Abstract
Intracellular delivery of functional macromolecules using peptide transduction domains (PTDs) is an exciting technology with both experimental and therapeutic applications. Recent data indicate that PTD-mediated transduction occurs via fluid-phase macropinocytosis involving an intracellular pH drop to ∼5. Nitrilotriacetic acid (NTA)-coordinated metals avidly bind hexahistidine-tagged macromolecules, including peptides and proteins. Histidine’s imidazole ring has a pKa of 6, making this an attractive target for the biological pH drop of PTD-mediated macropinocytotic delivery. The objective of this study was to develop a pH-sensitive PTD delivery peptide (NTA3-PTD). We demonstrate the in vitro function of this novel peptide by delivering fluorescently labeled peptides (1.6 kDa) and functional enzymes, β-galactosidase (119 kDa) and Cre recombinase (37 kDa). Furthermore, the NTA3-PTD peptide was able to deliver functional Cre recombinase in an in vivo mouse model.
Intracellular delivery of functional macromolecules using peptide transduction domains is an exciting technology with both bench and bedside applications. Recent data1−3 indicate that transduction occurs via fluid-phase endocytosis involving an intracellular pH drop. Nitrilotriacetic acid (NTA) is a pH-sensitive moiety capable of binding hexahistidine (6xHis)-tagged proteins via a coordinate covalent bond for pH >6. The objective of this study was to develop a pH-sensitive peptide transduction domain (NTA-PTD, Figure 1A) capable of (1) forming a noncovalent bond with 6xHis-tagged cargo, (2) inducing cellular uptake, and (3) dissociating from the cargo following internalization via the intracellular endosomal pH drop. We demonstrate the in vitro function of this novel peptide by delivering fluorescently labeled peptides (1.6 kDa) and functional enzymes (β-galactosidase and Cre recombinase, 119 and 37 kDa, respectively.) Furthermore, this peptide was able to deliver functional Cre recombinase in an in vivo mouse model.
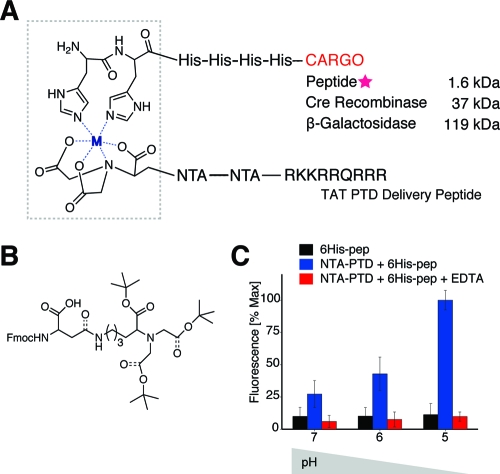
Figure 1.
Cargo delivery via pH-sensitive NTA-PTD. (A) NTA3-PTD forms a coordinate covalent bond with metal ions and the imidazole ring on histidine residues. (B) Fmoc-protected lysine derivative. (C) pH-sensitive release of 6xHis-rhodamine via NTA3-PTD.
Intracellular transduction via PTDs occurs via macropinocytosis, a specialized form of fluid-phase endocytosis.1−6 Cationic PTDs bind the cell surface and become internalized within macropinosomes.3,6 Once internalized, these endosomes undergo an intracellular pH drop to pH ∼4.5.7,8 Previous studies have utilized NTA, an aminopolycarboxylic acid capable of sequestering all metal ions,(9) in a variety of bioengineering.10−14 In the presence of metal ion, a coordinate bond can be formed between one molecule of NTA and two imidazole rings on adjacent histidine residues in a protein or other macromolecular cargo.(15) Thousands of recombinant proteins have been constructed for purification using 6xHis tags. However, for the vast majority of these proteins, intracellular bioavailability is limited due to size restrictions for diffusion through the plasma membrane.(4) Therefore, we sought to create a pH-sensitive noncovalent link between NTA and the PTD TAT for intracellular delivery of 6xHis-tagged proteins.
To facilitate synthesis of NTA-PTD, we synthesized monomeric Fmoc-protected NTA-derivatized aspartic acid(16) (Figure 1B and Supporting Information) by ring-opening condensation of (N-(9-fluorenyl)methoxycarbonyl (Fmoc)-aspartic anhydride(17) with an NTA derivative of lysine.(14) This NTA fragment was incorporated into the PTD using Fmoc solid-phase peptide synthesis with a Symphony Quartet peptide synthesizer (Rainin) by SPPS using l-amino acids with Fmoc protection on an amidated support (EMD). Crude products were purified by reversed-phase HPLC and confirmed by MALDI mass spectroscopy (Supporting Information). Rhodamine-labeled 6xHis peptide (Rhod-GGSGG-HHHHHH-G) was prepared by reaction with tetramethylrhodamine while on solid support. The sequence of the NTA3-PTD peptide is (NTA)-G-(NTA)-G-(NTA)-GGSGG-RKKRRQRRRG.
To assess the pH-sensitivity of NTA3-PTD, 2.2 nmol of 6xHis-rhodamine and NTA3-PTD plus 3 equiv of NiSO4 (1:1 molar ratio with single NTA3-PTD peptide) were mixed and added to washed ion-exchange S-Beads. Bead−peptide mixtures were washed three times and aliquoted into separate tubes before addition of 10 mM HEPES with 400 mM NaCl, pH 5−7. Beads were pelleted via centrifugation, and supernatant fluorescence was measured. We found pH-sensitive release of 6xHis-rhodamine (Figure 1C) in the physiologically relevant range of pH 5−7.
We tested the ability of NTA3-PTD to deliver rhodamine-labeled 6xHis-peptide to cells. NTA3-PTD-6xHis-rhodamine complexes were prepared and added to cells for 30 min, followed by three washes with PBS and heparin sulfate (0.5 mg/mL) and trypsinization.(3) Flow cytometry (Figure 2A) demonstrates robust cell association of 6xHis-rhodamine via NTA3-PTD, but not with 6xHis-rhodamine either alone or in trans with TAT peptide.
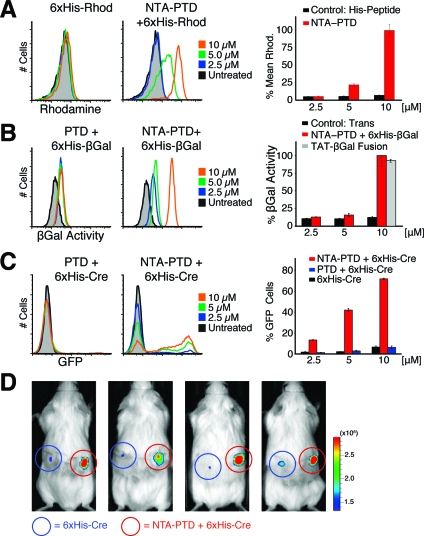
Figure 2.
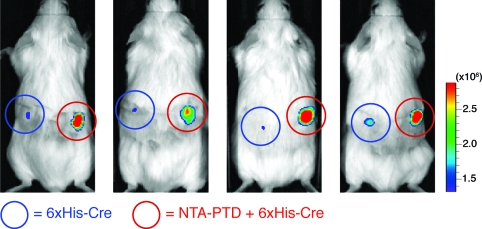
In vitro and in vivo function of NTA3-PTD. (A) H1299 cells were treated with NTA3-PTD + Ni plus 6xHis-rhodamine peptide or 6xHis-rhodamine peptide alone for 30 min, washed, and assayed by flow cytometry. (B) H199 cells were treated with NTA3-PTD plus 6xHis-β-Gal, 6xHis-β-Gal alone, or positive control TAT-β-Gal fusion protein for 2 h, washed, and incubated with C12FDG substrate for 45 min, followed by flow cytometry. (C) CHO-ZEG cells were treated with NTA3-PTD plus 6xHis-Cre or 6xHis-Cre alone for 30 min, washed, trypsinized, and replated. After 24 h, cells were assayed for GFP expression via flow cytometry. (D) ROSA-26R-LoxP-STOP-LoxP-luciferase mice were subcutaneously injected with 50 μL of 20 μM NTA3-PTD plus 6xHis-Cre or 6xHis-Cre alone and then assayed for luciferase expression at 4 days.
NTA3-PTD delivered the functional enzyme β-galactosidase (β-Gal, 119 kDa) into H1299 cells. Recombinant 6xHis-tagged β-Gal was prepared,(18) and cells were treated for 2 h. After washing and trypsinization, β-Gal activity was detected using the fluorescent substrate C12FDG, which yields FITC fluorescence after cleavage. Substantial β-Gal activity was observed (Figure 2B) following coordination with NTA3-PTD but not with control β-Gal in trans with TAT peptide.
In vitro delivery of 6xHis-Cre recombinase was facilitated by NTA3-PTD, resulting in genomic recombination of loxP sites to excise a stop sequence and initiate GFP expression (Figure 2C) in CHO reporter cells(6) in a dose-dependent manner. 6xHis-Cre alone or in trans with TAT peptide induced negligible GFP expression. We did not detect any cytotoxicity with NTA-TAT delivery of peptides or proteins.
Finally, we observed in vivo delivery of Cre recombinase via NTA3-PTD in a luciferase-reporter mouse model.(19) Mice (transgenic ROSA26 loxP-Stop-loxP luciferase) were injected with either 6xHis-Cre (left side) or NTA3-PTD 6xHis-Cre (right side) and assayed for luciferase expression after 4 days. NTA3-PTD coordination resulted in genomic recombination and substantial luciferase activity (Figure 2D).
We developed a pH-sensitive protein transduction domain, NTA3-PTD, capable of delivering functional 6xHis-tagged proteins into cells both in vitro and in vivo. This peptide immediately makes possible the intracellular delivery of thousands of existing 6xHis-tagged proteins.(15) Future application of this technology may address both scientific and therapeutic challenges.
Acknowledgments
We thank B. Meade for critical input. This work was supported by a Specialized Center of Research (SCOR) grant from the Leukemia & Lymphoma Society, the California Institute for Regenerative Medicine, a grant from an anonymous donor, and the Howard Hughes Medical Institute (all to S.F.D.).
Supporting Information Available
Detailed monomer and peptide synthesis and purification methods; experimental methods; and Scheme S1. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Nakase I.; Niwa M.; Takeuchi T.; Sonomura K.; Kawabata N.; Koike Y.; et al. Mol. Ther. 2004, 10, 1011–1022. [DOI] [PubMed] [Google Scholar]
- Wadia J. S.; Stan R. V.; Dowdy S. F. Nat. Med. 2004, 10, 310–315. [DOI] [PubMed] [Google Scholar]
- Kaplan I. M.; Wadia J. S.; Dowdy S. F. J. Controlled Release 2005, 102, 247–253. [DOI] [PubMed] [Google Scholar]
- Gump J. M.; Dowdy S. F. Trends Mol. Med. 2007, 13, 443–448. [DOI] [PubMed] [Google Scholar]
- Nakase I.; Takeuchi T.; Tanaka G.; Futaki S. Adv. Drug Delivery Rev. 2008, 60, 598–607. [DOI] [PubMed] [Google Scholar]
- Gump J. M.; June R. K.; Dowdy S. F. J. Biol. Chem. 2010, 285, 1500–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaughnessy L. M.; Hoppe A. D.; Christensen K. A.; Swanson J. A. Cell Microbiol. 2006, 8, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang A. W.; Oestergaard K.; Myers J. T.; Swanson J. A. J. Leuk. Biol. 2000, 68, 487–494. [PubMed] [Google Scholar]
- IUPAC Pure Appl. Chem. 1982, 54, 2693–2758. [Google Scholar]
- Boonyarattanakalin S.; Athavankar S.; Sun Q.; Peterson B. R. J. Am. Chem. Soc. 2006, 128, 386–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith C. R.; Jaworski J.; Sheng M.; Lippard S. J. J. Am. Chem. Soc. 2006, 128, 418–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein W. M.; Ross B. P.; Landsberg M. J.; Levy D.; Hankamer B.; McGeary R. P. J. Org. Chem. 2009, 74, 1473–1479. [DOI] [PubMed] [Google Scholar]
- Kim S. H.; Jeyakumar M.; Katzenellenbogen J. A. J. Am. Chem. Soc. 2007, 129, 13254–13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S.; Reichel A.; Brock R.; Tampe R.; Piehler J. J. Am. Chem. Soc. 2005, 127, 10205–10215. [DOI] [PubMed] [Google Scholar]
- Crowe J.; Dobeli H.; Gentz R.; Hochuli E.; Stuber D.; Henco K. Methods Mol. Biol. 1994, 31, 371–387. [DOI] [PubMed] [Google Scholar]
- Morgan J. R.; Lyon R. P.; Maeda D. Y.; Zebala J. A. Nucleic Acids Res. 2008, 36, 3522–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper W. J.; Waters M. L. Org. Lett. 2005, 7, 3825–3828. [DOI] [PubMed] [Google Scholar]
- Schwarze S. R.; Ho A.; Vocero-Akbani A.; Dowdy S. F. Science 1999, 285, 1569–1572. [DOI] [PubMed] [Google Scholar]
- Safran M.; Kim W. Y.; Kung A. L.; Horner J. W.; DePinho R. A.; Kaelin W. G. Mol. Imaging 2003, 2, 297–302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.