Abstract
Endoplasmic reticulum (ER)-associated degradation (ERAD) is a quality control system for newly synthesized proteins in the ER; nonfunctional proteins, which fail to form their correct folding state, are then degraded. The cytoplasmic peptide:N-glycanase is a deglycosylating enzyme that is involved in the ERAD and releases N-glycans from misfolded glycoproteins/glycopeptides. We have previously identified a mutant plant toxin protein, RTA (ricin A-chain nontoxic mutant), as the first in vivo Png1 (the cytoplasmic peptide:N-glycanase in Saccharomyces cerevisiae)-dependent ERAD substrate. Here, we report a new genetic device to assay the Png1-dependent ERAD pathway using the new model protein designated RTL (RTA-transmembrane-Leu2). Our extensive studies using different yeast mutants identified various factors involved in RTL degradation. The degradation of RTA/RTL was independent of functional Sec61 but was dependent on Der1. Interestingly, ER-mannosidase Mns1 was not involved in RTA degradation, but it was dependent on Htm1 (ERAD-related α-mannosidase in yeast) and Yos9 (a putative degradation lectin), indicating that mannose trimming by Mns1 is not essential for efficient ERAD of RTA/RTL. The newly established RTL assay will allow us to gain further insight into the mechanisms involved in the Png1-dependent ERAD-L pathway.
Keywords: Endoplasmic Reticulum (ER), Glycoprotein, Proteasome, Ubiquitin, Yeast, ERAD, PNGase
Introduction
In eukaryotes, the folding and assembly of nascent polypeptide chains in the endoplasmic reticulum (ER)3 are closely monitored by an ER quality control system, which ensures that only correctly folded/assembled proteins are delivered to their respective destinations (1). Terminally misfolded proteins are destroyed by a process called ER-associated degradation (ERAD) (2, 3). ERAD involves the extraction of proteins from the ER to the cytosol followed by proteasome-mediated degradation. Depending on the position of the misfolded region, the recognition step can occur on the luminal side (ERAD-L), the cytosolic side (ERAD-C), or in the ER membrane itself (ERAD-M) (4). Several key components of the ER quality control and ERAD systems have been identified to date, but the mechanisms by which the substrates are selectively recognized remain elusive.
The cytoplasmic peptide:N-glycanase (PNGase; Png1 in yeast) is a deglycosylating enzyme involved in the ERAD process (5). Many ERAD substrates are presumed to be deglycosylated by PNGase during the degradation process (6, 7), and the involvement of PNGase activity in the antigen presentation process has been suggested in mammalian cells (8). However, most of the tested ERAD substrates considered susceptible for deglycosylation were efficiently degraded, even when PNGase activity was inhibited (9), making it difficult to assess the biological significance of this enzyme in the ERAD process. Nevertheless, our finding that the ricin A-chain nontoxic mutant (RTA), an ERAD-L substrate, is degraded in a Png1-dependent fashion in Saccharomyces cerevisiae (10) prompted us to further characterize the mechanisms involved in the PNGase-dependent ERAD pathway.
To gain further insight into the Png1-dependent ERAD, a new genetic device was developed using the new model protein designated RTL. RTL is a membrane protein that consists of a luminal RTA, a type I transmembrane domain, and a cytoplasmic Leu2 protein. Using this system, we can assay the efficiency of RTL degradation by measuring the growth of cells in synthetic media containing limited amounts of leucine, i.e. only when the degradation of RTL is compromised, as cells contain sufficient quantities of Leu2 protein to support growth in media with low leucine concentrations. In our assay, RTL was, as with RTA, degraded in a Png1-dependent fashion, indicating that RTL is a good model substrate to study the PNGase-dependent ERAD pathway. Moreover, RTL lacking the first glycosylation site (RTL(N10Q)) did not require Png1 for its efficient degradation, indicating that the effect of Png1 was N-glycan-dependent.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
The yeast strains used in this study are listed in Table 1. The deletion of the RAD23 and DSK2 genes using the NATR gene was carried out with pAG25, as described previously (11). pAG25 was kindly provided by Dr. Claude A. Jakob (ETH, Zurich, Switzerland). Standard yeast media and genetic techniques were used (12, 13). The sec18-1 strains and isogenic wild-type cells were kindly provided by Dr. Yoshifumi Jigami (AIST, Japan).
TABLE 1.
Yeast strains used in this study
| Strain | Harboring plasmid | Genotype | Source |
|---|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Laboratory stock | |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Laboratory stock | |
| W303-1a | MATaleu2-3,112 his3-11 ade2-1 ura3-1 trp1-1 can1-100 | Laboratory stock | |
| W303-1b | MATα leu2-3,112 his3-11 ade2-1 ura3-1 trp1-1 can1-100 | Laboratory stock | |
| KA31-1a | MATα ura3 leu2 trp1 his3 | Dr. Yoshifumi Jigami | |
| Sec61 | pDQ1-TRP | MATasec61::HIS3 leu2-3,112 his3-11 ade2-1 ura3-1 trp1-1 can1-100 | This study |
| sec61-41 | psec61-41-TRP | MATasec61::HIS3 leu2-3,112 his3-11 ade2-1 ura3-1 trp1-1 can1-100 | This study |
| CMY765 | MATacim5-1 ura3-52 leu2Δ1 his3Δ200 | 80 | |
| cim5-1 png1Δ | MATacim5-1 png1::URA3 ura3-52 leu2Δ1 his3Δ200 FOAR | 81 | |
| png1Δ | png1::KanMX4 BY4741 | 16 | |
| rad23Δ | rad23::NatMX BY4741 | This study | |
| dsk2Δ | dsk2::NatMX BY4741 | This study | |
| cue1Δ | cue1::KanMX6 BY4741 | Open Biosystems | |
| der1Δ | der1::KanMX6 BY4742 | Open Biosystems | |
| der1Δ | der1::KanR W303-1a | 81 | |
| doa10Δ | doa10::KanMX6 BY4741 | Open Biosystems | |
| hrd1Δ | hrd1::KanMX6 BY4741 | Open Biosystems | |
| hrd3Δ | hrd3::KanMX6 BY4741 | Open Biosystems | |
| htm1Δ | htm1::KanMX6 BY4741 | Open Biosystems | |
| ubx2Δ | ubx2::KanMX6 BY4741 | Open Biosystems | |
| ufd2Δ | ufd2::KanMX6 BY4741 | Open Biosystems | |
| usa1Δ | usa1::KanMX6 BY4741 | Open Biosystems | |
| yos9Δ | yos9::KanMX6 BY4741 | Open Biosystems | |
| mns1Δ | mns1::KanMX6 BY4741 | Open Biosystems | |
| sec18-1 | Sec18-1 KA31-1a | Dr. Yoshifumi Jigami |
Plasmids
To generate pDQ1-TRP and psec61-41-TRP, the SEC61 and sec61-41 regions of pDQ1 and psec61-41 (14) were cloned into pRS314 as follows. pDQ1 and psec61-41 were kindly provided by Dr. Randy W. Schekman (University of California, Berkeley). The SEC61 and sec61-41 regions of pDQ1 and psec61-41 were cut out by HindIII/EcoRI digestion and were cloned into the equivalent site of pBluescript. These plasmids thus obtained were digested with SalI/EcoRI, and fragments were cloned into the equivalent sites of pRS314 vector.
To generate pRS316-GAL4RTL, the CPY* region of pRS316-GAL4CTL* (15) was replaced by the RTA as follows. pRS316-GAL4CTL* was a kind gift from by Dr. Dieter H. Wolf (University of Stuttgart). The RTA fragment was amplified from pRS315-GPDKar2-RTA (7) using the following primers: RTL-forward, 5′-TTTCAGCTTCATCTCCAGATTGTGTCTACGTAATGCACGCCATCATTTTAAGAGAGGACAGAGAAGCAAGCCTCCTGAAAGATGTTTTTCAACAGACTAAG-3′; and RTL-reverse, 5′-GAATTGACATTAGCTAAGTAATCATTGGTTGTAGATATTTGACAGAAACTACAGGTGTCAGTGGCATTTTCATCCTCGAGATGATGATGATGATGATGAAA-3′. This PCR product was cotransformed in yeast with the linearized pRS316-GAL4CTL* and digested by BglII and Bsu36I, and the transformants were grown on an SC-Ura plate to select the yeast strain. Colonies with the correct homologous recombination were selected with the colony PCR, and the plasmids isolated from the colonies were sequenced. DNA sequencing was carried out using PCR-based dideoxy termination methods using BigDye version 3.1 and the 3100 DNA sequencer (ABI). pRS316-GAL4RTL(N10Q), pRS316-GAL4RTL(N10Q/N236Q), pRS315-GPDRTA(N10Q), pRS315-GPDRTA(N236Q), pRS315-GPDRTA(N10Q/N236Q), and pRS315-GPDFLAG-RTA were generated using site-directed mutagenesis (QuikChange, Stratagene) according to the manufacturer's instructions.
To generate pRS313-PNG1Png1, PNG1 allele, including its promoter, was cut out from pRS316-PNG1 by XhoI/NotI digestion (16) and was cloned into the equivalent site of pRS313. To generate pRS313-GAL1Png1, the PNG1 allele, including GAL1 promoter region, was cut out from pRD53-PNG1 (17) with XhoI/NotI digestion and was cloned into the equivalent site of pRS313.
To generate pRS316-MET25RTL, the MET25 promoter and CYC1 terminator derived from pGFP-C-FUS (18) and RTL derived from pRS316-GAL4RTL were cloned into pRS316 as follows. The MET25 promoter region of pGFP-C-FUS was isolated by SacI/HindIII digestion, and the fragment was cloned into the equivalent site of pRS316, yielding pRS316-MET25 promoter. The CYC1 terminator in pGFP-C-FUS was then isolated by SalI/KpnI digestion and was cloned into the equivalent site of pRS316-MET25 promoter, yielding pRS316-MET25. The RTL fragment was amplified from pRS316-GAL4RTL using the following primers: Hind-KAR2-N-forward, 5′-GTTTAAGCTTATGTTTTTCAACAGACTAAGCGCTGGCAAG-3′; Sal-LEU2-C-reverse, 5′-GTTTGTCGACTTAAGCAAGGATTTTCTTAACTTCTTCGGC-3′. This PCR product was digested with SalI and HindIII and was cloned into the equivalent site of pRS316-MET25 to generate pRS316-MET25RTL.
To generate pRS313-Htm1, HTM1, including the promoter region, was amplified from yeast wild-type genome using the following primers: Htm1(−776/−747), 5′-GTGGTGTATCTATTGTTATACTTAGTCGAC-3′; Htm1-C-BglII, 5′-AGATCTTCATACAATAAATAAGTTGATGATGGGCG-3′. The PCR product was digested with SacI/BglII and was cloned into the equivalent site of pRS313. pRS313-Htm1(E222Q) was generated from pRS313-Htm1 using site-directed mutagenesis (QuikChange, Stratagene) according to the manufacturer's instructions.
Spotting Assay
Strains harboring the RTL expression plasmid were grown in SC−uracil liquid medium containing 2% raffinose. 4-Fold serial dilutions of each strain harboring the RTL plasmid were spotted onto SC−uracil medium (complete media in this assay) or SC−uracil−leucine medium containing galactose, and cells were incubated at 30 °C for the indicated times. To improve cell growth, the plates were supplemented with additional histidine (20 mg/liter). Photographs of the gels were taken using FUJIFILM LAS-3000mini (Fujifilm Co., Tokyo, Japan).
Cycloheximide Decay Assay
Strains harboring the RTA expression plasmid were grown in SC−leucine liquid medium. Then cycloheximide was added to the cultures (final concentration, 250 μg/ml), and the cultures were collected at the indicated times.
Cell Extracts and Western Blotting
Unless noted, all experiments were carried out at either 4 °C or on ice. Protein extraction was carried out essentially as described previously (19). In brief, about 108 yeast cells were harvested, and the pellets were suspended in 0.1 m NaOH and incubated at room temperature for 10 min. After the cells were collected by centrifugation at 3000 × g for 1 min, the pellet was resuspended in 100 μl of SDS sample buffer (62.5 mm Tris-HCl (pH 6.8), 5% β-mercaptoethanol, 2% SDS, 5% sucrose, 0.04% bromphenol blue) and boiled at 95 °C for 5 min. Cell debris was then removed by centrifugation at 10,000 × g for 3 min. Equal amounts of each sample were analyzed by 10% SDS-polyacrylamide gel and then transferred to BiotraceTM polyvinylidene difluoride membranes (PALL Co.). To detect RTA, the membranes were incubated with an anti-ricin monoclonal antibody (CP75 or CP37) (Abcam) diluted 1:5000 in 2% skimmed milk in TBS-T (25 mm Tri-HCl (pH 8.0), 137 mm NaCl, 0.05% Tween 20), washed, and then incubated with horseradish peroxidase-conjugated anti-mouse secondary antibody diluted 1:5000 in 2% skimmed milk in TBS-T. The secondary antibodies were detected with the ImmobilonTM Western blot chemiluminescent horseradish peroxidase substrate system in accordance with the manufacturer's instructions (Millipore).
N-Glycan Structure Analysis on RTA
Strains harboring the FLAG-RTA expression plasmid pRS315-GPDFLAG-RTA were grown in SC−leucine liquid medium. Cultured cells (4 g) were washed twice with TNE buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 5 mm EDTA, and protease inhibitor mixture), resuspended with 4 ml of TNE buffer, and were lysed with glass beads. The lysate was centrifuged for 5 min at 300 × g to remove cell debris. Triton X-100 was added to the supernatant to the final 1% concentration to solubilize the membrane fraction, and the solution was incubated at 4 °C for 1 h. The unsolubilized membranes were removed by centrifugation at 15,000 × g, 4 °C, for 15 min. 20 μl of anti-FLAG M2 affinity resin (Sigma) was added to the supernatant, and the sample was incubated at 4 °C for 1 h with a rocking movement. The resin were then washed four times with TNE buffer, 1% Triton X-100, and the bound RTA was eluted with 200 μl of 0.1 m glycine-HCl buffer (pH 3.5). 40 μl of 0.5 m sodium acetate buffer (pH 5.5) and 20 milliunits of endo-β-N-acetylglucosaminidase H (Seikagaku Kogyo Co., Tokyo, Japan) was added to the eluted solution. Incubation was carried out at 37 °C overnight. 5 milliunits of endo-β-N-acetylglucosaminidase H was added to the sample, and it was incubated for another 4 h to complete deglycosylation. To stop the enzyme reaction, 1.5 volumes of ethanol were added to the reaction mixture, and the oligosaccharide released was recovered by centrifugation at 15,000 × g, 4 °C for 5 min. Supernatant thus obtained was dried up, resuspended with 1 ml of distilled H2O, and purified using InertSep GC column (150 mg/3 ml; GL-Science, Tokyo, Japan) as described previously (20). Labeling of oligosaccharides with the pyridylamino group and size fractionation of pyridylamino-labeled glycans were performed as described previously (20). Separation and structural identification of the oligosaccharides were carried out using reversed-phase high pressure liquid chromatography according to the method reported previously (21).
Free Oligosaccharide Analysis
Isolation and structural analysis of free oligosaccharides from mns1Δ cells were carried out as described previously (20).
RESULTS
RTL Spotting Assay
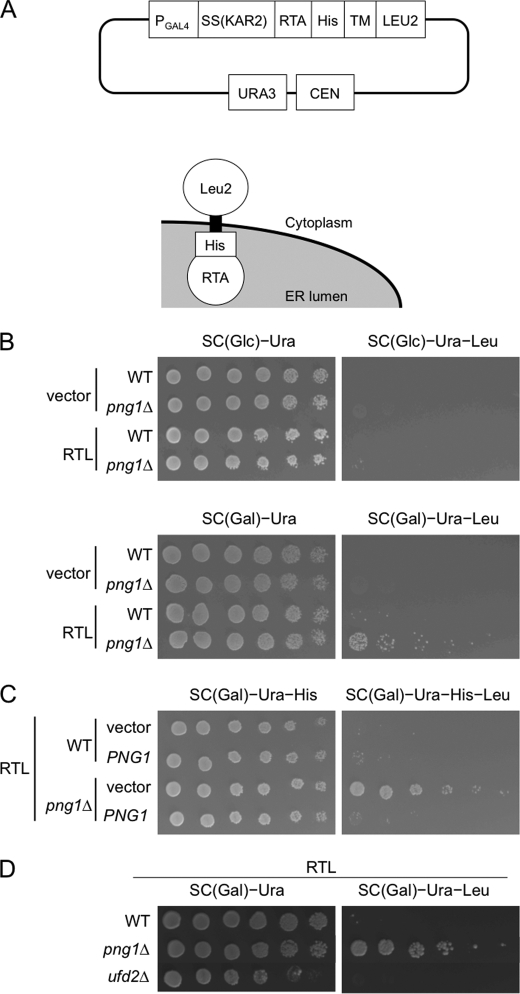
RTA (ricin A-chain nontoxic mutant) was previously identified as a Png1-dependent ERAD substrate in yeast (7, 10). Interestingly, some of the N-glycosylated substrates did not require Png1 for efficient degradation (10), and the detailed mechanism of Png1-dependent degradation remains unknown. Because RTA has a very short half-life in most strains, we found that it can be difficult to identify subtle differences in RTA stability by pulse-chase or cycloheximide decay experiments (7, 10). Therefore, we developed a new genetic device using the new substrate designated RTL. RTL was composed of luminal RTA, a transmembrane domain of Pdr5 and the cytoplasmic Leu2, and was constructed by remodeling CTL* (22). The main advantage of this assay system, as with the CTL* assay (22), is its simplicity and reproducibility, because this method uses cell growth, one of the most sensitive indicators of alterations in cell physiology, to assess the protein stability. The logic behind this screening is that strains with leu2 auxotrophy are unable to grow in media lacking leucine, whereas mutants with defective RTL degradation can support cell growth under such conditions because such yeast cells contain sufficient Leu2 proteins on RTL. Therefore, we can assess the efficiency of RTL degradation in various ERAD-related strains by examining the growth of cells incubated in media with limited or no leucine. Schematic views of RTL and the plasmids expressing this protein are shown in Fig. 1A.
FIGURE 1.
Characterization of the RTL assay. A, schematic view of the RTL expression vector and topology of RTL. SS(KAR2), signal peptide from Kar2; His, His6 epitope; TM, transmembrane domain of Pdr5; CEN, centromere in S. cerevisiae. B, png1Δ cells with stabilized RTL. BY4741 WT and png1Δ cells were plated on SC glucose or SC galactose medium with or without leucine. The plates were incubated for 3 days at 30 °C. The leucine auxotrophic WT strain expressing RTL cannot grow in media without leucine. The leucine deficiency is complemented by RTL in the RTA degradation-defective png1Δ cells (bottom row). C, expression of Png1 rescued the defective RTL degradation in png1Δ cells. WT and png1Δ cells carrying the plasmids expressing Png1 and RTL were plated on SC galactose medium with or without leucine. The plates were incubated for 3 days at 30 °C. D, Ufd2 was not required for RTL degradation. BY4741 WT and ufd2Δ cells carrying the plasmids expressing RTL were plated on SC galactose medium with or without leucine. The plates were incubated for 3 days at 30 °C.
RTL is expressed under the control of the weak galactose-inducible GAL4 promoter. Thus, the expression of RTL is moderately induced in galactose-containing medium (but not enough to support its growth in media without leucine content) and is suppressed in glucose-containing medium.
To examine whether the newly developed method can differentiate the stability of RTL between wild-type and isogenic png1Δ cells, a spotting assay was performed. As shown in Fig. 1B, png1Δ cells, but not wild-type cells, can only grow when the cells were transfected with the RTL plasmid, and galactose was used as the only carbon source (i.e. the expression of the RTL protein was induced) (Fig. 1B, compare the top panel and the bottom panel, bottom row). These results indicate the following: (a) RTL expression cannot support growth in media lacking leucine unless the stability of RTL was compromised, and (b) there are clear differences in the stability of RTL between wild-type cells and png1Δ cells; this faithfully reproduces the results obtained using RTA (10). Furthermore, the growth of png1Δ cells expressing RTL was suppressed by PNG1 expression (Fig. 1C, compare the 3rd and 4th row), indicating the growth observed was not an indirect effect.
Ufd2, a ubiquitin elongation factor, was previously shown to bind to Rad23 and was involved in the degradation of ubiquitin fusion degradation substrates (23). This protein, however, was not involved in RTA degradation (10). Because Rad23-Ufd2 was found to form a distinct complex with Png1-Rad23 (10), it was hypothesized that these two complexes are involved in distinct ERAD pathways (10). Therefore, we tested the effect of Ufd2-deletion (ufd2Δ) on the degradation of RTL. Consistent with the results with RTA, RTL was also degraded in ufd2Δ cells (Fig. 1D). Taking all of the results together, this newly established RTL assay was found to be a good system to examine the molecular mechanism of the PNG1-dependent ERAD pathway.
N-Glycosylation at Asn-10 Is Critical for the Png1-dependent ERAD Pathway
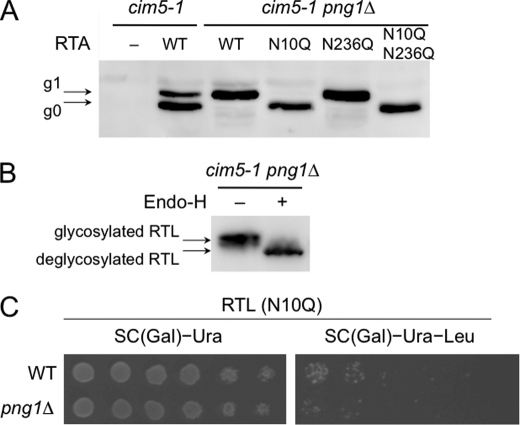
Next, we examined the role of N-glycan in the degradation of RTA and RTL. It was previously shown that the cytosolic version of RTA was degraded through a Png1-independent pathway (10). However, the N-glycan dependence of RTA degradation has not been rigorously tested. Although RTA has two potential glycosylation sites, Asn-10 and Asn-236 (24), we mainly find the g1 form, RTA with one N-glycan (7, 10). To identify which of the glycosylation sites were involved, glycosylation site mutants of RTA were generated, namely RTA(N10Q), RTA(N236Q), and RTA(N10Q/N236Q). These mutant proteins were then expressed in cim5-1 png1Δ cells, and the presence of N-glycans on these mutants was analyzed. As shown in Fig. 2A, almost all RTA(N10Q) and RTA(N10Q/N236Q) were found to be in the g0 form. On the other hand, RTA(N236Q) remained in the g1 form, indicating that the primary glycosylation site for the g1 form is Asn-10. We then carried out the RTL assay for RTL(N10Q) mutants. As shown in Fig. 2B, Endo-H digestion revealed that RTL was also N-glycosylated. Consistent with the RTA assay (data not shown), RTL(N10Q) was also efficiently degraded in png1Δ cells (Fig. 2C), which strongly suggests that N-glycosylation at Asn-10 in RTL(N10Q) was required for Png1-dependent degradation of RTL. These results clearly demonstrate the N-glycan dependence on RTL degradation and further validate the effectiveness of the RTL assay.
FIGURE 2.
N-Glycan-dependent degradation of RTA/RTL. A, Western blotting analysis of RTA and its glycosylation mutants. RTA, RTA(N10Q), RTA(N236Q), and RTA(N10Q/N236Q) were expressed in cim5-1 or cim5-1 png1Δ cells. Extracts from these cells were resolved by SDS-PAGE, and RTA was visualized by immunoblotting using anti-ricin antibody. The arrows indicate RTA modified with one (g1) or no (g0) glycan. B, RTL was N-glycosylated. RTL was expressed using pRS316-MET25RTL in cim5-1 png1Δ cells. Cell extracts were mock-treated (−) or digested (+) with Endo-H and resolved by SDS-PAGE, and RTL was visualized by immunoblotting using anti-ricin antibody. C, RTL lacking the first glycosylation site was degraded in a Png1-independent manner. BY4741 WT and png1Δ cells carrying the plasmids expressing RTL(N10Q) were plated on SC galactose medium with or without leucine. The plates were incubated for 3 days at 30 °C.
Role of the Png1-Rad23 Complex on RTL Degradation
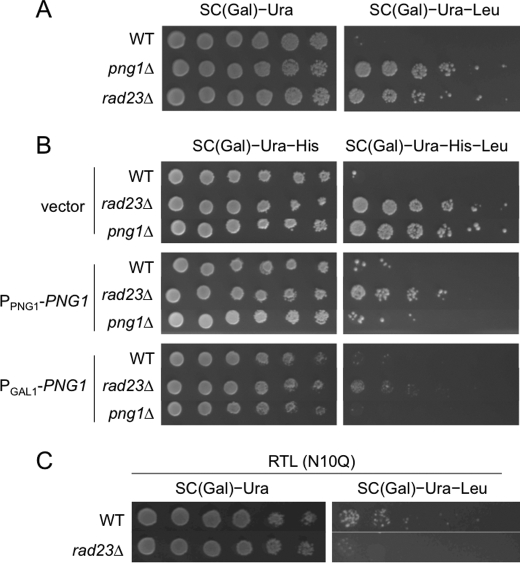
Rad23 is thought to be important for ERAD because it has the ability to bind to ubiquitinated substrates (through the UBA domain) (25) and to the 26 S proteasome (through the UBL domain) (26). Therefore, Rad23 links the substrate destined for degradation with the proteasome, the protein-destruction machinery. Rad23 was previously identified as a cytoplasmic PNGase-binding protein (17, 27). RTA degradation was also shown to only be moderately affected in rad23Δ cells (10). It was also found that the Rad23 mutant, which fails to bind to Png1, also delayed the degradation of RTA, suggesting that the Png1-Rad23 complex is required for efficient degradation of RTA. Therefore, we examined whether the same is true for RTL substrates. As shown in Fig. 3A, RTL was also significantly stabilized in rad23Δ cells (Fig. 3A), suggesting that Rad23 is also required for efficient RTL degradation. Therefore, we next examined whether the expression of Png1 suppresses the RTL degradation defect in rad23Δ cells. As shown in Fig. 3B, the RTL degradation defect in rad23Δ cells was significantly suppressed, but not completely, by the expression of Png1 (Fig. 3B). The enhanced expression of Png1 (GAL promoter-driven compared with the endogenous promoter) increased the suppression of the defect (Fig. 3B, compare 2nd, 5th, and 8th rows), further supporting the idea that defects in the rad23Δ cells are largely dependent on the function of Png1. The fact that there was no complete recovery after Png1 overexpression indicated that, although the role of Rad23 in RTL degradation was dependent on PNGase enzyme activity, Rad23 acts as a scaffold protein and supports the full functionality of Png1 in this process. Consistent with this observation, the defects in rad23Δ cells also exhibited N-glycan dependence because RTL(N10Q) (Fig. 3C) and RTA(N10Q) (data not shown) did not require Rad23 for efficient degradation. Moreover, Dsk2, another UBL-UBA protein, was not involved in the degradation of RTA and RTL (data not shown). This result contrasts with that for CTL* (22), a Leu2 fusion protein derivative of CPY*, which indicates that Rad23 and Dsk2 are both required for optimal degradation. This result further supports the hypothesis that the importance of Rad23 is its ability to bind to Png1, rather than its activity as a UBL-UBA protein.
FIGURE 3.
Png1 overexpression suppressed the degradation defect of RTL in png1Δ and rad23Δ cells. A, Rad23 was required for RTL degradation. BY4741 WT and rad23Δ cells carrying the plasmids expressing RTL were plated on SC galactose medium with or without leucine. The plates were incubated for 3 days at 30 °C. B, effects of Png1 expression on RTL degradation in png1Δ and rad23Δ cells. WT, rad23Δ, and png1Δ cells carrying the plasmids expressing RTL and PPNG1-PNG1 or PGAL1-PNG1 were plated on SC galactose medium with or without leucine. The plates were incubated for 3 days at 30 °C. C, absence of Rad23 dependence on degradation of nonglycosylated RTL. BY4741 WT and rad23Δ cells carrying the plasmids expressing RTL(N10Q) were plated on SC galactose medium with or without leucine. The plates were incubated for 3 days at 30 °C.
Taking all of these results together, it appears that the effect of rad23Δ is largely dependent on PNGase function. This result was consistent with previous studies of RTA (10), indicating that RTA and RTL appear to have similar requirements for ERAD components.
Der1 but Not Sec61 Is Required for RTL and RTA Degradation
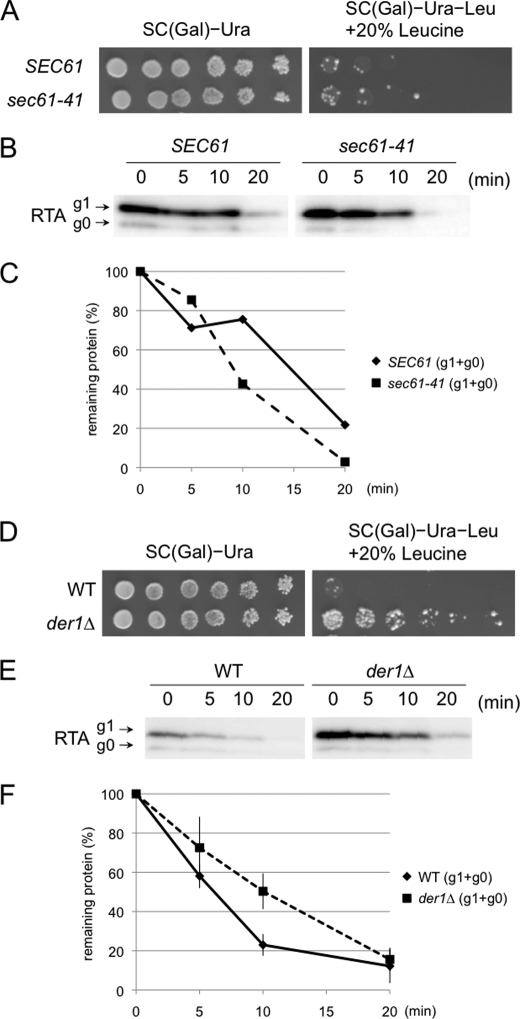
Having established RTL as a new ERAD substrate, various mutants bearing defects in the ERAD pathway were examined to characterize the RTL degradation pathway. It was previously reported that functional Sec61, a component of the protein translocation channel on the ER membrane, was required for efficient degradation of RTA(E177D), a less toxic RTA mutant (28). Therefore, we assessed RTL degradation in sec61-41 cells, an allele reported to show defective RTA(E177D) degradation (28). It is important to note that, compared with nonessential genes, one must take precautions when applying the RTL assay to essential genes because the stability of the ERAD substrate was assessed by the growth of cells. Therefore, the overall growth effects should be assessed as a combination of their intrinsic growth defect as well as the RTL-stabilizing effect, and unless one can identify the conditions under which significant growth can be observed for such mutants, it is difficult to draw conclusions regarding the involvement of such essential proteins with the RTL assay. Using sec61-41, an obvious degradation defect was not detected at several temperature conditions tested (Fig. 4A) (data not shown), raising the possibility that Sec61 might, compared with RTA(E177D), be dispensable for RTA degradation. To further confirm this observation, the stability of RTA in sec61-41 cells was also examined. To impair the degradation of RTA, the cells were treated at 24 °C for 1 h, conditions that reveal the defects in the ER export of Δgpαf (14). As shown in Fig. 4, B and C, no defects in RTA degradation were observed, even at 24 °C for 1 h in which a clear defect in RTA(E177D) degradation was observed (28). On the other hand, Der1, another putative component of the retrotranslocation channel in the ERAD process (29, 30), was found to be important in RTL degradation (Fig. 4D). Consistent with this result, delayed degradation in Der1 cells was also observed for RTA (Fig. 4, E and F). These data suggest that Der1 but not Sec61 is required for degradation of RTA.
FIGURE 4.
Der1 but not Sec61 is required for degradation of RTL/RTA. A, RTL assay for sec61-41 and its isogenic WT. SEC61 and sec61-41 cells carrying the plasmids expressing RTL were plated on SC galactose medium with 100 or 20% leucine (low leucine media, optimal leucine concentration for RTL assay using this strain background). The plates were incubated for 3 days at 30 °C. B, cycloheximide decay assay for RTA in sec61-41 and SEC61 WT. Experiments were performed at 24 °C, at which clear inhibition of RTA(E177D) was observed (28). Cycloheximide was added at t = 0 min, and the samples were collected at the indicated time points and subjected to SDS-PAGE, followed by immunoblotting using anti-ricin antibody. C, quantitation of data in B (g1 + g0 RTA). D, RTL assay for der1Δ and its isogenic WT. W303-1a WT and der1Δ cells carrying the plasmids expressing RTL were plated on SC galactose medium with 100 or 20% leucine (low leucine media, optimal leucine concentration for RTL assay using this strain background). The plates were incubated for 3 days at 30 °C. E, cycloheximide decay assay for RTA in der1Δ and its isogenic WT. The cycloheximide decay assays were performed in WT (BY4742) and der1Δ cells at 30 °C. Cycloheximide was added at t = 0 min, and the samples were collected at the indicated times and subjected to SDS-PAGE, followed by immunoblotting using anti-ricin antibody. F, quantitation of cycloheximide decay assay for RTA in der1Δ and its isogenic WT. The graph represents the mean of three independent experiments including one shown in E.
Other Components Involved in RTL Degradation
Next, we performed a comprehensive analysis of the various mutants reported to be involved in ERAD (31) using the RTL and CTL* assays (22). As shown in Fig. 5A and Table 2, RTL stabilization was observed for various mutants involved in ERAD. Most of these proteins were identified as a component required for degradation of ERAD-L substrates, with mutations in the luminal regions (4). For example, Hrd1 is an E3 ligase for the ERAD-L substrate (32–36), and Hrd3 is an Hrd1-interacting protein (32, 37). Both Hrd1 and Hrd3 are reported to be required for the degradation of RTA (10). On the other hand, Doa10, an E3 ligase required for ERAD-C substrates, carries a mutation on the cytosolic side of the membrane proteins (38, 39) and was not required for RTL degradation. Consistent with the results for RTA, Ufd2 was not required for RTL degradation (Fig. 1D and Table 2). These results indicate that RTL, as expected, can be classified as an ERAD-L substrate, and the component requirements are similar to the original RTA (10).
FIGURE 5.
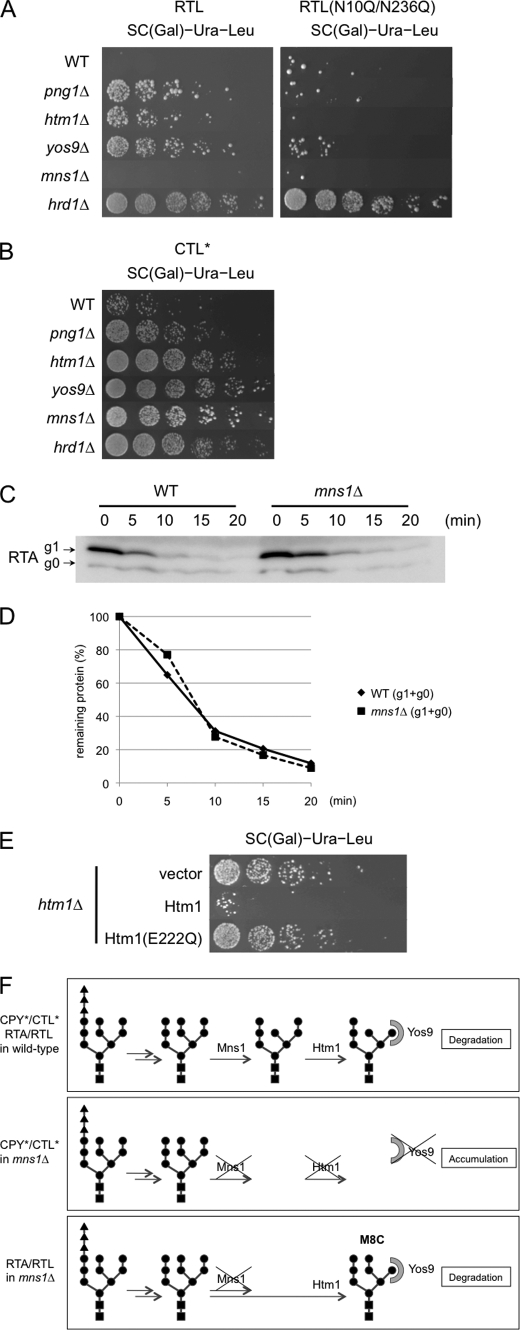
RTL assay for mutants involved in ERAD. A, RTL (left) and RTL(N10Q/N236Q) (right) assay for various mutants known to be required for N-glycan-specific ERAD machinery. Wild-type and various ERAD-related mutants cells carrying the plasmids expressing RTL or RTL(N10Q/N236Q) were subjected to the spotting assay on SC galactose medium without leucine. The plates were incubated for 3 days at 30 °C. B, CTL* assay for various mutants known to be required for N-glycan-specific ERAD machinery. Wild-type and various ERAD-related mutant cells carrying the plasmids expressing CTL* were subjected to spotting assay on SC galactose medium without leucine. The plates were incubated for 2 days at 30 °C. C, cycloheximide decay assay for RTA in mns1Δ and isogenic MNS1 (WT) cells as shown Fig. 4E. D, quantitation of data in C. E, effects of α-mannosidase activity of Htm1 on RTL degradation. htm1Δ cells carrying the plasmids expressing RTL and Htm1 or Htm1(E222Q) were plated on SC galactose medium without leucine. The plates were incubated for 3 days at 30 °C. No growth difference was observed for these three strain on SC galactose medium with leucine. F, schematic representation of the N-glycan processing for distinct ERAD substrates. Top panel, N-glycan processing in wild-type cells. Both CPY*/CTL* and RTA/RTL undergo successive processing by Mns1 and Htm1, followed by the recognition by Yos9 (50, 51). Middle panel, N-glycan processing of CPY*/CTL* in mns1Δ cells. As Htm1 requires prior Mns1 action, the degradation of these proteins is impaired (50, 51). Bottom panel, N-glycan processing of RTA/RTL in mns1Δ cells. As Htm1 can act on N-glycans in an Mns1-independent fashion, proteins are degraded as efficiently as in wild-type cells.
TABLE 2.
Comparison of the components involved in each ERAD substrate
| Function | RTA/RTL | CPY* | |
|---|---|---|---|
| Png1a | Peptide:N-glycanase | + | − (16)b |
| Dsk2 | UBL-UBA protein | − | + (22) |
| Sec61 | Translocon | − | + (82) |
| Mns1 | α-Mannosidase | − | + (53) |
| Rad23 | UBL-UBA protein | + | + (22) |
| Hrd1 | E3 ubiquitin ligase | + | + (33) |
| Doa10 | E3 ubiquitin ligase | − | − (38) |
| Ufd2 | E4 ubiquitin ligase | − | − (83) |
| Cue1 | Ubc7-binding protein | + | + (56) |
| Ubc7 | E2 ubiquitin-conjugating enzyme | + | + (55) |
| Ubx2 | UBX protein | + | + (61) |
| Hrd3 | Component of Hrd1 complex | + | + (34) |
| Usa1 | Component of Hrd1 complex | + | + (39) |
| Der1 | Candidate for translocon | + | + (29) |
| Htm1 | α-Mannosidase | + | + (40) |
| Yos9 | ER lectin | + | + (15) |
a Proteins in boldface indicate the ones exhibiting a different requirement between degradation of RTA/RTL and CPY*.
b Delay in degradation of CPY* was observed while the deglycosylated form of intact CPY* was not detected (16).
RTL stabilization was also observed in htm1Δ and yos9Δ cells (Fig. 5A, right). Htm1 is an ER degradation-enhancing α-mannosidase-like protein homolog containing an α-mannosidase-like domain and was reported to be involved in N-glycan-dependent ERAD (40–43). Yos9, on the other hand, is an OS-9 homolog containing a mannose 6-phosphate receptor homology domain (44), which might serve as a glycan-recognition protein (15, 45–47). Indeed, EDEMs and Htm1 were recently found to possess α-mannosidase activity (48–50), whereas OS-9 and Yos9 were shown to be lectins involved in the recognition of the N-glycosylated substrate (51, 52). We found that the effects of Yos9, Htm1, and Png1 were N-glycan-dependent because they were not involved in the degradation of nonglycosylated RTL (RTL(N10Q/N236Q); Fig. 5A, right).
Interestingly, RTL stabilization was not observed in mns1Δ cells (Fig. 5A). Mns1 is an α1,2-mannosidase localized in the ER and is required for CPY* and CTL* degradation as Htm1 and Yos9 (22, 42, 53, 54). Consistent with these observations, we also observed Mns1 dependence for CTL* degradation (Fig. 5B), suggesting that the lack of an effect of Mns1 was specific for RTL. It was also confirmed that Mns1 was dispensable for the degradation of RTA (Fig. 5, C and D).
To get more insight into the glycan-processing event, FLAG-tagged RTA protein was isolated from wild-type cells as well as mns1Δ cells, and glycan structures on this protein were compared. FLAG-RTA was successfully isolated by immunoprecipitation. FLAG-RTA was obtained as the major band as judged by SDS-PAGE, and the glycan was released from the purified sample by endo-β-N-acetylglucosaminidase H digestion. We did not use PNGase F to release N-glycans to avoid contamination of those from anti-FLAG antibody. The glycan released was labeled with pyridylamino group and initially separated with a size fractionation column. We did not detect any N-glycans from the immunoisolated samples without FLAG-RTA expression, strongly indicating that the glycans obtained are released from FLAG-RTA (data not shown).
As shown in Table 3, the sizes of the glycans from FLAG-RTA were sifted to one hexose unit between wild-type and mns1Δ cells, suggesting that almost all glycans on RTA were modified by Mns1 in wild-type cells. Next, we tried to determine the isomeric structure of these glycans. For this purpose, we utilized reversed-phase high pressure liquid chromatography (21) to separate isomers of various high mannose-type glycans with Gn1 glycans, which bear only a single GlcNAc residue at their reducing termini. Unfortunately, we could not determine the precise structures for all structural isomers, probably because of the presence of unusual Och1-modified glycans (20). We therefore focused on the occurrence of M8C glycans, which is most likely formed by the action of Htm1 in the absence of Mns1 (50). As expected, there are no M8C glycans observed on RTA isolated from wild-type cells. On the other hand, M8C glycans were observed on RTA from mns1Δ cells, suggesting that, at least on RTA, Htm1 is capable of acting on N-glycans in the absence of Mns1. Moreover, structure analysis of free oligosaccharides (derived from misfolded glycoproteins (20)) revealed that the content of M8C was much higher in mns1Δ cells (39%), when compared with that of wild-type cells (0.5%) (20). Therefore, low content of Hex8GlcNAc2 glycans on RTA is most likely due to the fact that Htm1 action is a rate-limiting step for subsequent proteolysis. Taken the results together, we could safely conclude that Htm1 can act on N-glycans on RTA/RTL to generate M8C structure in the absence of Mns1.
TABLE 3.
Composition of N-glycans on FLAG-RTA isolated from WT and isogenic mns1Δ cells
a Number represents that of hexose (Glc/Man) residues, based on the size fractionation high performance liquid chromatography analysis (20).
b ND means not detected.
We also tested the possibility that Htm1, for RTA/RTL degradation, may also function in an enzyme-independent fashion, i.e. lectin-like molecule as one possibility. To validate this hypothesis, we generated the enzyme-inactive Htm1 mutant, Htm1(E222Q). It was previously shown that this mutant did not support efficient degradation of CPY* (50). To examine the effect of this mutation on RTL degradation, this mutant, or wild-type Htm1, was expressed in htm1Δ cells, and RTL assay was carried out. As shown in Fig. 5E, the defect of RTL degradation in htm1Δ cells was only suppressed by wild-type Htm1 but not by Htm1(E222Q) (Fig. 5E). Because this mutant was previously shown to express well in yeast (50), this result suggested that α-mannosidase activity of Htm1 appears essential for RTA/RTL degradation. Taking these results together, it can be concluded that Mns1 is not required for RTA/RTL degradation, clearly suggesting that essentiality of Mns1 on ERAD is substrate-specific (Fig. 5F).
The other proteins found to be required for RTL degradation were Ubc7/Cue1 (55–59) and Ubx2 (60–62). These proteins were reported to be involved in both ERAD-L and ERAD-C. Usa1 is a component of the Hrd1 complex, is involved in ERAD-L (39), and was shown to be required for RTL degradation. Quite recently, this protein was found to be required for the degradation of RTA (63).
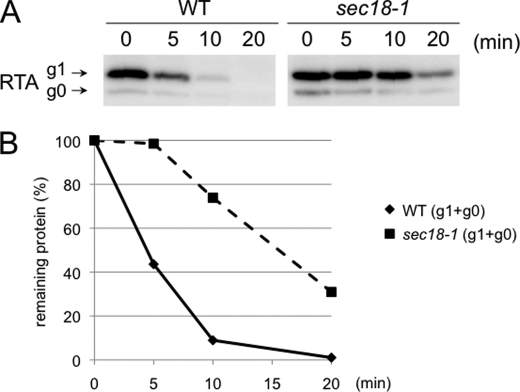
A sec18-1 mutant was previously shown to exhibit compromised degradation of soluble ERAD-L substrates (58, 64–67). This suggests the involvement of the ER-Golgi transport system in the ERAD machinery. Sec18 is a homolog of the mammalian N-ethylmaleimide-sensitive fusion protein (68). Because sec18-1 cells show severe growth defects when incubated in galactose-containing media, even at the permissive temperature (25 °C), we examined RTA degradation in this mutant. As shown in Fig. 6, A and B, RTA stabilization was also observed in sec18-1 cells. This result indicates that RTA degradation also requires functional ER-Golgi transport.
FIGURE 6.
Degradation of RTL/RTA requires Sec18. A, cycloheximide decay assay for RTA in sec18-1 and its isogenic WT. Cycloheximide decay experiments were performed in WT and sec18-1 cells grown at 25 °C and sifted at 37 °C for 30 min. Cycloheximide was added at t = 0 min, and the samples were collected at the indicated times and subjected to SDS-PAGE, followed by immunoblotting. The immunoblots were analyzed with the anti-ricin antibody. B, quantitation of data in A.
DISCUSSION
The process of the ERAD pathway has been extensively studied for at least 1 decade, and many components involved in this pathway have now been identified. However, our knowledge is still limited because most of the studies were carried out using “model” ERAD substrates. This is particularly true for Png1-dependent degradation; a Png1-dependent model ERAD substrate was not available until our recent identification of RTA (7, 10). This prevented us from gaining more insight into the Png1-dependent ERAD pathway and its relationship with the pathways already characterized. At present, the structural feature of RTA/RTL that renders their degradation Png1p-dependent remains to be determined.
In this study, we have established a new genetic device, the RTL assay, to characterize the Png1-dependent ERAD pathway in yeast. Our results indicate that RTL, as is the case with RTA, is a typical ERAD-L substrate. From our study, it is evident that the effect of Png1 is N-glycan-specific, because RTL/RTA lacking the first N-glycosylation site lost the Png1 dependence.
Rad23 is implicated in various ERAD pathways, and Ufd2, an E4 enzyme (ubiquitin-polymerization factor), was shown to bind with this protein (23). Interestingly, Ufd2 was not involved in the degradation of RTA, whereas Deg1-Sec62, a Ufd2-dependent ERAD substrate, did not require Png1 for efficient degradation (10), indicating that the protein complex involving Rad23 has marked substrate specificity. Consistent with this observation, we showed that the Rad23-Ufd2 complex is a distinct complex from Rad23-Png1 (10). We have previously shown that Rad23 is also involved in the degradation of RTA, but the effect was mild compared with that in png1Δ cells (10). Our current results indicate that the effects of rad23Δ on RTL degradation were largely dependent on PNGase function but not with its activity as a UBL-UBA protein, because of the following: (a) Png1 overexpression can suppress the defects in Rad23, and (b) the requirement for Rad23 in RTL degradation is N-glycan-dependent. We have previously shown that Rad23 is not involved in the stability of PNGase (10), and consistent with these results, rad23Δ did not decrease the PNGase activity in the cell extract in the in vitro enzyme assay (17). However, the reduced efficiency of RTA/RTL in terms of Png1-dependent degradation implies that, to optimize the activity of PNGase, PNGase must form a complex with Rad23. Nevertheless, the mechanism by which Rad23 regulates PNGase activity in vivo remains unclear.
With respect to the requirement for other ERAD-related components, the RTA/RTL degradation exhibited a similarity with other ERAD-L substrates, such as CPY*, which is the most studied example (Fig. 7). There are, however, some differences, other than the Png1 dependence, between the degradation of RTA/RTL and CPY*/CTL*, including the dependence for another UBL-UBA protein, Dsk2 (Table 2). Dsk2 does not contain the XPCB domain required for Png1 binding and was not required for RTA/RTL degradation (Table 2) (10).
FIGURE 7.
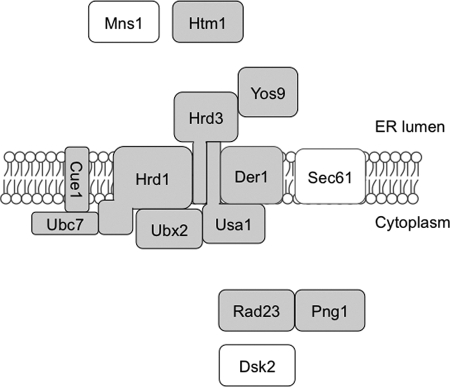
Schematic representation of components required for degradation of RTL. Components reported to be required for CPY* degradation were shown. The shaded components are required for degradation of RTL, although the other components are not required.
Htm1, an EDEM ortholog in yeast, was recently shown to be an α-mannosidase that is required for glycoprotein ERAD (50). This protein and Yos9, a putative degradation lectin that recognizes the product of Htm1 (50, 51), was also found to be required for degradation of RTL but not for nonglycosylated mutant of RTL(N10Q/N236Q) (Fig. 5A). These results clearly suggest that the RTL also utilizes Htm1/Yos9-dependent glycoprotein ERAD machinery. Most surprising finding, however, was the independency of Mns1 for RTA/RTL degradation (Fig. 5). To our knowledge, this was the first example of Htm1-dependent yet Mns1-independent ERAD substrate. It was previously suggested that demannosylation by Mns1 may be critical for subsequent Htm1 action (50), but our results clearly suggest the function of Htm1 does not always depend on prior Mns1 action. How Htm1 distinguishes between Mns1-dependent substrates and Mns1-independent substrates remains unknown.
It is also noteworthy that significant and reproducible CTL* stabilization was observed in png1Δ cells (Fig. 5B). Accumulation of CPY* in png1Δ cells has been reported, whereas the deglycosylated form of intact CPY* was not detected (16). One plausible explanation for these observations is that PNGase might be critical for the efficient degradation of proteolytic degradation intermediates, which could not be detected by pulse-chase analysis, possibly because of inefficient immuno-isolation of CPY*.
It is interesting to note that a cytosolic chaperone Hsc70, a member of Hsp70 family, interacts with the wild-type RTA in vitro and the implicated cytosolic co-chaperone activity determines the fate of the dislocated RTA (69). Therefore, dislocated RTA might be recognized by the Ssa family (Hsp70) of chaperones in yeast, although the Ssa family of chaperones is not required for CPY* degradation (70). This could represent another difference between the degradation of CPY* and RTA.
One puzzling observation in this study was that the degradation of RTA and RTL was not dependent on functional Sec61. Instead, we found a clear dependence of RTA and RTL degradation on Der1, a putative component of the retro-translocation channel. These results contrast markedly with the previous observations, which suggest that the retro-translocation of the ricin A-chain (or its less toxic mutant) was dependent on Sec61 but not on Der1 in yeast and in mammalian cells (28, 71). The simplest explanation for this apparent discrepancy is that we used nontoxic RTA and its derivatives, although other researchers used a toxic ricin protein. The export of toxins from the ER to the cytosol had unique characteristics compared with the ERAD substrates (72, 73); therefore, the loss of toxicity might have caused a change in the export mechanism, including Der1 dependence.
The dependence of RTL on Der1 also appears to be distinct from that of CTG*, a green fluorescent protein-fused transmembrane protein version of CPY*, because the degradation of CTG* was not Der1-dependent (70), although for RTA, the transmembrane protein derivative (RTL) was still Der1-dependent. It should be noted that recent studies revealed that the degradation of a model transmembrane protein or an ERAD-C transmembrane protein substrate could be dependent on Der1 or its mammalian ortholog Derlin-1 (64, 74–79).
In summary, we have successfully developed a new ERAD assay, the RTL assay, to gain further insight into the Png1-dependent ERAD pathway. We have provided evidence that RTL is an ERAD-L substrate. We also found some notable differences between the degradation of RTL/RTA and that of CPY*. We also showed some novel findings, including the Mns1-independent and Der1-dependent degradation of RTL. This study shows a novel Mns1-independent ERAD for a subset of glycoprotein ERAD substrates. Further studies will clarify the details of this ERAD pathway.
Acknowledgments
We thank Drs. Dieter H. Wolf, Yoshifumi Jigami, Takehiko Yoko-o, Claude A. Jakob, and Randy W. Schekman for kindly providing strains and reagents. We thank Drs. Yoshiki Yamaguchi and Tadashi Satoh (RIKEN, Wako, Japan), and the members of the Glycometabolome Team for helpful discussions.
This work was supported in part by the Global Center of Excellence Program (to A. H. and T. S.), grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and RIKEN President's Discretionary Fund (Strategic Programs for R&D) (to T. S.).
- ER
- endoplasmic reticulum
- CPY*
- carboxypeptidase Y(G255R) mutant
- CTL*
- CPY*-transmembrane-Leu2
- ERAD
- ER-associated degradation
- PNGase
- peptide:N-glycanase
- RTA
- ricin toxin A-chain nontoxic mutant
- RTL
- RTA-transmembrane-Leu2
- SC
- synthetic complete
- WT
- wild type.
REFERENCES
- 1.Anelli T., Sitia R. (2008) EMBO J. 27, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Römisch K. (2005) Annu. Rev. Cell Dev. Biol. 21, 435–456 [DOI] [PubMed] [Google Scholar]
- 3.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sayeed A., Ng D. T. (2005) Crit. Rev. Biochem. Mol. Biol. 40, 75–91 [DOI] [PubMed] [Google Scholar]
- 5.Suzuki T. (2007) Semin. Cell Dev. Biol. 18, 762–769 [DOI] [PubMed] [Google Scholar]
- 6.Suzuki T., Park H., Lennarz W. J. (2002) FASEB J. 16, 635–641 [DOI] [PubMed] [Google Scholar]
- 7.Tanabe K., Lennarz W. J., Suzuki T. (2006) Methods Enzymol. 415, 46–55 [DOI] [PubMed] [Google Scholar]
- 8.Kario E., Tirosh B., Ploegh H. L., Navon A. (2008) J. Biol. Chem. 283, 244–254 [DOI] [PubMed] [Google Scholar]
- 9.Blom D., Hirsch C., Stern P., Tortorella D., Ploegh H. L. (2004) EMBO J. 23, 650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim I., Ahn J., Liu C., Tanabe K., Apodaca J., Suzuki T., Rao H. (2006) J. Cell Biol. 172, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Science 294, 2364–2368 [DOI] [PubMed] [Google Scholar]
- 12.Rose M. D., Winston F., Hieter P. (1990) Methods in Yeast Genetics, pp. 177–186, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 13.Sherman F. (1991) Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 14.Pilon M., Schekman R., Römisch K. (1997) EMBO J. 16, 4540–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buschhorn B. A., Kostova Z., Medicherla B., Wolf D. H. (2004) FEBS Lett. 577, 422–426 [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T., Park H., Hollingsworth N. M., Sternglanz R., Lennarz W. J. (2000) J. Cell Biol. 149, 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki T., Park H., Kwofie M. A., Lennarz W. J. (2001) J. Biol. Chem. 276, 21601–21607 [DOI] [PubMed] [Google Scholar]
- 18.Niedenthal R. K., Riles L., Johnston M., Hegemann J. H. (1996) Yeast 12, 773–786 [DOI] [PubMed] [Google Scholar]
- 19.Kushnirov V. V. (2000) Yeast 16, 857–860 [DOI] [PubMed] [Google Scholar]
- 20.Hirayama H., Seino J., Kitajima T., Jigami Y., Suzuki T. (2010) J. Biol. Chem. 285, 12390–12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki T., Matsuo I., Totani K., Funayama S., Seino J., Taniguchi N., Ito Y., Hase S. (2008) Anal. Biochem. 381, 224–232 [DOI] [PubMed] [Google Scholar]
- 22.Medicherla B., Kostova Z., Schaefer A., Wolf D. H. (2004) EMBO Rep. 5, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim I., Mi K., Rao H. (2004) Mol. Biol. Cell 15, 3357–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolliffe N. A., Di Cola A., Marsden C. J., Lord J. M., Ceriotti A., Frigerio L., Roberts L. M. (2006) J. Biol. Chem. 281, 23377–23385 [DOI] [PubMed] [Google Scholar]
- 25.Chen L., Madura K. (2002) Mol. Cell. Biol. 22, 4902–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. (2004) J. Biol. Chem. 279, 26817–26822 [DOI] [PubMed] [Google Scholar]
- 27.Park H., Suzuki T., Lennarz W. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11163–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson J. C., Roberts L. M., Römisch K., Davey J., Wolf D. H., Lord J. M. (1999) FEBS Lett. 459, 80–84 [DOI] [PubMed] [Google Scholar]
- 29.Knop M., Finger A., Braun T., Hellmuth K., Wolf D. H. (1996) EMBO J. 15, 753–763 [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 31.Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. (2008) Cell 132, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hampton R. Y., Gardner R. G., Rine J. (1996) Mol. Biol. Cell 7, 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bordallo J., Plemper R. K., Finger A., Wolf D. H. (1998) Mol. Biol. Cell 9, 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plemper R. K., Bordallo J., Deak P. M., Taxis C., Hitt R., Wolf D. H. (1999) J. Cell Sci. 112, 4123–4134 [DOI] [PubMed] [Google Scholar]
- 35.Deak P. M., Wolf D. H. (2001) J. Biol. Chem. 276, 10663–10669 [DOI] [PubMed] [Google Scholar]
- 36.Bays N. W., Gardner R. G., Seelig L. P., Joazeiro C. A., Hampton R. Y. (2001) Nat. Cell Biol. 3, 24–29 [DOI] [PubMed] [Google Scholar]
- 37.Gardner R. G., Swarbrick G. M., Bays N. W., Cronin S. R., Wilhovsky S., Seelig L., Kim C., Hampton R. Y. (2000) J. Cell Biol. 151, 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson R., Locher M., Hochstrasser M. (2001) Genes Dev. 15, 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 40.Nakatsukasa K., Nishikawa S., Hosokawa N., Nagata K., Endo T. (2001) J. Biol. Chem. 276, 8635–8638 [DOI] [PubMed] [Google Scholar]
- 41.Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., Nagata K. (2001) EMBO Rep. 2, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jakob C. A., Bodmer D., Spirig U., Battig P., Marcil A., Dignard D., Bergeron J. J., Thomas D. Y., Aebi M. (2001) EMBO Rep. 2, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kostova Z., Wolf D. H. (2005) J. Cell Sci. 118, 1485–1492 [DOI] [PubMed] [Google Scholar]
- 44.Friedmann E., Salzberg Y., Weinberger A., Shaltiel S., Gerst J. E. (2002) J. Biol. Chem. 277, 35274–35281 [DOI] [PubMed] [Google Scholar]
- 45.Bhamidipati A., Denic V., Quan E. M., Weissman J. S. (2005) Mol. Cell 19, 741–751 [DOI] [PubMed] [Google Scholar]
- 46.Spear E. D., Ng D. T. (2005) J. Cell Biol. 169, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim W., Spear E. D., Ng D. T. (2005) Mol. Cell 19, 753–764 [DOI] [PubMed] [Google Scholar]
- 48.Hirao K., Natsuka Y., Tamura T., Wada I., Morito D., Natsuka S., Romero P., Sleno B., Tremblay L. O., Herscovics A., Nagata K., Hosokawa N. (2006) J. Biol. Chem. 281, 9650–9658 [DOI] [PubMed] [Google Scholar]
- 49.Olivari S., Cali T., Salo K. E., Paganetti P., Ruddock L. W., Molinari M. (2006) Biochem. Biophys. Res. Commun. 349, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 50.Clerc S., Hirsch C., Oggier D. M., Deprez P., Jakob C., Sommer T., Aebi M. (2009) J. Cell Biol. 184, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quan E. M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J. S. (2008) Mol. Cell 32, 870–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hosokawa N., Kamiya Y., Kamiya D., Kato K., Nagata K. (2009) J. Biol. Chem. 284, 17061–17068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knop M., Hauser N., Wolf D. H. (1996) Yeast 12, 1229–1238 [DOI] [PubMed] [Google Scholar]
- 54.Jakob C. A., Burda P., Roth J., Aebi M. (1998) J. Cell Biol. 142, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hiller M. M., Finger A., Schweiger M., Wolf D. H. (1996) Science 273, 1725–1728 [DOI] [PubMed] [Google Scholar]
- 56.Biederer T., Volkwein C., Sommer T. (1997) Science 278, 1806–1809 [DOI] [PubMed] [Google Scholar]
- 57.Wang Q., Chang A. (1999) EMBO J. 18, 5972–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vashist S., Kim W., Belden W. J., Spear E. D., Barlowe C., Ng D. T. (2001) J. Cell Biol. 155, 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. (2004) J. Biol. Chem. 279, 38369–38378 [DOI] [PubMed] [Google Scholar]
- 60.Schuberth C., Buchberger A. (2005) Nat. Cell Biol. 7, 999–1006 [DOI] [PubMed] [Google Scholar]
- 61.Neuber O., Jarosch E., Volkwein C., Walter J., Sommer T. (2005) Nat. Cell Biol. 7, 993–998 [DOI] [PubMed] [Google Scholar]
- 62.Wilson J. D., Liu Y., Bentivoglio C. M., Barlowe C. (2006) Traffic 7, 1213–1223 [DOI] [PubMed] [Google Scholar]
- 63.Kim I., Li Y., Muniz P., Rao H. (2009) PLoS One 4, e7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vashist S., Ng D. T. (2004) J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caldwell S. R., Hill K. J., Cooper A. A. (2001) J. Biol. Chem. 276, 23296–23303 [DOI] [PubMed] [Google Scholar]
- 66.Taxis C., Vogel F., Wolf D. H. (2002) Mol. Biol. Cell 13, 1806–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujita M., Yoko-O T., Jigami Y. (2006) Mol. Biol. Cell 17, 834–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steel G. J., Laude A. J., Boojawan A., Harvey D. J., Morgan A. (1999) Biochemistry 38, 7764–7772 [DOI] [PubMed] [Google Scholar]
- 69.Spooner R. A., Hart P. J., Cook J. P., Pietroni P., Rogon C., Höhfeld J., Roberts L. M., Lord J. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17408–17413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taxis C., Hitt R., Park S. H., Deak P. M., Kostova Z., Wolf D. H. (2003) J. Biol. Chem. 278, 35903–35913 [DOI] [PubMed] [Google Scholar]
- 71.Slominska-Wojewodzka M., Gregers T. F., Wälchli S., Sandvig K. (2006) Mol. Biol. Cell 17, 1664–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heiligenstein S., Eisfeld K., Sendzik T., Jimenéz-Becker N., Breinig F., Schmitt M. J. (2006) EMBO J. 25, 4717–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kothe M., Ye Y., Wagner J. S., De Luca H. E., Kern E., Rapoport T. A., Lencer W. I. (2005) J. Biol. Chem. 280, 28127–28132 [DOI] [PubMed] [Google Scholar]
- 74.Hill K., Cooper A. A. (2000) EMBO J. 19, 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lilley B. N., Ploegh H. L. (2004) Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 76.Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. (2006) Cell 126, 571–582 [DOI] [PubMed] [Google Scholar]
- 77.Baker B. M., Tortorella D. (2007) J. Biol. Chem. 282, 26845–26856 [DOI] [PubMed] [Google Scholar]
- 78.Huttunen H. J., Guénette S. Y., Peach C., Greco C., Xia W., Kim D. Y., Barren C., Tanzi R. E., Kovacs D. M. (2007) J. Biol. Chem. 282, 28285–28295 [DOI] [PubMed] [Google Scholar]
- 79.Schwieger I., Lautz K., Krause E., Rosenthal W., Wiesner B., Hermosilla R. (2008) Mol. Pharmacol. 73, 697–708 [DOI] [PubMed] [Google Scholar]
- 80.Ghislain M., Udvardy A., Mann C. (1993) Nature 366, 358–362 [DOI] [PubMed] [Google Scholar]
- 81.Suzuki T., Lennarz W. J. (2002) Glycobiology 12, 803–811 [DOI] [PubMed] [Google Scholar]
- 82.Plemper R. K., Böhmler S., Bordallo J., Sommer T., Wolf D. H. (1997) Nature 388, 891–895 [DOI] [PubMed] [Google Scholar]
- 83.Ye Y., Meyer H. H., Rapoport T. A. (2001) Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]