association in haplotype block 1 became weaker (P = 0.027) when dropping the sum of rare haplotypes (i.e. combining them with the reference haplotype, 1A). The PROGINS haplotypes (1D, 2C, 3C and 4D) were associated with 34–54% increased risk compared with the most common haplotypes in each block (Table II). In addition, haplotypes 1C, 2D, 3D, 3F, 4C and 4E were also associated with 38–77% increased risk of endometrial cancer. Compared with the long-range haplotype of 1A-2A-3B-4A, the long-range combination of PROGINS haplotypes (1D-2C-3C-4D) was associated with 49% increased risk (P = 0.016), and the long-range haplotype containing the 4E haplotype (1C-2D-3D-4E) was associated with 98% increased risk (P = 0.002; Table II).
Table II.
Association between PGR gene haplotypes and endometrial cancer risk among whitesa
| Haplotypes | Global model |
Single-haplotype model |
P for heterogeneityb | |||
| OR (95% CI) | P | OR (95% CI) | P | |||
| Block 1 | 0.010 (4 df) | |||||
| 1A | AAC | 1 (ref) | 0.80 (0.66–0.97) | 0.020 | 0.45 | |
| 1B | GAT | 1.10 (0.88–1.38) | 0.40 | 0.92 (0.76–1.11) | 0.37 | 0.69 |
| 1C | AAT | 1.38 (1.07–1.77) | 0.012 | 1.22 (0.98–1.51) | 0.077 | 0.34 |
| 1Dc | GTT | 1.42 (1.08–1.86) | 0.013 | 1.25 (0.99–1.57) | 0.067 | 0.34 |
| Block 2 | 0.013 (4 df) | |||||
| 2A | CGC | 1 (ref) | 0.87 (0.72–1.05) | 0.15 | 0.43 | |
| 2B | CAT | 0.99 (0.80–1.23) | 0.94 | 0.85 (0.71–1.02) | 0.084 | 0.44 |
| 2Cc | GGT | 1.37 (1.05–1.79) | 0.021 | 1.28 (1.01–1.61) | 0.042 | 0.12 |
| 2D | CGT | 1.40 (1.06–1.86) | 0.020 | 1.29 (1.00–1.66) | 0.048 | 0.19 |
| Block 3 | 0.005 (6 df) | |||||
| 3A | TAATGC | 1 (ref) | 0.86 (0.72–1.04) | 0.110 | 0.42 | |
| 3B | TGACGC | 0.86 (0.66–1.12) | 0.27 | 0.74 (0.58–0.95) | 0.014 | 0.83 |
| 3Cc | CGATGC | 1.34 (1.03–1.73) | 0.029 | 1.23 (0.98–1.56) | 0.079 | 0.050 |
| 3D | TGACGT | 1.47 (1.11–1.95) | 0.007 | 1.35 (1.05–1.74) | 0.020 | 0.029 |
| 3E | TAGTAC | 1.02 (0.72–1.46) | 0.90 | 0.91 (0.65–1.27) | 0.575 | 0.97 |
| 3F | TAGTGC | 1.40 (1.02–1.93) | 0.039 | 1.31 (0.97–1.75) | 0.079 | 0.27 |
| Block 4 | 0.020 (5 df) | |||||
| 4A | GCTAA | 1 (ref) | 0.73 (0.59–0.91) | 0.005 | 0.51 | |
| 4B | ACTAG | 1.22 (0.95–1.56) | 0.12 | 0.94 (0.78-1.14) | 0.55 | 0.88 |
| 4C | ACTAA | 1.40 (1.06–1.84) | 0.017 | 1.09 (0.88–1.36) | 0.41 | 0.83 |
| 4Dc | GATTA | 1.54 (1.15–2.06) | 0.004 | 1.22 (0.97–1.55) | 0.099 | 0.040 |
| 4E | GCCTA | 1.77 (1.21–2.59) | 0.004 | 1.40 (1.00–1.96) | 0.055 | 0.45 |
| Long-range | 0.040 (7 df) | |||||
| 1A-2A-3B-4A | AACCGCTGACGCGCTAA | 1 (ref) | 0.75 (0.59–0.96) | 0.020 | 0.93 | |
| 1C-2D-3D-4E | AATCGTTGACGTGCCTA | 1.98 (1.28–3.06) | 0.002 | 1.59 (1.09–2.32) | 0.018 | 0.40 |
| 1C-2A-3F-4C | AATCGCTAGTGCACTAA | 1.46 (0.99–2.14) | 0.055 | 1.17 (0.85–1.62) | 0.34 | 0.42 |
| 1A-2A-3E-4A | AACCGCTAGTACGCTAA | 1.00 (0.64–1.57) | 0.99 | 0.77 (0.51–1.15) | 0.19 | 0.88 |
| 1A-2B-3A-4B | AACCATTAATGCACTAG | 1.08 (0.67–1.74) | 0.75 | 0.84 (0.55–1.28) | 0.40 | 0.79 |
| 1B-2B-3A-4B | GATCATTAATGCACTAG | 1.25 (0.93–1.67) | 0.14 | 0.97 (0.79–1.20) | 0.79 | 0.85 |
| 1D-2C-3C-4Dc | GTTGGTCGATGCGATTA | 1.49 (1.08–2.06) | 0.016 | 1.18 (0.92–1.51) | 0.19 | 0.07 |
df, degrees of freedom.
All models were stratified by age (50 to <60, 60 to <70 and ≥70) and study group (MEC whites, CTS whites). Global model refers to a model containing all terms of haplotypes within a block except the reference haplotype. Sum of all rare haplotypes in each block was entered in each global model, but their estimates are not shown. Single-haplotype models refer to a model where one specific haplotype term is entered at a time, and therefore estimates the OR per copy of each haplotype compared with all other haplotypes combined. P-values are based on likelihood ratio tests.
Heterogeneity tests among whites, based on likelihood ratio tests (1 df) of product terms between each haplotype and study group (MEC whites and CTS whites) using single-haplotype model.
Haplotypes where PROGINS allele is located.
We next examined whether each haplotype association in whites differed between the two studies (MEC and CTS). The magnitude of association for haplotype 3D was larger among MEC whites [OR among MEC whites = 2.07; OR among CTS whites = 1.31 (supplementary Table 3 is available at Carcinogenesis Online), P for interaction = 0.029 (Table II)]. However, when we limited the analyses to Type I endometrial cancer cases, this heterogeneity was no longer statistically significant (P for interaction = 0.11; OR among MEC whites = 1.77; OR among CTS whites = 1.28). The associations of PROGINS haplotypes were stronger in CTS whites, and for one of the PROGINS haplotypes (4D), the P for interaction between study groups was 0.040 (Table II). Otherwise, the associations were similar in the two studies.
Consistent with the haplotype-based analyses, several htSNPs that exclusively represent one specific risk-associated haplotype in each block were also associated with endometrial cancer risk in overall analyses or in analyses restricted to whites (Table III). These include rs499590, rs516693 and rs561650, each representing 1A, 3D and 4E, respectively, as well as markers of the PROGINS allele, although these associations were not statistically significant after correcting for multiple tests. Another SNP, rs608995, in block 4 is partially linked to the PROGINS allele (rs1042838) in whites (r2 = 0.6) and Latinas (r2 = 0.4) and is shared by the risk haplotypes 4D (PROGINS haplotype) and 4E. The OR per variant allele of rs608995 was 1.16 (95% CI = 0.99–1.36; P = 0.068) in overall analyses and was slightly stronger among whites (OR = 1.30, 95% CI = 1.06–1.59; P = 0.012). When the PROGINS marker (rs1048238) and rs608995 were included in the same model, the ORs for rs608995 changed little, whereas the ORs for rs10482838 became attenuated (data not shown). Other htSNPs listed above (rs499590, rs516693 and rs561650) were not linked to the PROGINS allele (r2 < 0.1 in each ethnic group).
Table III.
Association between PGR gene polymorphisms and endometrial cancer riska
| Genotype | All combined |
Whites (MEC and CTS whites) |
|||||||
| N (cases/control) | OR (95% CI) | P | Pb | N (cases/control) | OR (95% CI) | P | Pc | ||
| Block 1 | |||||||||
| rs568157 | A/A | 229/899 | 1 (ref) | 98/227 | 1 (ref) | ||||
| A/G | 240/770 | 0.85 (0.67–1.08) | 0.19 | 190/428 | 0.97 (0.72–1.31) | 0.85 | |||
| G/G | 102/254 | 0.94 (0.69–1.28) | 0.70 | 90/176 | 1.08 (0.76–1.55) | 0.66 | |||
| per G | 0.95 (0.82–1.11) | 0.54 | 0.47 | 1.04 (0.87–1.24) | 0.68 | 0.51 | |||
| rs474320d | A/A | 423/1556 | 1 (ref) | 252/599 | 1 (ref) | ||||
| A/T | 139/342 | 1.19 (0.94–1.52) | 0.14 | 115/215 | 1.29 (0.98–1.70) | 0.072 | |||
| T/T | 14/29 | 1.33 (0.68–2.61) | 0.40 | 13/20 | 1.58 (0.76–3.30) | 0.22 | |||
| per T | 1.18 (0.96–1.45) | 0.11 | 0.47 | 1.28 (1.01–1.61) | 0.038 | 0.73 | |||
| rs499590 | T/T | 328/1121 | 1 (ref) | 195/359 | 1 (ref) | ||||
| T/C | 206/588 | 0.97 (0.78–1.2) | 0.76 | 155/353 | 0.82 (0.63–1.07) | 0.14 | |||
| C/C | 42/135 | 0.89 (0.61–1.32) | 0.57 | 29/96 | 0.63 (0.40–1.00) | 0.048 | |||
| per C | 0.95 (0.81–1.12) | 0.57 | 0.010 | 0.80 (0.66–0.98) | 0.027 | 0.79 | |||
| Block 2 | |||||||||
| rs3740753d | C/C | 425/1577 | 1 (ref) | 254/612 | 1 (ref) | ||||
| C/G | 136/323 | 1.25 (0.99–1.60) | 0.066 | 112/205 | 1.32 (1.00–1.74) | 0.050 | |||
| G/G | 16/30 | 1.58 (0.83–2.99) | 0.16 | 13/21 | 1.57 (0.76–3.26) | 0.22 | |||
| per G | 1.25 (1.03–1.54) | 0.028 | 0.40 | 1.30 (1.03–1.64) | 0.028 | 0.31 | |||
| rs481775 | G/G | 336/1228 | 1 (ref) | 173/355 | 1 (ref) | ||||
| G/A | 185/564 | 0.81 (0.65–1.02) | 0.077 | 154/361 | 0.85 (0.65–1.11) | 0.22 | |||
| A/A | 47/129 | 0.73 (0.50–1.07) | 0.11 | 45/112 | 0.78 (0.52–1.16) | 0.22 | |||
| per A | 0.84 (0.71–0.99) | 0.038 | 0.86 | 0.87 (0.72–1.05) | 0.14 | 0.38 | |||
| rs529359 | T/T | 184/515 | 1 (ref) | 159/316 | 1 (ref) | ||||
| T/C | 270/839 | 1.09 (0.87–1.37) | 0.44 | 184/382 | 0.99 (0.76–1.28) | 0.91 | |||
| C/C | 124/545 | 1.05 (0.78–1.42) | 0.75 | 36/115 | 0.67 (0.44–1.04) | 0.072 | |||
| per C | 1.03 (0.89–1.2) | 0.66 | 0.031 | 0.87 (0.72–1.05) | 0.16 | 0.53 | |||
| Block 3 | |||||||||
| rs565186d | T/T | 422/1562 | 1 (ref) | 253/606 | 1 (ref) | ||||
| T/C | 138/321 | 1.28 (1.00–1.62) | 0.048 | 113/205 | 1.32 (1.00–1.74) | 0.050 | |||
| C/C | 14/31 | 1.27 (0.66–2.47) | 0.48 | 11/22 | 1.20 (0.56–2.55) | 0.64 | |||
| per C | 1.22 (1.00–1.50) | 0.051 | 0.43 | 1.24 (0.98–1.56) | 0.072 | 0.062 | |||
| rs613120 | A/A | 188/709 | 1 (ref) | 96/222 | 1 (ref) | ||||
| A/G | 267/849 | 0.99 (0.79–1.25) | 0.96 | 192/410 | 1.07 (0.79–1.44) | 0.66 | |||
| G/G | 123/333 | 1.11 (0.84–1.48) | 0.47 | 92/186 | 1.16 (0.82–1.66) | 0.40 | |||
| per G | 1.05 (0.91–1.21) | 0.52 | 0.46 | 1.08 (0.90–1.29) | 0.40 | 0.84 | |||
| rs543215 | A/A | 299/830 | 1 (ref) | 251/557 | 1 (ref) | ||||
| A/G | 199/679 | 1.15 (0.92–1.44) | 0.21 | 120/251 | 1.12 (0.86–1.47) | 0.41 | |||
| G/G | 83/401 | 1.22 (0.84–1.77) | 0.30 | 10/19 | 1.20 (0.54–2.66) | 0.65 | |||
| per G | 1.12 (0.95–1.33) | 0.19 | 0.65 | 1.11 (0.88–1.41) | 0.37 | 0.44 | |||
| rs572483 | T/T | 294/1036 | 1 (ref) | 179/394 | 1 (ref) | ||||
| T/C | 225/717 | 0.99 (0.80–1.21) | 0.90 | 159/343 | 1.00 (0.77–1.30) | 0.99 | |||
| C/C | 54/143 | 1.15 (0.81–1.64) | 0.44 | 37/86 | 0.98 (0.64-1.51) | 0.93 | |||
| per C | 1.04 (0.89–1.21) | 0.64 | 0.36 | 0.99 (0.82–1.20) | 0.95 | 0.14 | |||
| rs601040 | G/G | 490/1627 | 1 (ref) | 332/714 | 1 (ref) | ||||
| G/A | 75/271 | 0.99 (0.74–1.33) | 0.97 | 44/113 | 0.86 (0.59–1.26) | 0.44 | |||
| A/A | 11/29 | 1.71 (0.81–3.62) | 0.16 | 4/7 | 1.22 (0.35–4.31) | 0.76 | |||
| per A | 1.09 (0.86–1.40) | 0.48 | 0.38 | 0.91 (0.66–1.28) | 0.60 | 0.90 | |||
| rs516693 | C/C | 411/1386 | 1 (ref) | 274/654 | 1 (ref) | ||||
| C/T | 152/494 | 1.13 (0.91–1.42) | 0.27 | 99/166 | 1.40 (1.04–1.87) | 0.024 | |||
| T/T | 16/46 | 1.43 (0.78–2.61) | 0.25 | 8/13 | 1.45 (0.58–3.59) | 0.42 | |||
| per T | 1.15 (0.95–1.39) | 0.14 | 0.16 | 1.34 (1.04–1.73) | 0.023 | 0.039 | |||
| Block 4 | |||||||||
| rs660541 | A/A | 170/599 | 1 (ref) | 105/215 | 1 (ref) | ||||
| A/G | 255/852 | 0.94 (0.74–1.19) | 0.61 | 180/396 | 0.92 (0.68–1.24) | 0.59 | |||
| G/G | 154/458 | 1.13 (0.85–1.49) | 0.40 | 96/217 | 0.93 (0.66–1.30) | 0.66 | |||
| per G | 1.06 (0.92–1.22) | 0.43 | 0.31 | 0.96 (0.81–1.14) | 0.65 | 0.84 | |||
| rs1042838d | C/C | 429/1569 | 1 (ref) | 259/615 | 1 (ref) | ||||
| C/A | 134/313 | 1.25 (0.98–1.59) | 0.073 | 109/199 | 1.29 (0.98–1.71) | 0.074 | |||
| A/A | 14/31 | 1.26 (0.65–2.43) | 0.50 | 11/22 | 1.19 (0.56–2.52) | 0.66 | |||
| per A | 1.20 (0.98–1.48) | 0.075 | 0.45 | 1.22 (0.96–1.54) | 0.10 | 0.038 | |||
| rs561650 | T/T | 429/1569 | 1 (ref) | 326/740 | 1 (ref) | ||||
| T/C | 134/313 | 1.21 (0.93–1.58) | 0.15 | 53/90 | 1.34 (0.92–1.94) | 0.13 | |||
| C/C | 14/31 | 2.27 (0.97–5.27) | 0.058 | 4/4 | 2.70 (0.65-11.24) | 0.17 | |||
| per C | 1.28 (1.02–1.62) | 0.036 | 0.76 | 1.39 (1.00–1.94) | 0.051 | 0.45 | |||
| rs608995 | A/A | 315/1100 | 1 (ref) | 203/509 | 1 (ref) | ||||
| A/T | 218/729 | 1.08 (0.88–1.32) | 0.46 | 145/290 | 1.24 (0.95–1.61) | 0.11 | |||
| T/T | 42/99 | 1.55 (1.04–2.30) | 0.032 | 28/39 | 1.85 (1.09–3.12) | 0.022 | |||
| per T | 1.16 (0.99–1.36) | 0.068 | 0.53 | 1.30 (1.06–1.59) | 0.012 | 0.65 | |||
| rs1870019 | A/A | 347/1256 | 1 (ref) | 182/389 | 1 (ref) | ||||
| A/G | 183/538 | 0.85 (0.68–1.07) | 0.17 | 157/354 | 0.91 (0.70–1.19) | 0.50 | |||
| G/G | 42/100 | 0.92 (0.61–1.37) | 0.67 | 39/85 | 0.97 (0.63–1.48) | 0.88 | |||
| per G | 0.91 (0.77–1.08) | 0.28 | 0.74 | 0.96 (0.79–1.16) | 0.65 | 0.67 | |||
| Inter-block | |||||||||
| rs10895068 (+331 C/T) | C/C | 520/1769 | 1 (ref) | 328/726 | 1 (ref) | ||||
| C/T | 55/129 | 1.09 (0.77–1.55) | 0.61 | 51/90 | 1.20 (0.83–1.75) | 0.33 | |||
| T/T | 3/3 | 2.80 (0.54–14.5) | 0.22 | 2/2 | 2.18 (0.30–15.9) | 0.44 | |||
| per T | 1.16 (0.84–1.60) | 0.37 | 0.79 | 1.23 (0.87–1.75) | 0.25 | 0.19 | |||
All models were stratified by age (50 to <60, 60 to <70 and ≥70) and ethnic/study groups. P-values are based on Wald’s test.
P for heterogeneity (5 df) across ethnic/study groups on allele-dosage models.
P for heterogeneity among whites (1 df) across study group on allele-dosage models.
All are tightly linked with the PROGINS allele.
We assessed whether these associations varied across environmental risk factors using the entire study population. No significant interaction with use of postmenopausal HT (none, ever use), oral contraceptive use (none, ever use) or BMI (<30, ≥30 kg/m2) was observed. However, when we limited this evaluation to postmenopausal women, the interaction between BMI and haplotype 1A was statistically significant: the OR for women with BMI ≥30 kg/m2 was 1.57 (95% CI: 1.11–2.23), whereas the OR for women with BMI < 30 kg/m2 was 0.85 (95% CI: 0.69–1.05, P for interaction = 0.015). However, this heterogeneity was not observed when analyses were limited to whites. Instead, among all white women (premenopausal and postmenopausal), the effect of haplotype 3D was heterogeneous across BMI categories: the OR for haplotype 3D was 2.65 (95% CI = 1.40–5.00) in women with BMI ≥ 30 kg/m2, whereas the OR for women with BMI < 30 kg/m2 was 1.18 (95% CI = 0.89–1.58, P for interaction = 0.027).
Discussion
In this study, we utilized a haplotype-based approach to examine whether genetic variation at the PGR locus is associated with endometrial cancer risk. Haplotype frequencies within each ethnic group were similar to what has been reported previously (12). The PROGINS allele and the haplotypes containing the PROGINS allele (1D, 2C, 3C and 4D) were associated with increased endometrial cancer risk. Haplotypes 3D and 3F were also associated with endometrial cancer risk. Haplotype 4E had the largest OR estimate; although the global model for haplotype block 4 was not statistically significant after Bonferroni adjustment. When combining the block-specific haplotypes, the long-range haplotype 1C-2D-3D-4E showed the strongest association. rs608995, one of the SNPs shared by the haplotype 4E and the PROGINS haplotype, had an OR of 1.30 per T allele (P = 0.012) among whites.
In vitro studies have suggested that the function of the PROGINS allele differs depending on cell and tissue type (8,9). In ovarian cancer cell lines, but not in breast cancer cell lines, the Alu insertion element of the PROGINS allele reduced stability of the PGR transcript, and the V660L amino acid substitution reduced transactivation activity of the progesterone receptor (8), although the data are not entirely consistent (9). In the human endometrium, however, one study reported that transcription of PGR in women carrying the PROGINS allele was similar to the level in other women (8), although it is not known whether the PROGINS allele affects stability or signaling of progesterone receptor. We observed in this study that the PROGINS allele association was weakened when another SNP in block 4 (rs608995) was included in the same model. It suggests that the risk association of the PROGINS allele or the PROGINS haplotypes could have been driven by rs608995 (or a causal allele that rs608995 is tagging).
Further, considering that haplotypes across blocks were tightly correlated (12), the effect of several haplotypes could be explained by haplotypes in other blocks. In particular, haplotypes 1C-2D-3D-4E are tightly correlated, and therefore the risk associations of 1C, 2D and 3D observed among whites could be explained by 4E, which contains the rs608995 variant allele. In fact, the observed association of other htSNPs as well became weaker after adjusting for rs608995. Our observation that the combination of 1C-2D-3D-4E showed the stronger association in whites (OR = 1.98), than another haplotype containing 1C (1C-2A-3F-4C; OR = 1.46), is compatible with this possibility, although the scarcity of other combinations of 1C, 2D, 3D and 4E precludes a definitive conclusion.
Our data suggest that the association between PGR and endometrial cancer risk is heterogeneous across ethnic groups. This may be due to statistical fluctuation because of the small numbers in all groups except whites. However, it could also be because the haplotypes or htSNPs in this study were markers of other causal alleles in this gene (or outside this gene) and that these linkage patterns differ across ethnic groups. In addition, the heterogeneity might have been observed because unidentified environmental effect modifiers might have different population distributions across ethnic/study groups.
Across the entire study population, we did not observe any statistically significant interaction between genetic variation in PGR and several environmental factors such as use of exogenous hormones (menopausal HT or OCs) and BMI. However, the main mechanism by which BMI increases endometrial cancer risk depends on the woman’s menopausal status (2,35,36). Prior to menopause, obesity is associated with irregular or anovulatory menstrual cycles, which may result in insufficient progesterone to oppose estrogen-driven proliferation of endometrium (2,35,36). After menopause, obese women have higher postmenopausal estrogen levels than non-obese women due to peripheral conversion of androgens to estrogens in fat tissue (37). In this study, analyses among postmenopausal women showed that haplotype 1A was associated with increased risk only in obese women (BMI ≥ 30 kg/m2). Thus, our results suggest that obese women with certain PGR haplotypes are more susceptible to the otherwise subtle effect of PGR genetic variation (for example haplotype 1A after menopause).
While a case–control study nested in the Nurses Health Study has reported a statistically significant increased risk of endometrial cancer associated with rs10895068, a putatively functional SNP in the promoter region of the PGR (+331 C/T) (19), we and others (23,24) did not replicate this finding. This SNP has been associated with increased ovarian cancer risk in young women (≤50 years) in one study (13), but not in the large replication effort of Ovarian Cancer Association Consortium (17).
Results from a pooled analyses of two population-based ovarian cancer case–control studies suggest that PGR haplotypes are associated with ovarian cancer risk (12). Given that progesterone, an established protective factor for endometrial cancer, is hypothesized to also reduce risk of ovarian cancer (38), it is plausible that the role of the PGR in these cancers is similar. Consistent with this hypothesis, our risk estimates for each PGR haplotype are in the same direction as that which Pearce et al. (12) observed in their phased haplotype analyses. While the large Ovarian Cancer Association Consortium effort did not observe an overall association with a PROGINS allele or rs608995 (not investigated as haplotypes), they did find an association limited specifically to the endometrioid type of ovarian cancer (17). Considering the shared histological characteristics of this subtype of ovarian cancer with endometrial cancer, and that ∼10% of endometrioid ovarian cancer is associated with concurrent endometrial carcinoma (39–41), our finding that genetic variation in the PGR is associated with the risk of endometrial cancer warrants further investigation.
Conclusions
Our findings suggest that genetic variation in the PGR locus is associated with endometrial cancer risk. While we cannot identify the true causal allele in this study, our results indicate that overall genetic variation in the PGR locus may influence endometrial cancer risk.
Supplementary material
Supplementary Tables 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
MEC supported by National Cancer Institute (CA54281, CA63464); National Cancer Institute Career Development Award grant (CA116543 to V.W.S.); CTS supported by National Cancer Institute (R01 CA91019, R01 CA77398); Contract 97-10500 from the California Breast Cancer Research Fund; The collection of cancer incidence data used in this study was supported by the California Department of Health Services as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885, the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract N01-PC-35136 awarded to the Northern California Cancer Center, contract N01-PC-35139 awarded to the University of Southern California and contract N02-PC-15105 awarded to the Public Health Institute; Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement #U55/CCR921930-02 awarded to the Public Health Institute.
Supplementary Material
Acknowledgments
For the MEC: We are most indebted to the participants of the MEC for their participation and commitment to the study. We thank Loreall Pooler, David Wong for their laboratory assistance and Dr Kristine Monroe, Grace Sheng and Hank Huang for their support with the data management.
For the CTS: The funding sources did not contribute to the design or conduct of the study, nor to the writing or submission of this manuscript. The ideas and opinions expressed herein are those of the authors and endorsement by the State of California, Department of Health Services, the National Cancer Institute and the Centers for Disease Control and Prevention or their contractors and subcontractors is not intended nor should be inferred.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BMI
body mass index
- CI
confidence interval
- CTS
California Teachers Study
- HWE
Hardy–Weinberg equilibrium
- htSNP
haplotype-tagging SNP
- HT
hormone therapy
- LD
linkage disequilibrium
- MAF
minor allele frequency
- MEC
multiethnic cohort
- OR
odds ratio
- PGR
progesterone receptor
- SNP
Single nucleotide polymorphism
References
- 1.Ries LAG, et al. SEER Cancer Statistics Review, 1975–2005. Bethesda, MD: National Cancer Institute; 2008. [Google Scholar]
- 2.Key TJ, et al. The dose-effect relationship between ‘unopposed’ oestrogens and endometrial mitotic rate: its central role in explaining and predicting endometrial cancer risk. Br. J. Cancer. 1988;57:205–212. doi: 10.1038/bjc.1988.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olson SH, et al. Variants in estrogen biosynthesis genes, sex steroid hormone levels, and endometrial cancer: a HuGE review. Am. J. Epidemiol. 2007;165:235–245. doi: 10.1093/aje/kwk015. [DOI] [PubMed] [Google Scholar]
- 4.King RJ, et al. Assessment of oestrogen and progestin effects on epithelium and stroma from pre- and postmenopausal endometria. J. Steroid Biochem. 1981;15:175–181. doi: 10.1016/0022-4731(81)90273-9. [DOI] [PubMed] [Google Scholar]
- 5.Pike MC, et al. Progestins and menopause: epidemiological studies of risks of endometrial and breast cancer. Steroids. 2000;65:659–664. doi: 10.1016/s0039-128x(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 6.Pike MC, et al. Estrogen-progestin replacement therapy and endometrial cancer. J. Natl Cancer Inst. 1997;89:1110–1116. doi: 10.1093/jnci/89.15.1110. [DOI] [PubMed] [Google Scholar]
- 7.Rowe SM, et al. Ovarian carcinoma-associated TaqI restriction fragment length polymorphism in intron G of the progesterone receptor gene is due to an Alu sequence insertion. Cancer Res. 1995;55:2743–2745. [PubMed] [Google Scholar]
- 8.Romano A, et al. The PROGINS polymorphism of the human progesterone receptor diminishes the response to progesterone. J. Mol. Endocrinol. 2007;38:331–350. doi: 10.1677/jme.1.02170. [DOI] [PubMed] [Google Scholar]
- 9.Agoulnik IU, et al. A germline variation in the progesterone receptor gene increases transcriptional activity and may modify ovarian cancer risk. J. Clin. Endocrinol. Metab. 2004;89:6340–6347. doi: 10.1210/jc.2004-0114. [DOI] [PubMed] [Google Scholar]
- 10.Leite DB, et al. Progesterone receptor (PROGINS) polymorphism and the risk of ovarian cancer. Steroids. 2008;73:676–680. doi: 10.1016/j.steroids.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 11.McKenna NJ, et al. A germline TaqI restriction fragment length polymorphism in the progesterone receptor gene in ovarian carcinoma. Br. J. Cancer. 1995;71:451–455. doi: 10.1038/bjc.1995.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearce CL, et al. Clarifying the PROGINS allele association in ovarian and breast cancer risk: a haplotype-based analysis. J. Natl Cancer Inst. 2005;97:51–59. doi: 10.1093/jnci/dji007. [DOI] [PubMed] [Google Scholar]
- 13.Romano A, et al. Two functionally relevant polymorphisms in the human progesterone receptor gene (+331 G/A and progins) and the predisposition for breast and/or ovarian cancer. Gynecol. Oncol. 2006;101:287–295. doi: 10.1016/j.ygyno.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 14.Tong D, et al. Analysis of the human progesterone receptor gene polymorphism progins in Austrian ovarian carcinoma patients. Int. J. Cancer. 2001;95:394–397. doi: 10.1002/1097-0215(20011120)95:6<394::aid-ijc1070>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 15.Lancaster JM, et al. No relationship between ovarian cancer risk and progesterone receptor gene polymorphism in a population-based, case-control study in North Carolina. Cancer Epidemiol. Biomarkers Prev. 2003;12:226–227. [PubMed] [Google Scholar]
- 16.Manolitsas TP, et al. No association of a 306-bp insertion polymorphism in the progesterone receptor gene with ovarian and breast cancer. Br. J. Cancer. 1997;75:1398–1399. doi: 10.1038/bjc.1997.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearce CL, et al. Progesterone receptor variation and risk of ovarian cancer is limited to the invasive endometrioid subtype: results from the Ovarian Cancer Association Consortium pooled analysis. Br. J. Cancer. 2008;98:282–288. doi: 10.1038/sj.bjc.6604170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pijnenborg JM, et al. Aberrations in the progesterone receptor gene and the risk of recurrent endometrial carcinoma. J. Pathol. 2005;205:597–605. doi: 10.1002/path.1738. [DOI] [PubMed] [Google Scholar]
- 19.De Vivo I, et al. A functional polymorphism in the promoter of the progesterone receptor gene associated with endometrial cancer risk. Proc. Natl Acad. Sci. USA. 2002;99:12263–12268. doi: 10.1073/pnas.192172299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Junqueira MG. Progesterone receptor (PROGINS) polymorphism and the risk of endometrial cancer development. Int. J. Gynecol. Cancer. 2007;17:229–232. doi: 10.1111/j.1525-1438.2006.00767.x. [DOI] [PubMed] [Google Scholar]
- 21.Mulac-Jericevic B, et al. Reproductive tissue selective actions of progesterone receptors. Reproduction. 2004;128:139–146. doi: 10.1530/rep.1.00189. [DOI] [PubMed] [Google Scholar]
- 22.Mulac-Jericevic B, et al. Subgroup of reproductive functions of progesterone mediated by progesterone receptor-B isoform. Science. 2000;289:1751–1754. doi: 10.1126/science.289.5485.1751. [DOI] [PubMed] [Google Scholar]
- 23.Dossus L, et al. No association between progesterone receptor gene +331G/A polymorphism and endometrial cancer. Cancer Epidemiol. Biomarkers Prev. 2006;15:1415–1416. doi: 10.1158/1055-9965.EPI-06-0215. [DOI] [PubMed] [Google Scholar]
- 24.Rebbeck TR, et al. Estrogen sulfation genes, hormone replacement therapy, and endometrial cancer risk. J. Natl Cancer Inst. 2006;98:1311–1320. doi: 10.1093/jnci/djj360. [DOI] [PubMed] [Google Scholar]
- 25.Kolonel LN, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Setiawan VW, et al. Two estrogen-related variants in CYP19A1 and endometrial cancer risk: a pooled analysis in the Epidemiology of Endometrial Cancer Consortium. Cancer Epidemiol. Biomarkers Prev. 2009;18:242–247. doi: 10.1158/1055-9965.EPI-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haiman CA, et al. A comprehensive haplotype analysis of CYP19 and breast cancer risk: the Multiethnic Cohort. Hum. Mol. Genet. 2003;12:2679–2692. doi: 10.1093/hmg/ddg294. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein L, et al. High breast cancer incidence rates among California teachers: results from the California Teachers Study (United States) Cancer Causes Control. 2002;13:625–635. doi: 10.1023/a:1019552126105. [DOI] [PubMed] [Google Scholar]
- 29.Razavi P, et al. Long-term postmenopausal hormone therapy and endometrial cancer. Cancer Epidemiol. Biomarkers Prev. 2010;19:475–483. doi: 10.1158/1055-9965.EPI-09-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 31.Excoffier L, et al. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 32.Stram DO, et al. Choosing haplotype-tagging SNPS based on unphased genotype data using a preliminary sample of unrelated subjects with an example from the Multiethnic Cohort Study. Hum. Hered. 2003;55:27–36. doi: 10.1159/000071807. [DOI] [PubMed] [Google Scholar]
- 33.Amant F, et al. Endometrial cancer. Lancet. 2005;366:491–505. doi: 10.1016/S0140-6736(05)67063-8. [DOI] [PubMed] [Google Scholar]
- 34.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 35.Hale GE, et al. Endometrial cancer: hormonal factors, the perimenopausal “window of risk,” and isoflavones. J. Clin. Endocrinol. Metab. 2002;87:3–15. doi: 10.1210/jcem.87.1.8132. [DOI] [PubMed] [Google Scholar]
- 36.Kaaks R, et al. Obesity, endogenous hormones, and endometrial cancer risk: a synthetic review. Cancer Epidemiol. Biomarkers Prev. 2002;11:1531–1543. [PubMed] [Google Scholar]
- 37.Key TJ, et al. Body mass index, serum sex hormones, and breast cancer risk in postmenopausal women. J. Natl Cancer Inst. 2003;95:1218–1226. doi: 10.1093/jnci/djg022. [DOI] [PubMed] [Google Scholar]
- 38.Risch HA. Hormonal etiology of epithelial ovarian cancer, with a hypothesis concerning the role of androgens and progesterone. J. Natl Cancer Inst. 1998;90:1774–1786. doi: 10.1093/jnci/90.23.1774. [DOI] [PubMed] [Google Scholar]
- 39.Storey DJ, et al. Endometrioid epithelial ovarian cancer: 20 years of prospectively collected data from a single center. Cancer. 2008;112:2211–2220. doi: 10.1002/cncr.23438. [DOI] [PubMed] [Google Scholar]
- 40.Kline RC, et al. Endometrioid carcinoma of the ovary: retrospective review of 145 cases. Gynecol. Oncol. 1990;39:337–346. doi: 10.1016/0090-8258(90)90263-k. [DOI] [PubMed] [Google Scholar]
- 41.Soliman PT, et al. Synchronous primary cancers of the endometrium and ovary: a single institution review of 84 cases. Gynecol. Oncol. 2004;94:456–462. doi: 10.1016/j.ygyno.2004.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.