Abstract
Radiation affects several cellular and molecular processes, including double strand breakage and modifications of sugar moieties and bases. In outer space, protons are the primary radiation source that poses a range of potential health risks to astronauts. On the other hand, the use of proton irradiation for tumor radiation therapy is increasing, as it largely spares healthy tissues while killing tumor tissues. Although radiation-related research has been conducted extensively, the molecular toxicology and cellular mechanisms affected by proton irradiation remain poorly understood. Therefore, in this study, we irradiated rat lung epithelial cells with different doses of protons and investigated their effects on cell proliferation and death. Our data show an inhibition of cell proliferation in proton-irradiated cells with a significant dose-dependent activation and repression of reactive oxygen species and antioxidants glutathione and superoxide dismutase, respectively, compared with control cells. In addition, the activities of apoptosis-related genes such as caspase-3 and -8 were induced in a dose-dependent manner with corresponding increased levels of DNA fragmentation in proton-irradiated cells compared with control cells. Together, our results show that proton irradiation alters oxidant and antioxidant levels in cells to activate the apoptotic pathway for cell death.
Keywords: Antioxidant, Cell Death, DNA Damage, Oxidative Stress, Reactive Oxygen Species (ROS)
Introduction
The mechanism by which radiation causes damage to human tissue is by ionization of atoms. Radiation is known to interfere with cellular functions at all levels of cell organization. Studies of people exposed to high doses of radiation have shown that there is a risk of cancer associated with high doses. The specific types of cancers associated with radiation exposure include leukemia, multiple myeloma, breast cancer, lung cancer, and skin cancer. Animals exposed to 55-MeV protons had a high incidence of malignant brain tumors (1). Accumulating evidence suggests that radiation-induced genomic instability is a nontargeted phenomenon triggered by radiation that may initiate and likely contribute to radiation-induced carcinogenesis (2). In outer space, protons are the primary radiation source. They pose a range of potential health risks to astronauts, including work performance and psychological as well as somatic functions (3). Literature on this subject has shown that space radiation induces oxidative stress-mediated cell damage in astronauts after space flight (4). On the other hand, proton therapy is the most precise, efficient, and advanced form of radiation treatment today. It irradiates primarily the tumor site, leaving surrounding healthy tissue and organs intact. Cell death induced by a proton beam has also been identified as apoptosis (5). Irradiation studies on neural cells showed the depletion of precursor cells in vivo, and reductions of these critical cells are believed to impair neurogenesis and cognition (6).
Radiation-induced DNA damage investigation is one of the most important areas of study in modern biology, but the information available on the effects of ionizing radiation, particularly protons, is still very limited. Our recently published and previous observations on proton-irradiated mouse brain showed an alteration of oxidative stress-mediated apoptosis-inducing genes and a differential expression pattern of DNA damage- and oxidative stress-related genes (7, 8). In this study, we developed an in vitro system using cultured rat lung epithelial (LE)2 cells and studied proton-mediated cell killing. We observed an increased level of reactive oxygen species (ROS) and lipid peroxidation (LPO), followed by inhibition of antioxidants glutathione (GSH) and superoxide dismutase (SOD) in proton-irradiated cells compared with control cells. In addition, a significant activation of cell death-related genes such as caspase-3 and -8 was detected in these cells. Together, these observations suggest that in both in vivo and in vitro model systems, proton irradiation causes similar effects by inducing oxidative stress, which in turn activates the signaling cascade for DNA and cell damage.
EXPERIMENTAL PROCEDURES
Cell Line and Proton Exposure
Rat LE cells (RL-65, CRL-10354) were purchased from American Type Culture Collection (Manassas, VA); cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin; and incubated at 37 °C in a humidified chamber with 5% CO2. Exponentially growing LE cells were split and reseeded 1 day prior to irradiation. LE cells were irradiated with 250-MeV protons at different doses (0.1, 1, 2, and 4 gray (Gy)) at the Loma Linda Radiation Facility, cultured, and harvested at different time points depending on the experimental procedure. For comparative purposes, control cells were cultured similarly and harvested along with irradiated cells.
Cell Viability Assay
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) system is a simple, accurate, and reproducible means of measuring the activity of living cells via mitochondrial dehydrogenase activity. The key component is MTT. Solutions of MTT solubilized in tissue culture media or balanced salt solutions, without phenol red, are yellowish in color. Mitochondrial dehydrogenases of viable cells cleave the tetrazolium ring, yielding purple MTT formazan crystals, which are insoluble in aqueous solutions. The crystals can be dissolved in acidified isopropyl alcohol. The resulting purple solution is spectrophotometrically measured. An increase in cell number results in an increase in the amount of MTT formazan formed and an increase in absorbance. The cytotoxicity assay was performed using MTT as described previously (9). LE cells grown overnight were irradiated with different doses of protons and cultured for 36 h. The irradiated LE cells were washed with phosphate-buffered saline (PBS), MTT was added to a final concentration of 125 μg/ml, and incubation was continued for another 3 h. The formazan formed inside the cells was extracted using acidic methanol, and the absorbance was measured at 570 nm. Live/dead cell assays were performed essentially as described previously (9). Briefly, 105 LE cells were cultured for 24 h, irradiated with different doses of protons, and incubated for another 24 h. The irradiated cells were stained with 5 μm ethidium homodimer and 5 μm calcein-AM (Molecular Probes, Eugene, OR) and incubated for 1 h at 37 °C. The stained cells were analyzed under a Zeiss fluorescence microscope and photographed.
Detection of ROS
The measurement of intracellular ROS was performed as described previously (10). Briefly, equal numbers of rat LE cells (2000 cells/well) were seeded in 96-well plates and grown for 24 h. The cells were then incubated with 10 μm dichlorofluorescein(5,6)carboxy-2,7′-dichlorodihydroxy fluorescein diacetate (H2DCF-DA) for 3 h, washed with PBS, and exposed to different doses of protons, and the intensity of fluorescence was measured at excitation and emission wavelengths of 485 and 527 nm, respectively. The readings were taken immediately after irradiation and continued up to 3 h. The 2.5 h of post-irradiated readings shown in Fig. 2A are expressed as fluorescence units.
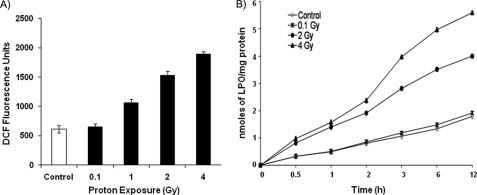
FIGURE 2.
Activation of ROS and LPO in proton-irradiated LE cells. A, equal numbers of rat LE cells were seeded in 96-well plates and cultured for 24 h. Cells were then incubated with 10 μm H2DCF-DA for 3 h in Hanks' balanced salt solution and exposed to different doses of protons (0.1, 1, 2 and 4 Gy), and ROS dichlorofluorescein (DCF) fluorescence was measured. The 2.5 h of post-irradiated values are expressed as dichlorofluorescein fluorescence units. B, LE cells were equally seeded, grown for 24 h, and irradiated with protons (0.1, 1, 2, and 4 Gy), and proteins were extracted at different time points as shown. Fifty microgram of protein from each dose and time point was used to measure LPO levels. Values are means ± S.D. of three experiments performed independently.
Assay for LPO
Proton-induced LPO was determined using a kit from Cayman Chemical as described previously (11). Equal numbers of LE cells (4 × 105 cells/well) were seeded in 6-well plates and grown for 24 h. Following incubation, cells were washed with PBS, exposed to different doses of protons, and incubated for 12 h. The cells were then scraped with PBS and sonicated. Fifty micrograms of cell lysate and methanol were mixed and centrifuged after adding precooled chloroform at 1500 × g for 10 min. The supernatant containing hydroperoxides was collected and used for the estimation of thiobarbituric acid-reactive malondialdehyde. The chromogen formed was detected at a wavelength of 500 nm.
Detection of Glutathione
Glutathione is the key antioxidant present in most cells (12). The reduced intracellular GSH activity was measured using a glutathione assay kit following the instructions provided by the manufacturer (Cayman Chemical Co., Ann Arbor, MI). In brief, LE cells (4 × 105 cells/well) were seeded in a 6-well plate and grown for 24 h. Next, cells were irradiated with different doses of protons, and incubation was continued for 12 h. The cells were then scraped and homogenized using PBS. Fifty micrograms of protein was deproteinized using 5% 5-sulfosalicylic acid dihydrate solution and 400 mm sodium carbonate, followed by 1:8 dilutions with phosphate/EDTA buffer and incubation for 10 min at room temperature. The supernatant was then treated with 5,5′-dithiobis(2-nitrobenzoic acid), and incubation was continued for another 10 min. The GSH activity was measured at an absorbance of 415 nm.
SOD Assay
The assay was performed using the superoxide dismutase kit from Trevigen, Inc. (catalog no. 7500-100-K; Gaithersburg, MD). Fifty micrograms of protein extracts was used to assay total SOD activities following the manufacturer's protocol. Briefly, SOD reaction buffer was mixed with xanthine solution followed by nitro blue tetrazolium solution, the sample proteins isolated 12 h after proton irradiation were added, and the absorbance was set to zero at 550 nm. Finally, xanthine oxidase (XOD) solution was added to each sample, and readings were taken at 550 nm every 30 s for a period of 5 min. The total SOD activity was calculated based on the manufacturer's formula.
Western Blotting
Whole cell extracts were prepared at different time points from proton-irradiated and control cells using mammalian cell extraction buffer (BioVision, Inc., Mountain View, CA) as described previously (7). Equal amounts of proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membrane. The membrane was blocked in 5% nonfat dry milk powder in PBS containing 0.1% IGEPAL, probed with the appropriate primary antibody followed by secondary antibody conjugated to horseradish peroxidase, and developed using Pierce detection solution (Thermo Scientific, Rockford, IL). The following antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA): caspase-3 (sc-7272), caspase-8 (sc-7890), SOD-1 (sc-11407), and SOD-2 (sc-18503). Anti-β-actin antibody was from Sigma (A5441).
Caspase-3 and Caspase-8 Activity Assay
The cleavage activities of caspase-3 and -8 substrates DEVD-AFC and IETD-AFC (Santa Cruz Biotechnology) were measured according to the manufacturer's protocol. Briefly, protein extracts were prepared from 24-h proton-irradiated and control cells, followed by estimation using Coomassie Plus protein assay reagent (catalog no. 1856210, Thermo Fisher Scientific). The DEVD-AFC and IETD-AFC substrates were added to 50 μg of protein extract for detecting caspase-3 and -8, respectively, and incubated for 1 h at 37 °C. The formation of free AFC in the extract was measured at an excitation wavelength of 400 nm and an emission wavelength of 495 nm. The values of experimental samples were compared with those of control samples and expressed as fluorescence units.
Genomic DNA Isolation and DNA Fragmentation
Genomic DNA was isolated from proton-irradiated (1, 2, and 4 Gy) and control cells using the ApoTarget quick apoptotic DNA ladder kit (catalog no. SKU-KHO1021, Invitrogen) according to the procedure specified by the manufacturer. Briefly, equal numbers of 24-h post-irradiated and control cells were homogenized in Tris/EDTA buffer, followed by mixing with Enzyme A solution and incubation at 37 °C for 10 min. Enzyme B solution was added to the Enzyme A solution and incubated for an additional 30 min at 50 °C. To this, 0.1 volume of ammonium acetate and 2.5-fold cold ethanol were added and precipitated at −20 °C for 15 min, followed by centrifugation to obtain the DNA pellet. The pellet was washed with 70% cold ethanol and centrifuged again. Finally, the DNA was air-dried, resuspended in DNA suspension buffer, and analyzed on 1.2% agarose gel.
Statistical Analysis
Data are expressed as means ± S.D., and statistical significance was analyzed by Student's t test. A p value <0.05 was considered statistically significant.
RESULTS
Proton Irradiation Inhibits Cell Viability in LE Cells
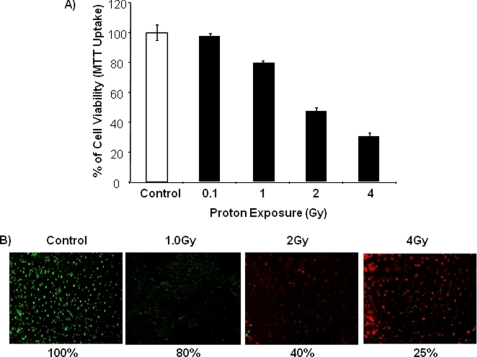
Previous observations from our group have shown the induction of DNA and tissue damage in 2-Gy proton-irradiated mouse brain tissues (7, 8). Therefore, in this study, we tested the effect of protons on the in vitro system of rat LE cells. Here, we observed a significant dose-dependent inhibition of cell proliferation in proton-irradiated cells compared with control cells, which was evident by standard MTT dye uptake cell viability assay (Fig. 1A). Notably, LE cells exposed to a lower dose (0.1 Gy) did not show any change in cell proliferation compared with untreated control cells. To reconfirm cell viability, a live/dead cell assay was performed, and the results showed a similar effect, with an increased numbers of dead cells in proton-irradiated cells compared with control cells in a dose-dependent manner (Fig. 1B).
FIGURE 1.
Proton exposure inhibits cell viability. A, equal numbers of rat LE cells were seeded and grown for 24 h. Cells were then exposed to different doses of protons (0.1, 1, 2, and 4 Gy) and cultured for 36 h. Cell viability was assayed based on MTT dye uptake (absorbance at 570 nm), and the values were calculated based on the control. The experiment was carried out in triplicate, and means ± S.D. are shown. B, LE cells were seeded equally, exposed to protons (1, 2, and 4 Gy), and cultured for 24 h, and live and dead cells were visualized based on the dye uptake method and photographed. The percentage of live cells is shown below.
Activation of ROS and LPO in Proton-irradiated Cells
Because we observed an inhibition of cell proliferation in proton-irradiated cells, we were interested in investigating whether proton irradiation alters oxidative stress to inhibit cell proliferation. As shown in Fig. 2A, proton irradiation activated ROS significantly in a dose-dependent manner compared with control cells. For example, ROS levels were 3-fold higher at 4 Gy, 2.5-fold higher at 2 Gy, and 1.8-fold higher at 1 Gy than in control cells. However, a lower dose of 0.1 Gy did not affect ROS production. It has been shown in the literature that LPO, a standard biomarker for oxidative stress, is activated during external stress (12). Therefore, we measured LPO levels at different time points in proton-irradiated cells and found a dose-dependent increased level of LPO in these cells that directly correlated with increased ROS levels observed under proton-irradiated conditions (Fig. 2B).
Inhibition of Antioxidants in Proton-irradiated Cells
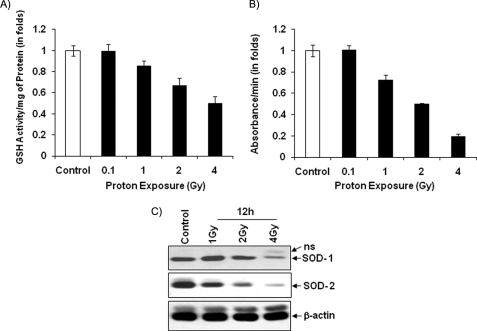
The presence of many antioxidants has been reported in cells as a protective mechanism during oxidative stress and apoptotic cell death (13). Glutathione and SOD are the major antioxidants in cells, and they balance ROS levels to maintain normal cellular functions when cells are under external stress (14). Because an alteration of oxidant levels has been observed in proton-irradiated cells, antioxidant levels were also analyzed. A significant dose-dependent inhibition of GSH and SOD activities was detected in proton-irradiated cells compared with control cells. (Fig. 3, A and B). Notably, 50 and 80% reductions of GSH and SOD activities, respectively, were detected in 4-Gy proton-irradiated cells compared with control cells. In addition, Fig. 3C shows the dose-dependent reduction of SOD-1 and -2 proteins level in 12-h post-irradiated cells compared with control cells.
FIGURE 3.
Inhibition of antioxidants in proton-exposed LE cells. A, proteins were extracted from 12-h post-irradiated and control LE cells, and 50 μg of total proteins was used to assay glutathione activity as described under “Experimental Procedures.” B, SOD activity was assayed by mixing SOD reaction buffer, xanthine, and nitro blue tetrazolium solution with 50 μg of proteins isolated from 12-h post-irradiated (different doses of protons) and control LE cells following the manufacturer's formula. The glutathione and SOD results are means ± S.D. of three independent experiments. C, protein extracts were prepared from 12-h post-irradiated (different doses) and control cells and analyzed for SOD-1 and -2 proteins using specific antibodies. β-Actin was used as an internal loading control. ns, nonspecific band.
Proton Irradiation Induces Caspase-3 and Caspase-8 in LE Cells
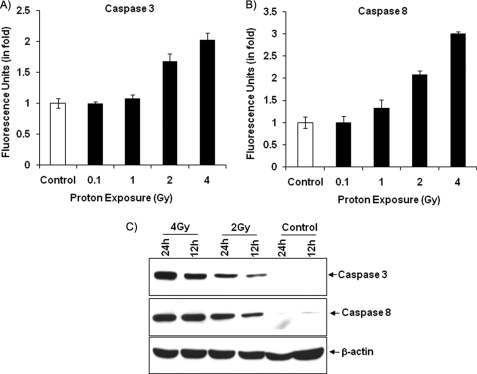
DNA damage is caused mainly by external factors, particularly radiation and chemicals (15). Our group has used external stress-inducing agents in an in vitro system and shown an induction of oxidative stress-mediated cell death through activation of caspase-3 and -8 (12, 16). Here, we performed the same strategy in proton-irradiated cells to determine whether caspase levels are affected by proton irradiation. Fig. 4A shows a significantly increased level of caspase-3 activities at higher proton doses (2 and 4 Gy) compared with control cells and at a lower dose (0.1 Gy). Furthermore, the activation of caspase-3 was dependent on the early activation of either caspase-8 or -9. We continued our investigation of caspase-8 activity and observed a dose-dependent increase in caspase-8 activity in 2-Gy (2-fold) and 4-Gy (3-fold) proton-irradiated cells compared with control cells (Fig. 4B). Our protein data for caspase-3 and -8 also show an increased level of these proteins in 2- and 4-Gy proton-irradiated cells compared with control cells (Fig. 4C).
FIGURE 4.
Induction of caspase-3 and -8 activities in proton-exposed cells. A and B, protein extracts isolated from 24-h post-irradiated (0.1, 1, 2, and 4 Gy) and control cells were mixed with DEVD-AFC and IETD-AFC for caspase-3 and -8 activities, respectively, and the formation of free AFC in the mixture was measured at an excitation wavelength of 400 nm and an emission wavelength of 495 nm. The experimental values were compared with the control and are expressed as fluorescence units. Values are means ± S.D. of three independent experiments. C, protein extracts were prepared from 12- and 24-h 2- and 4-Gy proton-irradiated and control cells and analyzed for caspase-3 and 8 proteins using specific antibodies. β-Actin was used as an internal loading control.
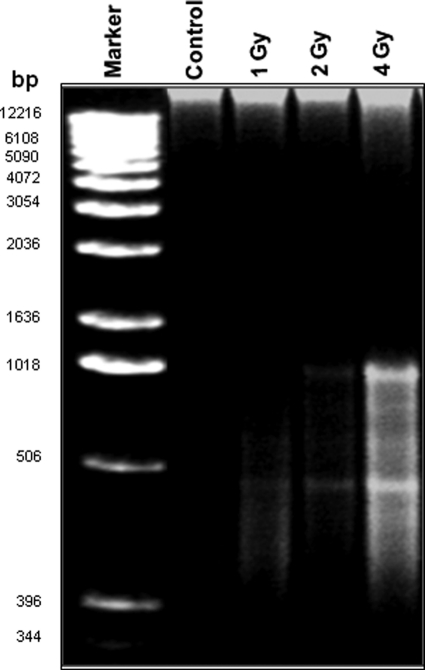
Proton-induced DNA Fragmentation in LE Cells
It has long been known that radiation induces DNA damage. Also, our recently published report on irradiated mouse brain showed significant DNA damage compared with the control brain (7). DNA fragmentation is a key feature of programmed cell death, and the process is characterized by the activation of endogenous endonucleases with subsequent cleavage of chromatin DNA into internucleosomal fragments of 180 bp and multiples thereof (15). There are many methods to assess the DNA fragmentation caused by the apoptosis event, and the most standard technique involves detection of DNA ladders using agarose gel electrophoresis. Fig. 5 shows an apoptotic ladder of genomic DNA isolated from different doses of proton-irradiated cells and control cells. Notably, we detected more DNA fragmentation in 4-Gy proton-irradiated cells than in control cells and at other lower doses.
FIGURE 5.
Proton irradiation induces DNA fragmentation. Genomic DNA was extracted from 24-h proton-irradiated (1, 2, and 4 Gy) and control cells using the quick apoptotic DNA ladder kit according to the manufacturer's instructions, analyzed on 1.2% agarose gel, and photographed.
DISCUSSION
Proton radiation therapy offers a number of potential advantages over conventional (photon) γ-radiation therapy for cancer because of a more localized delivery of the radiation dose. It is generally assumed that the relative biological effectiveness of protons is 1.1 cobalt Gy eq (sometimes referred to as Gy eq). At hospital-based proton facilities, a dose of 1.8–2 cobalt Gy eq is often used per fraction, with one fraction delivered per day over a period of 5 days/week for 5–7 weeks, depending on the type and location of the tumor as well as other considerations. Thus, the dose of 2 Gy used in this study is approximately equivalent to one fraction of proton irradiation delivered during therapy. However, it should be noted that although the total doses delivered to the intended target volume during treatment are much higher than those used here, normal cells located at a distance from the target may well be exposed to 0.1–4 Gy during or by the end of therapy. For example, at the Loma Linda University Medical Center, there are seven proton energies used for patient treatment, and all are within the range of 100–250 MeV. Protons are also the most abundant type of particles encountered by astronauts during missions both in low Earth orbit and to the Moon or Mars (16). The dose received by astronauts during a low Earth orbit mission that lasts for several months can be as high as 0.1 sievert. In space, proton energies have a much wider range and dependence on certain conditions. For example, proton energies could be up to several 100 MeV during a solar particle event and up to several 1000 MeV in galactic cosmic rays (17). Research on radiation has been ongoing for several decades, but the information available on protons is minimal. Currently, we lack critical knowledge of the molecular mechanism of proton irradiation to assess human radiation exposure, and this is a high priority, as it would enable better determination of health risks.
Recently, a published report on a mouse system showed an activation of DNA damage- and apoptosis-related genes in 2-Gy proton-irradiated brain tissues compared with control brain tissues (7). Therefore, in this work, we extended our radiation studies using different doses of protons in an in vitro cell line system, particularly rat LE cells, and investigated radiation-induced oxidative stress followed by cell death. Our cell viability observations from this study shows that proton irradiation inhibits cell proliferation in a dose-dependent manner in LE cells (Fig. 1). Several reports have shown that ROS could be activated through various stressors such as uranium, tobacco smoke, carbon nanotubes, high glucose, tumor necrosis factor-α, chemicals, radiation, etc., and the elevated levels of ROS regulate a broad array of signal transduction pathways that control various biological processes, including gene expression and cell growth, differentiation, and apoptosis (18, 19). In agreement with the above-mentioned ROS activation, our data from this study also show a dose-dependent activation of ROS in proton-irradiated cells compared with control cells (Fig. 2A).
In biological systems, oxidant and antioxidant levels are maintained in a balanced state. For example, when cells are under stress, antioxidants are produced at higher concentrations to neutralize the ROS level for normal cellular activities. Evidence in the literature also suggests that overexpression of SOD protects cells from ROS-mediated apoptosis (20, 21). Here, the data from proton-irradiated cells show an inhibition of antioxidant levels compared with control cells, suggesting that proton irradiation blocks the production of antioxidants through an unidentified mechanism to kill the irradiated cells.
The ultimate effect of radiation is to induce DNA damage and cell death (14). A recent report showed that cell death induced by protons is apoptosis rather than necrosis or autophagy, and the apoptosis process is mediated through the activation of caspases (6). Very recently, we also showed that mice exposed to 2-Gy protons demonstrated caspase-3 and -8 activation, fragmented DNA, and significant tissue damage with altered expression of DNA damage- and oxidative stress-signaling genes compared with control brain tissues (7, 8). Consistent with our earlier in vivo observation, the current in vitro cell line data also show a similar effect with an induction of caspase-3 and -8 and significant levels of fragmented DNA in proton-irradiated cells compared with control cells (Fig. 4, A and B), which confirms that proton-irradiated cells induce the apoptotic pathway through activation of caspase-3 and -8 for cell death. Even though we detected a 2–3-fold increase in caspase activity in 4-Gy proton-irradiated cells, the caspase-3 protein seemed to be increased much more from 12 to 24 h in 4-Gy proton-irradiated cells. At present, we do not know why there is a dramatic increase in caspase-3 at these time points. In addition, an apoptotic ladder study showed more DNA fragmentation at 4 Gy compared with control cells and at other lower doses, suggesting that the proton-mediated cell death is dependent on dose and exposure time. It is important to note here that our DNA fragmentation and caspase activation data correlate with our cell viability data showing more dead cells at 24 h of 4-Gy proton irradiation. Studies on proton-mediated cellular signaling pathways such as NF-κB, AP-1, etc., and their effect on oxidative stress-induced cell death are under way to dissect the exact molecular mechanism of proton-mediated cell killing.
Taken together, our findings show that there is an involvement of the oxidative stress pathway leading to functional activation of cell death-related caspase-3 and -8 following proton exposure that complements the response triggered by DNA damage. These data advance our knowledge of the cellular and molecular effects of proton irradiation and could be useful in improving current proton therapy protocols.
This work was supported, by NIH (1P20MD001822-1) and NASA Grants NNX08BA47A and NCC-1-02038.
- LE
- lung epithelial
- ROS
- reactive oxygen species
- LPO
- lipid peroxidation
- SOD
- superoxide dismutase
- Gy
- gray
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PBS
- phosphate-buffered saline
- AFC
- 7-amino-4-trifluoromethylcoumarin.
REFERENCES
- 1.Wood D. (1991) Radiat. Res. 126, 132–140 [PubMed] [Google Scholar]
- 2.Little J. B. (2000) Carcinogenesis 21, 397–404 [DOI] [PubMed] [Google Scholar]
- 3.Gridley D. S., Coutrakon G. B., Rizvi A., Bayeta E. J., Luo-Owen X., Makinde A. Y., Baqai F., Koss P., Slater J., Pecaut M. J. (2008) Radiat. Res. 169, 280–287 [DOI] [PubMed] [Google Scholar]
- 4.Stein T. P., Leskiw M. J. (2000) Am. J. Physiol. Endocrinol. Metab. 278, E375–E382 [DOI] [PubMed] [Google Scholar]
- 5.Di Pietro C., Piro S., Tabbì G., Ragusa M., Di Pietro V., Zimmitti V., Cuda F., Anello M., Consoli U., Salinaro E. T., Caruso M., Vancheri C., Crimi N., Sabini M. G., Cirrone G. A., Raffaele L., Privitera P., Pulvirenti A., Giugno R., Ferro A., Cuttone G., Lo Nigro S., Purrello R., Purrello F., Purrello M. (2006) Apoptosis 11, 57–66 [DOI] [PubMed] [Google Scholar]
- 6.Giedzinski E., Rola R., Fike J. R., Limoli C. L. (2005) Radiat. Res. 164, 540–544 [DOI] [PubMed] [Google Scholar]
- 7.Baluchamy S., Zhang Y., Ravichandran P., Ramesh V., Sodipe A., Hall J. C., Jejelowo O., Gridley D. S., Wu H., Ramesh G. T. (April11, 2010) Mol. Cell. Biochem. 10.1007/s11010-010-0451-4 [DOI] [PubMed] [Google Scholar]
- 8.Baluchamy S., Zhang Y., Ravichandran P., Ramesh V., Sodipe A., Hall J. C., Jejelowo O., Gridley D. S., Wu H., Ramesh G. T. (2010) In Vitro Cell. Dev. Biol. Anim. 10.1007/s11626-010-9330-2 [DOI] [PubMed] [Google Scholar]
- 9.Manna S. K., Sarkar S., Barr J., Wise K., Barrera E. V., Jejelowo O., Rice-Ficht A. C., Ramesh G. T. (2005) Nano Lett. 5, 1676–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar S., Sharma C., Yog R., Periyakaruppan A., Jejelowo O., Thomas R., Barrera E. V., Rice-Ficht A. C., Wilson B. L., Ramesh G. T. (2007) J. Nanosci. Nanotechnol. 7, 584–592 [PMC free article] [PubMed] [Google Scholar]
- 11.Wise K. C., Manna S. K., Yamauchi K., Ramesh V., Wilson B. L., Thomas R. L., Sarkar S., Kulkarni A. D., Pellis N. R., Ramesh G. T. (2005) In Vitro Cell. Dev. Biol. Anim. 41, 118–123 [DOI] [PubMed] [Google Scholar]
- 12.Ravichandran P., Periyakaruppan A., Sadanandan B., Ramesh V., Hall J. C., Jejelowo O., Ramesh G. T. (2009) J. Biochem. Mol. Toxicol. 23, 333–344 [DOI] [PubMed] [Google Scholar]
- 13.Sharma C. S., Sarkar S., Periyakaruppan A., Barr J., Wise K., Thomas R., Wilson B. L., Ramesh G. T. (2007) J. Nanosci. Nanotechnol. 7, 2466–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiseman H., Halliwell B. (1996) Biochem. J. 313, 17–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burrows C. J., Muller J. G. (1998) Chem. Rev. 98, 1109–1152 [DOI] [PubMed] [Google Scholar]
- 16.NCRP (1989) Guidance on Radiation Received in Space Activities (Report 98), National Council on Radiation Protection and Measurements, Bethesda, MD [Google Scholar]
- 17.Schimmerling W. (2003) Gravit. Space Biol. Bull. 16, 5–10 [PubMed] [Google Scholar]
- 18.Periyakaruppan A., Kumar F., Sarkar S., Sharma C. S., Ramesh G. T. (2007) Arch. Toxicol. 81, 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engel R. H., Evens A. M. (2006) Front. Biosci. 1, 300–312 [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y., Kiningham K. K., Lin S. M., Clair D. K. (2001) Antioxid. Redox Signal. 3, 375–386 [DOI] [PubMed] [Google Scholar]
- 21.Dominguez-Rodriguez J. R., Gomez-Contreras P. C., Hernandez-Flores G., Lerma-Diaz J. M., Carranco A., Cervantes-Munguia R., Orbach-Arbouys S., Bravo-Cuella A. (2001) Anticancer Res. 21, 1869–1872 [PubMed] [Google Scholar]