Abstract
Transport of acetyl-CoA between intracellular compartments is mediated by carnitine acetyltransferases (Cats) that reversibly link acetyl units to the carrier molecule carnitine. The genome of the opportunistic pathogenic yeast Candida albicans encodes several (putative) Cats: the peroxisomal and mitochondrial Cat2 isoenzymes encoded by a single gene and the carnitine acetyltransferase homologs Yat1 and Yat2. To determine the contributions of the individual Cats, various carnitine acetyltransferase mutant strains were constructed and subjected to phenotypic and biochemical analyses on different carbon sources. We show that mitochondrial Cat2 is required for the intramitochondrial conversion of acetylcarnitine to acetyl-CoA, which is essential for a functional tricarboxylic acid cycle during growth on oleate, acetate, ethanol, and citrate. Yat1 is cytosolic and contributes to acetyl-CoA transport from the cytosol during growth on ethanol or acetate, but its activity is not required for growth on oleate. Yat2 is also cytosolic, but we were unable to attribute any function to this enzyme. Surprisingly, peroxisomal Cat2 is essential neither for export of acetyl units during growth on oleate nor for the import of acetyl units during growth on acetate or ethanol. Oxidation of fatty acids still takes place in the absence of peroxisomal Cat2, but biomass formation is absent, and the strain displays a growth delay on acetate and ethanol that can be partially rescued by the addition of carnitine. Based on our results, we present a model for the intracellular flow of acetyl units under various growth conditions and the roles of each of the Cats in this process.
Keywords: Carnitine, Fatty Acid Metabolism, Membrane Function, Metabolism, Peroxisomes, Candida albicans, Carnitine Acetyltransferase, Membrane Transport
Introduction
Compartmentalization is one of the main characteristics of eukaryotic cells, and separation of metabolic pathways to different organelles is thought to convey an advantage over the unicompartmental system of bacteria. However, the consequence of compartmentalization is that the various pathways at the different locations must be interconnected, requiring the transport of metabolites over the organellar membranes. Acetyl-CoA is a central metabolite that is the product and substrate of many pathways that partake in carbon metabolism. When yeast cells are grown on glucose, acetyl-CoA is produced in the mitochondria, where it can directly enter the tricarboxylic acid cycle to be oxidized to CO2 and H2O. However, during growth on other carbon sources like fatty acids, ethanol, or acetate, acetyl-CoA is produced in different locations in the cell, requiring shuttling of acetyl units between compartments. Utilization of ethanol or acetate as sole carbon source results in the cytosolic production of acetyl-CoA, whereas during growth on fatty acids, acetyl-CoA is produced in peroxisomes, the sole site of β-oxidation of fatty acids in most yeasts (1). Acetyl-CoA cannot cross the organellar membranes without the aid of the carrier molecule carnitine (2). Acetyl units are reversibly bound to carnitine by carnitine acetyltransferases (Cats),4 forming acetylcarnitine, which can be transported over the membrane. In the target organelle, the reverse reaction also catalyzed by Cat takes place, converting acetylcarnitine to acetyl-CoA and carnitine.
We and others have shown previously that in the opportunistic fungal pathogen Candida albicans, acetyl unit transport is strictly dependent on the carrier molecule carnitine, which is synthesized endogenously (3), and the activity of the major carnitine acetyltransferase (Cat2) while growing on oleic acid, ethanol, or acetate (4, 5). In both C. albicans and Saccharomyces cerevisiae, the products of the CAT2 gene are dually localized to peroxisomes and mitochondria (5, 6). The mechanism of dual localization of Cat2 has been extensively studied in S. cerevisiae, and it was shown that CAT2 has two in-frame start codons that are regulated by carbon source-dependent transcription initiation (6). The longer transcript codes for a protein containing a mitochondrial targeting signal (MTS), and thus its translation product is targeted to mitochondria. The protein encoded by the shorter transcript lacks the MTS and is targeted to peroxisomes via its C-terminal peroxisomal targeting signal (PTS1). The presence of both a MTS and PTS1 in C. albicans Cat2 suggests that a similar mechanism may cause the dual localization in this organism.
It is thought that peroxisomal Cat2 is involved in export of acetyl-CoA produced in the peroxisomes during β-oxidation when cells are grown on fatty acids. The main function of the mitochondrial Cat2 is presumably to release acetyl-CoA from acetylcarnitine in the mitochondria. Growth on fatty acids, ethanol, or acetate as the sole carbon source requires the activity of the microorganism-specific glyoxylate cycle. The most important feature of the glyoxylate cycle is that it bypasses the decarboxylation steps of the tricarboxylic acid cycle, thereby enabling the synthesis of malate (C4) from acetyl-CoA (C2) and glyoxylate (C2) for gluconeogenesis. This key reaction of the glyoxylate cycle is catalyzed by malate synthase (Mls1). Both the tricarboxylic acid cycle and the glyoxylate cycle are, therefore, dependent on acetyl-CoA supply. Because Mls1 and the other key enzyme of the glyoxylate cycle (i.e. isocitrate lyase (Icl1)) are peroxisomal in C. albicans (7), growth on C2 carbon sources, such as ethanol and acetate, requires import of acetyl-CoA into peroxisomes. Whether this import is carnitine-dependent is not known.
Both S. cerevisiae and C. albicans encode two other (putative) Cats: Yat1 (also known as Ctn1 in C. albicans) and Yat2 (also known as Ctn3). Through phenotypic analyses of S. cerevisiae and C. albicans yat1 and yat2 null mutants, the Yats were hypothesized to function in transport of cytosolic acetyl-CoA produced during growth on ethanol and acetate to mitochondria and/or peroxisomes (4, 8, 9). However, whether YAT1 and YAT2 encode true Cats in both organisms remains to be established. An S. cerevisiae cat2 null mutant displays very little (<5%) residual Cat activity (5, 10), which can be completely ascribed to the YAT2 gene (9). In a C. albicans cat2 null mutant, Cat activity is undetectable using standard enzyme measurements (5). In S. cerevisiae, unmodified Yat1 was found to be associated with the outer mitochondrial membrane (11), whereas C-terminally tagged Yat2-GFP localized to the cytosol (12). The localization of Yat1 and Yat2 in C. albicans remains to be established.
Although it has been firmly established that the CAT2 gene is indispensable during growth of C. albicans on non-fermentable carbon sources (4, 5), our understanding of the flow of acetyl units between the peroxisomal, cytosolic, and mitochondrial compartments and the individual roles of mitochondrial and peroxisomal Cat2, Yat1, and Yat2 in this process is very limited. In this study, we use a variety of Cat2, Yat1, and Yat2 mutants to address the localization and function of the Cat2 isozymes and the carnitine acetyltransferase homologs Yat1 and Yat2. Based on the biochemical and phenotypic analyses of the constructed mutants, we present a model explaining the flow of acetyl units between the peroxisomal, mitochondrial, and cytosolic compartments in C. albicans and the role of each of the Cats in this process.
EXPERIMENTAL PROCEDURES
Media and Culture Conditions
C. albicans strains were grown at 28 °C unless otherwise stated. For routine non-selective culturing of C. albicans strains, YPD + Uri (2% bactopeptone, 1% yeast extract, 2% glucose and 80 μg/ml uridine) was used. C. albicans transformants were selected and grown on minimal solid medium containing 0.67% yeast nitrogen base (YNB) without amino acids (Difco), 2% glucose and amino acids as needed (20 μg/ml arginine, 20 μg/ml histidine, 80 μg/ml uridine). Transformants generated with the SAT1 marker were selected on YPD plates containing 200 μg/ml nourseothricin. For recycling of the URA3 marker, strains containing the URA3 cassette with the loxP recombination sites were plated on YPD plates, grown overnight at 28 °C, and replica-plated the next day on minimal uridine plates containing 1 mg/ml 5-fluoroorotic acid to select for ura− colonies (13). 5-Fluoroorotic acid-resistant colonies were restreaked on minimal uridine plates without 5-fluoroorotic acid. Removal of the URA3 cassette and stable integration of the other cassettes was confirmed by PCR analysis. Growth curves were performed in liquid YNB medium with glucose (2%), oleic acid/Tween 80 (0.12%/0.2%), sodium acetate (2% with 0.5% potassium phosphate buffer, pH 6.0), or ethanol (2%). For enzyme assays or measurements of metabolites, strains were pregrown for 16 h on minimal glucose medium (YNB with 2% glucose), inoculated at A600 0.2 in YNB 0.3% glucose medium, and grown for 8 h. Finally, the strains were inoculated at A600 0.005 into rich oleate medium (YPO; 2% bactopeptone, 1% yeast extract, 0.12% oleic acid, 0.2% Tween 80) or rich acetate medium (YPA; 2% bactopeptone, 1% yeast extract, 2% sodium acetate) and grown for 16 h. For subcellular fractionations, cells were pregrown on YNB 2% glucose and YNB 0.3% glucose as described above and subsequently inoculated at A600 0.005 in YPO or YPO/M (YPO with 0.5% maltose).
Spot Test
Cells were pregrown on minimal glucose medium as described above, spun down, washed with water twice, and resuspended to a concentration of about 2.7 × 107 cells/ml (A600 1.0) and serially diluted (1:10 dilutions). Four microliters of each dilution was spotted onto agar plates. Plates contained 0.67% YNB with glucose (2%), oleic acid/Tween 80 (0.12%/0.2%), ethanol (2%), sodium acetate (2% with 0.5% potassium phosphate buffer pH 6.0), lactate (2%, to pH 4.5 with NaOH), or citrate (2%) as a carbon source. The pictures were taken after 3–5 days of incubation at 28 °C.
Strains and Plasmids
C. albicans strains used in this study are listed in supplemental Table 1 and are derivatives of SN76 (14). Plasmids used in this study are listed in supplemental Table 2. Primers are listed in supplemental Table 3. The marker of the S. cerevisiae yeast two-hybrid plasmids containing the Gal4 transactivating domain (pGAD; Clontech) and Gal4 DNA-binding domain (pGBT9; Clontech) were swapped to create pGAD-TR with tryptophan (TR) and pGBT10-L with leucine (L) as selectable markers. The multiple cloning site (MCS) of plasmid pPC (15) was introduced in pGAD-TR by using double-stranded oligonucleotides MCS-pGAD-F and MCS-pGAD-R and in pGBT10-L by PCR with double-stranded oligonucleotides MCS-pGBT-F and MCS-pGBT-R. A PCR product containing the full-length C. albicans PEX5 gene was obtained with CaPEX5-F-ATG and CaPEX5-R-STOP and cloned with BglII/SphI into pSP73 (Promega). The insert was cloned with BglII/SacII into plasmid 21.29 (16) and pGAD-TR, resulting in pTi252 and pKa01, respectively. Primers KS59 and KS60 were used for site-directed mutagenesis on pKa01 to introduce the N376D mutation in PEX5, resulting in plasmid pKa07. PCRs were performed with primers KS191 and KS192 and primers KS191 and KS193 on genomic DNA to amplify the C. albicans CAT2 gene with and without the three C-terminal amino acids (CAT2ΔAKLCOOH). Both PCR products were cloned with BamHI/SpeI into pGBT10-L, creating pKa55 and pKa56.
Construction of the C. albicans prototrophic SN76 (wild type) and cat2Δ/Δ strains have been described previously (5). Plasmid pIS52-CAT2, containing the full-length CAT2 gene and an 800-bp promoter region, was used to complement the cat2 null strain (5). To create a CAT2 construct that lacks the mitochondrial targeting sequence (MTS), site-directed mutagenesis was performed on pIS52-CAT2 with primers KS132 and KS133 changing the first ATG of the CAT2 gene into a stop codon (M001*), resulting in pIS52-perCAT2. To create a construct that lacks the peroxisomal targeting signal (PTS1), a PCR was performed with primers KS128 and KS129 to amplify the CAT2 promoter and gene without the C-terminal three amino acids encompassing the PTS1 (-AKLCOOH). To prevent translation from the second ATG, the predicted translation initiation site of the peroxisomal Cat2, the codon was changed by site-directed mutagenesis to an alanine (M023A) with primers KS130 and KS131, resulting in plasmid pIS52-mitCAT2. CEM28 (cat2Δ/Δ) was transformed with plasmids pIS52-perCAT2 and pIS52-mitCAT2, resulting in strains CKS59 (perCAT2) and CKS61 (mitCAT2).
To construct perCAT2 tagged at its N terminus with 3×HA, primers KS304 and KS305 were used in a PCR on plasmid pKa32, and the PCR product was cloned with BamHI/SphI into pIS56, resulting in plasmid pMAL-3×HA-perCAT2. To construct C-terminally 3×HA-tagged mitCAT2, primers KS306 and KS307 were used in a PCR on plasmid pKa36. A second PCR was performed with KS308 and KS309 on pIS56 to amplify the 3×HA tag. Subsequently a fusion PCR was performed with KS309 and KS310 using the two purified PCR products as a template. The final PCR product was cloned with SacI/SphI into pIS56, resulting in plasmid pMAL-mitCAT2–3×HA. The pMAL-3×HA-perCAT2 and pMAL-mitCAT2–3×HA plasmids were linearized with PacI and transformed to CEM28 (cat2Δ/Δ) to create the 3×HA-perCAT2 and mitCAT2–3×HA strains.
To construct HA-tagged Yat1 and Yat2, primers KS196 and KS200 and primers KS198 and KS201 were used in a PCR on genomic DNA to obtain the YAT1 and YAT2 gene, and the PCR products were cloned with BamHI/SphI into pIS56. Plasmid pIS56 was previously constructed in our laboratory5 and encompasses the MAL2 promoter followed by a triple HA tag, a MCS, and the URA3/IRO fragment in the pSP73 backbone (Promega). The pMAL-3×HA-YAT1 and pMAL-3×HA-YAT2 plasmids were linearized with PacI or XhoI and transformed to SN76 and CEM28 (cat2Δ/Δ).
The cat2Δ/Δ/yat2Δ/Δ (c2/y2Δ/Δ) and cat2Δ/Δ/yat2Δ/Δ/yat1Δ/Δ (c2/y2/y1Δ/Δ) strains were made using the previously described PCR-based procedure (3). Primers KS260 and KS261 and primers KS262 and KS263 were used to amplify the flanking regions of the YAT2 gene. These flanking regions were used in a second PCR on linearized plasmids pFA-CaURA3-loxP and pFA-SAT1-loxP, creating CaURA3 and SAT1 disruption cassettes, respectively. Strain CEM28 was transformed with the YAT2-SAT1 cassette, and positive transformants were selected on plates containing 200 μg/ml nourseothricin and confirmed by PCR. Next, the cat2Δ/Δ/yat2Δ/YAT2 was transformed with the YAT2-URA3 disruption cassette to create the cat2Δ/Δ/yat2Δ/Δ double knock-out strain. The URA3 marker was removed by 5-fluoroorotic acid treatment. Similarly, primers KS250 and KS251 and primers KS252 and KS253 were used to create a YAT1-URA3 disruption cassette. Transformation of this cassette into cat2Δ/Δ/yat2Δ/Δ resulted in strain cat2Δ/Δ/yat2Δ/Δ/yat1Δ/YAT1. After removal of the URA3 marker by 5-fluoroorotic acid treatment, a second YAT1-URA3 disruption cassette was transformed to the strain, creating the cat2Δ/Δ/yat2Δ/Δ/yat1Δ/Δ strain (c2/y2/y1Δ/Δ). Removal of the URA3 marker resulted in the c2/y2/y1Δ/Δ ura3− strain that was transformed with linearized plasmids pLUBP-YAT1, pLUBP-YAT2, pIS52-CAT2, pIS52-perCAT2, pIS52-mitCAT2, pMAL-3×HA-YAT1, pMAL-3×HA-YAT2, and the empty pLUBP plasmid.
Transformation
C. albicans was transformed using a modified lithium acetate protocol (17). The heat shock was carried out at 44 °C for 15 min.
Yeast Two-hybrid Interactions
The constructed Y2H plasmids were transformed to the S. cerevisiae two-hybrid reporter strain PCY2, and transformants were selected on minimal glucose plates supplemented with 30 μg/ml lysine, 20 μg/ml adenine, 20 μg/ml uracil, and 20 μg/ml histidine. Interactions were assayed by staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside X-gal and quantified by determining β-galactosidase activity (18). Total β-galactosidase activity was determined by the formula, (1000 × A420)/(PVt), where P represents protein (mg/ml), V is volume (ml), and t is time (min).
Subcellular Fractionation and Density Gradient Analysis
The subcellular fractionation of C. albicans strains was performed as previously described (5). The homogenate (H), organellar pellet (P), and cytosolic supernatant (S) of the Yat1/Yat2 fractionation experiment were analyzed by immunoblotting. Gradient fractions of the Cat2 subcellular fractionation experiments were analyzed for the presence of enzymatic activity of the peroxisomal marker 3-hydroxyacyl-CoA dehydrogenase (19), the mitochondrial marker fumarase (20), and Cat (6).
Mass Spectrometric Measurements of Metabolites
For acyl-CoA measurements, strains were grown for 16 h on YPO and washed twice with water. Acyl-CoA measurements were performed as described by Hammond et al. (21) with some modifications that were described previously (5). For carnitine and acetylcarnitine measurements, cells were grown for 16 h on YPO or YPA with or without 2 mm carnitine and washed twice with water. 20 OD units were spun down and taken up in 500 μl of phosphate-buffered saline, and 200 μl of glass beads were added, after which the tubes were vortexed for 20 min at 4 °C. The supernatant was transferred to a new tube, and the glass beads were washed with an additional 200 μl. The pooled supernatants were centrifuged twice at high speed to remove cell debris and whole cells, resulting in the final lysates. The protein concentration of the lysates was determined by the method of Bradford using bovine serum albumin as a standard (22). Carnitine and acetylcarnitine levels in the lysates were determined by liquid chromatography-tandem mass spectrometry as described previously (23), using [2H3]carnitine and [2H3]C3-carnitine as internal standards.
Fatty Acid β-Oxidation and Acetate Oxidation Measurements
β-Oxidation activity in intact cells was measured as described before (24) except that the cells were resuspended at an A600 of 1. For acetate oxidation, cells were resuspended at an A600 of 0.5. The assay mixture contained 50 mm MES (pH 6.0), 0.9% NaCl, 10 μm [1-14C]acetate, and 20 μl of cell suspension in a total volume of 200 μl. A tube with the assay mixture and a tube with 500 μl of 2 m NaOH were enclosed in an air-tight vial and incubated at 28 °C for 2 h with gentle shaking. The NaOH-entrapped [14C]CO2 was determined by liquid scintillation counting.
Statistical Analysis
The S.D. value was determined for triplicate experiments and represented as error bars. p values between all samples were determined using the one-way analysis of variance test with Bonferroni correction for multiple comparisons. Relevant p values are depicted in the figures.
Electron Microscopy
Oleate-induced or oleate/maltose-induced cells were fixed with 2% (w/v) formaldehyde, and ultrathin sections were prepared as described previously (25). Immunolabeling was performed using mouse monoclonal 12CA5 antibody for detection of HA-tagged proteins or rabbit polyclonal antibody directed against S. cerevisiae 3-ketoacyl-CoA thiolase for labeling of peroxisomes. Immune complexes were detected with gold-conjugated protein A.
Isocitrate Lyase Enzyme Assay
Preparations of cell-free extracts and enzyme assays were performed essentially as described previously (26), except that extracts were freshly prepared, and the assays were carried out in a UVIKON 820 double beam spectrophotometer (Kontron) at room temperature.
Immunoblotting
Protein extracts were separated on a 10% SDS-polyacrylamide gel and blotted to nitrocellulose membrane using a semidry system. The following rabbit polyclonal antibodies directed against S. cerevisiae proteins were used: thiolase (Thiol), catalase (Cta1), malate synthase (Mls1) (27), peroxisomal membrane protein 35 (Pmp35/Ant1) (28), citrate synthase (Cit1) (29), hexokinase (Hxk1), isocitrate dehydrogenase (Idh1) (a kind gift of H. van der Spek, Faculteit der Natuurwetenschappen, Wiskunde en Informatica), and glucose-6-phosphate dehydrogenase (Zwf1) (Sigma). The isocitrate lyase (Icl1) antibody was directed against Ashbya gossypii Icl1 (30). α-Thiol, α-Cta1, α-Icl1, α-Mls1, and α-Cit1 antibodies were previously tested for specific cross-reactivity with the corresponding C. albicans proteins (5, 7, 31). In this study, α-Ant1 (1:2000), α-Hxk1 (1:1000), α-Zwf1 (1:500), and α-Idh1 (1:1000) antibodies were tested for cross-reactivity with C. albicans proteins. Each antibody detected a band of the predicted molecular weight in a total cell lysate (not shown). Mouse monoclonal 12CA5 antibody was used (1:10) for detection of 3×HA-tagged Yat1 and Yat2.
RESULTS
Acetylcarnitine Synthesis in the Carnitine Acetyltransferase Mutant Strains
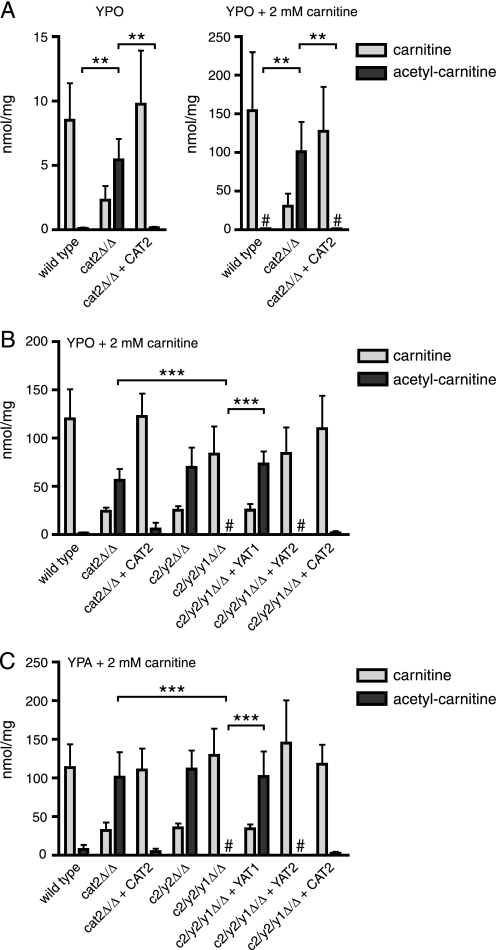
Previously, we have shown that no carnitine acetyltransferase activity could be detected in a C. albicans cat2 null strain lacking both peroxisomal and mitochondrial Cat2 (5), although two other proteins with homology to carnitine acetyltransferases are encoded in the genome: Yat1 and Yat2. In the absence of any Cat activity, it is expected that acetylcarnitine cannot be formed in the cell. To address this, we determined the intracellular carnitine and acetylcarnitine levels in the wild type, cat2 null, and CAT2 complemented strains grown overnight on rich oleate medium with or without additional carnitine (Fig. 1A). In all strains, total acetylcarnitine levels were much higher in the presence of additional carnitine compared with those grown without carnitine, suggesting that acetylcarnitine is synthesized intracellularly and not (only) taken up from the medium. Surprisingly, acetylcarnitine levels were found to be significantly higher in the cat2 null mutant compared with the wild type and complemented strains under both conditions (p < 0.01). These results show that in the absence of Cat2, acetylcarnitine is still synthesized but probably cannot be reconverted to acetyl-CoA and carnitine. In support of this, we found that carnitine levels in the cat2 null strain were relatively low compared with the wild type and complemented strain. To investigate the individual roles of Yat1 and Yat2 in acetylcarnitine accumulation in the cat2 null background, we constructed a cat2/yat2 double null (c2/y2Δ/Δ) and a cat2/yat2/yat1 triple null (c2/y2/y1Δ/Δ) strain and complemented the triple null strain with the CAT2, YAT1, or YAT2 open reading frames. We grew all strains on rich oleate medium with an additional 2 mm carnitine and determined the intracellular levels of carnitine and acetylcarnitine (Fig. 1B). Acetylcarnitine levels were found to be similar in the cat2 null and c2/y2Δ/Δ strain, but subsequent disruption of the YAT1 gene in the c2/y2Δ/Δ strain resulted in a complete reduction of acetylcarnitine levels to background values. Complementation of the c2/y2/y1Δ/Δ strain with the YAT1 gene restored acetylcarnitine levels to that of the cat2 null strain, whereas complementation with YAT2 did not yield any increase in acetylcarnitine levels. Very similar carnitine and acetylcarnitine levels were found in the same strains grown on rich acetate medium with 2 mm carnitine (Fig. 1C). Therefore, we conclude that the carnitine acetyltransferase homolog Yat1 contributes to acetylcarnitine synthesis in the cat2 null background, whereas the other carnitine acetyltransferase homolog, Yat2, does not.
FIGURE 1.
Carnitine and acetylcarnitine levels in the various carnitine acetyltransferase null strains. A, carnitine and acetylcarnitine levels in homogenates of the wild type, cat2Δ/Δ, and cat2Δ/Δ + CAT2 strains grown on rich oleate medium without or with 2 mm carnitine. B, carnitine and acetylcarnitine levels in homogenates of the wild type, cat2Δ/Δ, cat2Δ/Δ + CAT2, c2/y2Δ/Δ, c2/y2/y1Δ/Δ, c2/y2/y1Δ/Δ + YAT1, and c2/y2/y1Δ/Δ + YAT2 grown on rich oleate with 2 mm carnitine. C, same strains as in B grown on rich acetate medium with 2 mm carnitine. #, undetectable levels of the indicated metabolites. Data are represented as mean ± S.D. (error bars) from three independent experiments. Statistical analysis was performed, and relevant p values are indicated. **, p < 0.01; ***, p < 0.001.
N-terminally Tagged Yat1 and Yat2 Localize to the Cytosol
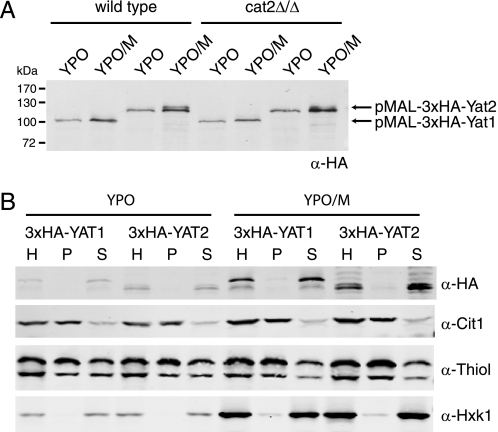
To study the carnitine acetyltransferase homologs YAT1 and YAT2 in more detail, we cloned both genes into an N-terminal 3×HA tagging plasmid. Both fusion proteins are under control of the MAL2 promoter, which is induced by maltose and repressed by glucose in the growth medium. The plasmids were linearized and transformed to the C. albicans wild type, the cat2 null strain, and the c2/y2/y1Δ/Δ strain. Western blot analysis of oleate-grown cells (YPO) revealed expression of both constructs that could be moderately induced by the addition of 0.5% maltose to the medium (YPO/M) (Fig. 2A). Expression of 3×HA-Yat1 in the wild type and cat2 null strains resulted in a single band of the predicted size. The 3×HA-Yat2 strain also showed a single band of the predicted size on YPO, but upon overexpression, an extra band was present that migrates slightly slower than the band visible on YPO. The nature of this extra, slower migrating band is currently unknown. To investigate whether the HA-tagged Yat1 and Yat2 are enzymatically active, we transformed the constructs to the c2/y2/y1Δ/Δ strain and determined carnitine and acetylcarnitine levels in cells grown on YPO or YPO/M. The HA-tagged Yat1, like the untagged protein (see Fig. 1A), restored acetylcarnitine levels in the c2/y2/y1Δ/Δ strain, whereas comparable expression of the tagged Yat2 did not (data not shown). These results show that the 3×HA-Yat1 fusion protein is functional but that the functionality of the 3×HA-Yat2 could not be assessed using acetylcarnitine accumulation as read out, as was also the case for the untagged protein. A direct enzyme assay for Cat activity in lysates of cat2 cells overexpressing either 3×HA-Yat1 or 3×HA-Yat2 from the MAL2 promoter failed to detect any activity (data not shown), an observation that is line with our previous results (5). Thus, although Yat1 contributes to acetylcarnitine formation in a cat2 null strain, direct evidence for carnitine acetyltransferase activity of Yat1, and even more so for Yat2, is still lacking.
FIGURE 2.
Expression and subcellular localization of tagged Yat1 and Yat2. A, immunoblot analysis with α-HA antibody of the wild type and cat2 null strain expressing pMAL2–3×HA-Yat1 and pMAL2–3×HA-Yat2 constructs. Strains were grown on rich oleate medium (YPO) or on YPO + 0.5% maltose (YPO/M) to induce expression from the MAL2 promoter. B, immunoblot analysis of a subcellular fractionation experiment of the 3×HA-Yat1 and 3×HA-Yat2 strains grown on YPO or YPO/M. The total homogenate (H) was fractionated into an organellar pellet fraction (P) and a cytosolic supernatant fraction (S). Thiol, thiolase (peroxisomal marker protein). Cit1, mitochondrial marker protein; Hxk1, cytosolic marker protein. All experiments were performed at least twice; representative experiments are shown.
To investigate the localization of Yat1 and Yat2, a subcellular fractionation experiment was performed on cells expressing the HA-tagged proteins. A total cellular homogenate (H) was separated into an organellar pellet (P) and a cytosolic supernatant (S) fraction, and equivalent volumes of the fractions were analyzed by immunoblotting (Fig. 2B). Both the 3×HA-Yat1 and 3×HA-Yat2 fusion proteins co-fractionated with the cytosolic marker hexokinase and were found almost exclusively in the supernatant fraction. The peroxisomal marker thiolase (Thiol) and the mitochondrial marker citrate synthase (Cit1) fractionated mainly in the pellet fraction, showing overall intactness of the organelles during the fractionation procedure. Immunoelectron microscopy of these strains with α-HA antibodies generally confirmed the subcellular fractionation results because very little gold particles were found associated with either peroxisomes or mitochondria (data not shown). Together, these data indicate that the tagged Yat1 and Yat2 are localized to the cytosol, although the possibility cannot be excluded that N-terminal tagging of the proteins influences their subcellular localization.
Construction and Validation of Strains Expressing either the Peroxisomal or Mitochondrial Form of Cat2
To dissect the individual roles of the peroxisomal and mitochondrial Cat2, we set out to construct strains expressing only one of the two isoenzymes. In S. cerevisiae, dual localization of Cat2 to peroxisomes and mitochondria is regulated at the transcriptional level, resulting in two proteins that either contain or lack the N-terminal MTS, whereas both proteins harbor the C-terminal PTS1. The C. albicans CAT2 gene encodes a (putative) MTS and PTS1, and two conserved in-frame ATGs are present. Because of this sequence similarity between S. cerevisiae and C. albicans CAT2, we predicted that transcriptional regulation and, thus, localization of Cat2 would be comparable between both organisms. First we addressed the question whether peroxisomal targeting of C. albicans Cat2 is dependent on its (putative) PTS1 (-AKLCOOH). In a two-hybrid setup, we confirmed that full-length Cat2, but not Cat2 lacking its C-terminal three amino acids (ΔAKLCOOH), interacts with the wild type PTS1 receptor Pex5. Full-length Cat2 did not interact in the two-hybrid with a mutant Pex5 receptor (CaPEX5N376D) that has previously been shown to abolish PTS1 interaction (data not shown) (32). These results show that the interaction between Pex5 and Cat2 is strictly dependent on the PTS1 of Cat2 and suggest that peroxisomal import of Cat2 can be abolished by deletion of the three C-terminal amino acids.
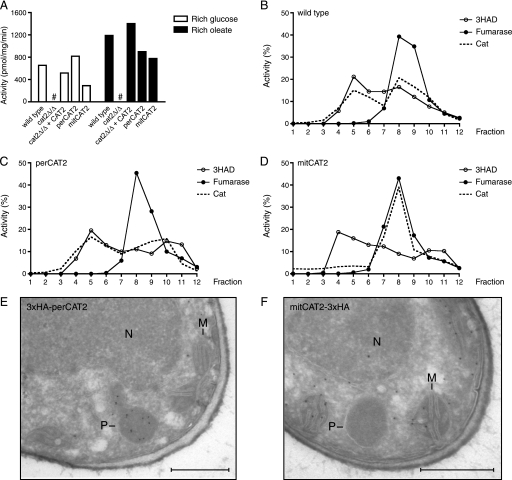
Next, we used the wild type Cat2 complementation construct, containing an 800-bp promoter region and the full-length gene (5), as a template to create constructs that either lack the predicted MTS (perCAT2 construct for expression of the peroxisomal Cat2) or the PTS1 (mitCAT2 construct for expression of mitochondrial Cat2). In the latter construct, also the second ATG was mutated to alanine (M023A), thereby preventing faulty translation of a protein lacking both the MTS and the PTS1 (see “Experimental Procedures” for details on the constructions). The cat2 null strain was transformed with the linearized perCAT2 or mitCAT2 plasmid, and PCR confirmed their correct integration in the URA3 locus. First, we determined total Cat2 activity in lysates of the wild type, cat2 null, CAT2 complemented, perCAT2, and mitCAT2 strains grown on rich glucose and rich oleate medium (Fig. 3A). Cat2 activity in all four strains was in a similar range of 300–800 pmol/mg/min on glucose and 800–1400 pmol/mg/min on oleate, showing that all constructs are enzymatically active and do not result in gross over- or underexpression of Cat2. Localization of Cat2 activity in the wild type, perCat2, and mitCat2 strains was investigated by subcellular fractionation and Nycodenz density gradient analysis. The wild type strain showed a dual distribution of Cat2 activity between peroxisomes and mitochondria (Fig. 3B). Cat2 activity in the perCAT2 strain colocalized with the peroxisomal marker 3-hydroxyacyl-CoA dehydrogenase. Two peaks are visible, one at the expected density of peroxisomes (fractions 3–5) and a second at the top of the gradient (fractions 10–12), most likely representing lysed peroxisomes (Fig. 3C). A single Cat2 activity peak is seen in the gradient of the mitCAT2 strain that shows a strict co-localization with the mitochondrial marker fumarase (Fig. 3D). To obtain independent evidence for the proper localization of the perCAT2 and mitCAT2 constructs, we tagged both proteins with a 3×HA tag and determined their subcellular distribution by immunoelectron microscopy. To prevent interference with targeting of the proteins, the perCAT2 construct was tagged at its N terminus, whereas the mitCAT2 construct was C-terminally tagged (see “Experimental Procedures” for details). The 3×HA-perCAT2 and mitCAT2–3×HA strains were grown on YPO/M to induce expression by the MAL2 promoter. Localization of the 3×HA-perCAT2 construct was found to be peroxisomal with negligible labeling of mitochondria and cytosol (Fig. 3E). The mitCAT2–3×HA protein localized to mitochondria, and some cytosolic labeling was observed, whereas no label was found in peroxisomes (Fig. 3F). Together, the fractionation and the electron microscopy data strongly suggest that Cat2 localizes to the designated organelles in the perCAT2 and mitCAT2 strains, although a low percentage of mislocalization cannot be ruled out.
FIGURE 3.
Subcellular distribution of Cat in wild type, perCAT2, and mitCAT2 strains. A, total Cat activity in homogenates of the wild type, cat2Δ/Δ, cat2Δ/Δ + CAT2, peroxisomal Cat2 (perCAT2), and mitochondrial Cat2 (mitCAT2) strain grown on rich glucose or rich oleate medium. All strains showed higher activity on oleate compared with glucose, except for the cat2Δ/Δ, in which no activity could be detected (#). B, distribution of Cat activity, the peroxisomal marker 3-hydroxyacyl-CoA dehydrogenase (3HAD), and the mitochondrial marker fumarase in a Nycodenz density gradient of the wild type strain, the perCAT2 strain (C), and the mitCAT2 strain (D). In each experiment, an organellar pellet fraction was loaded onto the gradient. Experiments were performed at least twice; representative experiments are shown. E and F, immunoelectron microscopy of 3×HA-perCAT2-expressing (E) and mitCAT2–3×HA-expressing (F) strains. Thin sections were prepared of cells grown on YPO/M medium and incubated with α-HA antibodies and protein A-conjugated gold particles. P, peroxisome; M, mitochondrion; N, nucleus. Bars, 0.5 μm.
Phenotypes of the Carnitine Acetyltransferase Mutant Strains
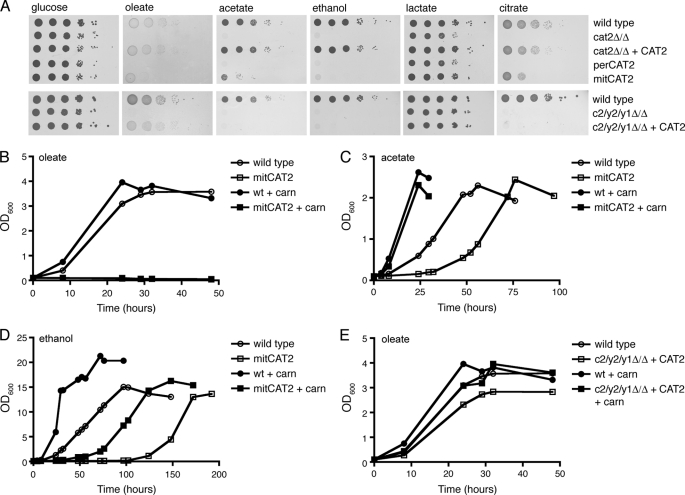
To assess the individual contribution of the peroxisomal and mitochondrial Cat2 isozymes to growth on various carbon sources, we performed spot assays and growth curves. Serial dilutions of the wild type, cat2 null mutant, CAT2 complemented strain, and perCAT2 and mitCAT2 strains were spotted on minimal YNB plates with glucose, oleate, acetate, ethanol, lactate, or citrate as the sole carbon source (Fig. 4A, top). All strains grew equally well on glucose. On non-fermentable carbon sources, the perCAT2 showed a similar growth defect as the cat2 null mutant; no growth was seen on oleate, citrate, acetate, and ethanol, and intermediate (reduced) growth was observed on lactate. The mitCAT2 strain on the other hand showed severely reduced growth on oleate, acetate, or ethanol plates, whereas growth on citrate was less affected, and growth on lactate was comparable with wild type. Although growth of the mitCAT2 strain on oleate, acetate, and ethanol was strongly reduced compared with the wild type, a few colonies did appear after several days in spots with the highest cell density.
FIGURE 4.
Growth phenotypes of the different carnitine acetyltransferase mutant strains. A, serial dilutions (1:10) of the indicated strains were spotted on minimal plates with glucose, oleate, lactate, citrate, acetate, or ethanol as a carbon source and incubated for 3–5 days at 28 °C before pictures were taken. B, growth curve of the wild type and mitCAT2 strains on minimal oleic acid medium without or with 10 μm l-carnitine. C, growth curve of the wild type and mitCAT2 strain on minimal acetate medium without or with 10 μm l-carnitine. D, growth curve of the wild type and mitCAT2 strain on minimal ethanol medium without or with 10 μm l-carnitine. E, growth curves of the wild type and c2/y2/y1Δ/Δ + CAT2 stains on minimal oleate medium without or with 10 μm l-carnitine. All experiments were performed at least twice; representative experiments are shown.
To investigate the intriguing growth phenotype of the mitCAT2 in more detail, we performed growth curves of the wild type and mitCAT2 strain on minimal oleate, acetate, and ethanol medium without or with the addition of 10 μm l-carnitine. In line with the spot assays, the mitCAT2 strain did not grow on liquid oleate medium, and the addition of 10 μm carnitine did not rescue this phenotype (Fig. 4B). Growth of the mitCAT2 strain on liquid acetate and ethanol was delayed compared with the wild type, but after a lag phase of 30 and 100 h, respectively, a nearly wild type growth rate was observed on both carbon sources. Microscopic examination of the cultures and plate assays confirmed that the observed increase in optical density was due to growth of the mitCAT2 strain and not caused by a contamination. The addition of 10 μm carnitine to the minimal acetate or ethanol media resulted in a shift of the mitCAT2 growth curves toward wild type (Fig. 4, C and D), indicating that the growth delay of the mitCAT2 is caused by carnitine depletion, most likely as a consequence of insufficient carnitine biosynthesis (3) and intracellular acetylcarnitine accumulation (data not shown). Together, these results show that enzymatic activity of peroxisomal Cat2 is essential for growth on oleate but dispensable during growth on lactate, citrate, ethanol, or acetate. The observation that growth on ethanol and acetate can occur independently of peroxisomal Cat2 implies that the cytosolic acetyl units can enter the peroxisomal glyoxylate cycle in a carnitine-independent manner. Entry of acetyl units into mitochondria, on the other hand, is strictly dependent on carnitine because the mitochondrial Cat2 is essential for growth on all non-fermentable carbon sources tested.
To investigate the contribution of Yat1 and/or Yat2 to acetyl unit transport on non-fermentable carbon sources, the c2/y2/y1Δ/Δ + CAT2 strain was spotted onto minimal plates containing glucose, oleate, acetate, ethanol, lactate, or citrate (Fig. 4A, bottom). In line with what was previously reported for a YAT1 knock-out strain (4), our strain lacking both Yat1 and Yat2 is unable to grow on ethanol, acetate, or citrate, indicating that under these growth conditions formation of acetylcarnitine in the cytosol is required. In contrast, the c2/y2/y1Δ/Δ + CAT2 strain showed a nearly wild type growth phenotype on oleate plates and in liquid minimal oleate medium, either in the absence or presence of additional carnitine (Fig. 4, A (bottom) and E). We therefore conclude that the cytosolic Yat1/Yat2 are not required for growth on oleate, suggesting that in the presence of both peroxisomal and mitochondrial Cat2, acetylcarnitine is formed in peroxisomes and directly exported to mitochondria without interference of either Yat1 or Yat2 in the cytosol.
Oxidation of Fatty Acids and Acetate in the Carnitine Acetyltransferase Mutant Strains
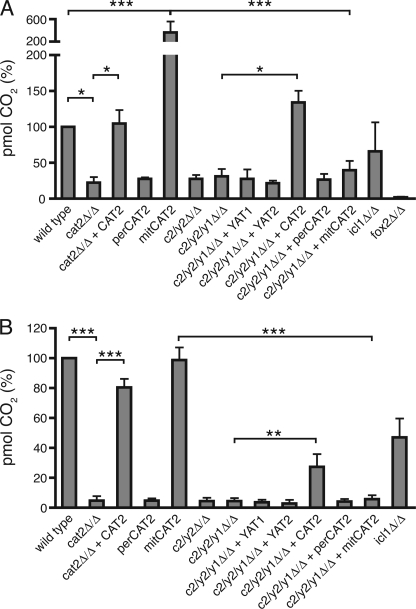
During growth on oleic acid or acetate, peroxisomal or cytosolic acetyl units need to be transported to the mitochondria, where they can be oxidized to H2O and CO2 in the tricarboxylic acid cycle. To directly determine the ability of the different strains to transport acetyl-CoA from peroxisomes or cytosol to the mitochondria, we incubated our strains for 2 h with 14C-labeled oleic acid or 14C-labeled acetate and measured the amount of labeled CO2 produced. Oxidation of oleic acid to CO2 was undetectable in the fox2 null mutant, which is blocked in fatty acid β-oxidation (31), and low in the cat2 null and the perCAT2 strains (Fig. 5A). Surprisingly, the mitCAT2 strain showed a 2–3-fold higher CO2 production than the wild type and complemented strain. The high β-oxidation in the mitCAT2 strain is lost when Yat1 and Yat2 are deleted (compare mitCAT2 with c2/y2/y1Δ/Δ + mitCAT2). These results strongly suggest that in C. albicans, export of peroxisomal acetyl units can occur independently of peroxisomal Cat2 and that under these conditions, cytosolic Yat activity becomes essential to supply acetyl units to the mitochondrial tricarboxylic acid cycle. Oxidation of labeled acetate was comparable between wild type, the complemented strain, and the mitCAT2 strain, showing that (as expected from the growth curves) peroxisomal Cat2 does not play a role in transport of acetyl units from cytosol to mitochondria (Fig. 5B). Finally, the acetate oxidation experiments support the role of Yat1/(Yat2) in the formation of acetylcarnitine in the cytosol when acetate (or ethanol) is the carbon source because CO2 production is almost completely abolished in the mitCAT2 strain when Yat1 and Yat2 are absent. Complementation of the c2/y2/y1Δ/Δ strain with full-length CAT2 slightly restores acetate oxidation, which might indicate that some Cat2 is present in the cytosol that enables transport to the mitochondria.
FIGURE 5.
Oxidation of fatty acids and acetate in the different carnitine acetyltransferase mutants. A, β-oxidation of 14C-labeled oleic acid in cells grown on rich oleate medium measured by production of labeled CO2. B, oxidation of 14C-labeled acetate in cells grown on rich oleate medium measured by production of labeled CO2. Data are represented as mean ± S.D. (error bars) from three independent experiments. Statistical analysis was performed, and relevant p values are indicated. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
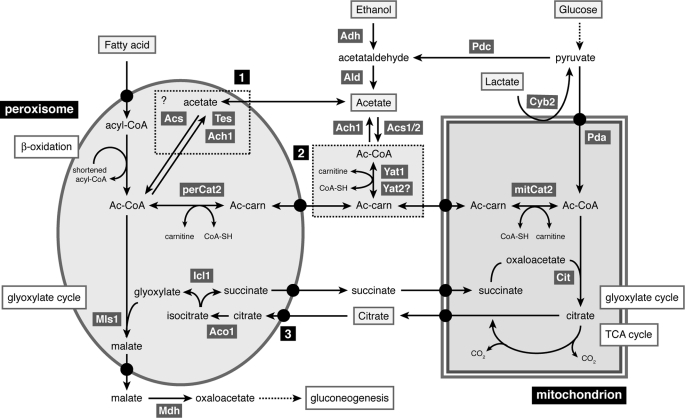
To unravel the mechanism of carnitine-dependent acetyl unit transport between organelles, we performed a detailed functional analysis of the three (putative) Cats of C. albicans, Cat2, Yat1, and Yat2. To dissect the individual roles of the peroxisomal and mitochondrial Cat2, we constructed mutant strains expressing only one of the two isoenzymes and strains lacking either Yat1 or Yat2 or both. We determined the localization of all proteins by HA tagging and subcellular fractionation. Based on detailed phenotypic and biochemical analyses of the various mutant strains, we present a model for carnitine-dependent transport between the peroxisomal, mitochondrial, and cytosolic compartments in C. albicans (Fig. 6).
FIGURE 6.
Model showing the interorganellar flow of acetyl units in C. albicans. Depicted biochemical pathways are β-oxidation of fatty acids, glyoxylate cycle, and tricarboxylic acid (TCA) cycle. Ach1, acetyl-CoA hydrolase; Aco, aconitase; Acs1/2, acetyl-CoA synthase; Adh, alcohol dehydrogenase; Ald, acetaldehyde dehydrogenase; Cit, citrate synthase; Cyb2, l-lactate dehydrogenase; Icl1, isocitrate lyase; mitCat2, mitochondrial Cat2; Mls1, malate synthase; Pda, pyruvate dehydrogenase complex; Pdc, pyruvate decarboxylase; perCat2, peroxisomal Cat2; Tes, thioesterase; Yat1/2, (putative) carnitine acetyltransferases. Box 1, alternative pathway for transport of acetyl units across the peroxisomal membrane in the form of acetate; Box 2, cytosolic carnitine acetyltransferases; 3, putative transport deficiency in the mitCAT2 strain. For details, see “Discussion.”
Formation of Acetylcarnitine in the Cytosol; Role of Yat1 and Yat2
During growth on non-fermentable carbon sources other than fatty acids, such as acetate and ethanol, acetyl-CoA is produced in the cytosol and needs to be transported to both peroxisomes, where the key enzymes of the glyoxylate cycle, Mls1 and Icl1, are localized (7) and to mitochondria to feed into the tricarboxylic acid cycle. Although both Yat1 and Yat2 are cytosolic (Fig. 2B), our data show that the enzymes do not have redundant functions because acetylcarnitine formation in the cytosol strictly depends on Yat1, whereas Yat2 does not contribute to this process (Fig. 1, B and C). Although we have been unable to directly measure Cat activity for either Yat1 or Yat2, the phenotypes of the various mutant strains strongly suggest that Yat1 is a true carnitine acetyltransferase. In the c2/y2/y1Δ/Δ + CAT2 strain, acetate oxidation is blocked, and this strain cannot grow on acetate or ethanol, whereas growth on oleate is unaffected (Fig. 4, A and B). These observations are in line with the results reported by Zhou and Lorenz (4) who showed that a yat1 null strain is unable to grow on acetate or ethanol. Intriguingly, a strain lacking the cytosolic Yats also cannot grow on citrate (Fig. 4A) (4). These results indicate that during growth on citrate, acetyl-CoA is generated in the cytosol, which is subsequently linked to carnitine by Yat1/(Yat2). In support of this, we show that the mitochondrial Cat2 and, to a lesser extent, peroxisomal Cat2 are required for citrate utilization (Fig. 4A).
Import of Acetyl Units into Mitochondria; Role of Mitochondrial Cat2
Activity of the mitochondrial Cat2 is very likely to be associated with the (intra)mitochondrial conversion of acetylcarnitine to acetyl-CoA, which feeds into the tricarboxylic acid cycle, generating reducing equivalents that can be used for ATP synthesis. Acetyl-CoA can be produced directly in the mitochondria from cytosolic pyruvate by the pyruvate dehydrogenase complex. Therefore, growth on glucose does not require acetyl unit transport, and likewise acetyl unit transport is not strictly required during growth on lactate. Indeed, none of the mutant stains showed a growth defect on glucose, and growth on lactate was only mildly affected in some mutants (Fig. 4A). Not surprisingly, we showed that the mitochondrial Cat2 is essential during growth on oleate, ethanol, and acetate because all of these carbon sources require import of acetyl units into the mitochondria. Accordingly, the perCAT2 strain that lacks the mitochondrial Cat2 cannot grow on or utilize these non-fermentable carbon sources (Figs. 4 and 5). The perCAT2 strain displays reduced growth on lactate, suggesting that not all pyruvate synthesized from lactate directly enters the mitochondria, but part is converted to acetyl-CoA in the cytosol via pyruvate decarboxylase (Pdc) (Fig. 6). That this pathway is operational in C. albicans has been suggested previously, based on the observation that an icl1 null mutant is unable to grow on lactate (31).
Export of Acetyl Units from Peroxisomes Is Not Strictly Dependent on Peroxisomal Cat2
In contrast to S. cerevisiae, C. albicans does not have an alternative, citrate synthase-dependent export pathway because it lacks the peroxisomal isoenzyme Cit2, and therefore this pathogenic yeast completely relies on the carnitine-dependent pathway (5). The activity of the peroxisomal Cat2 is thought to be involved in the export of acetyl units produced during β-oxidation of fatty acids. Peroxisomal Cat2 links the acetyl units to carnitine, and acetylcarnitine can cross the membrane and be transported further to the mitochondria. Our data show that although peroxisomal Cat2 is essential for growth on oleate (Fig. 4), the mitCAT2 strain is still able to oxidize fatty acids (Fig. 5). The β-oxidation assay used measures the production of labeled CO2 from 14C-labeled oleic acid in whole cells and therefore requires a functional peroxisomal β-oxidation, transport of acetyl units from peroxisomes to mitochondria, and a functional tricarboxylic acid cycle in which CO2 can be produced. The mitCAT2 strain shows a very high production of CO2 and incorporation of label in acid-soluble intermediates (Fig. 5) (data not shown). These results strongly suggest that under these conditions, acetyl units are transported from peroxisomes to mitochondria, even in the absence of peroxisomal Cat2. A possible way to export acetyl units independently of peroxisomal Cat2 is the conversion of acetyl-CoA to acetate by a peroxisomal thioesterase or a putative peroxisomal acetyl-CoA hydrolase (Fig. 6, dashed box 1). In mammals, peroxisomal thioesterases and carnitine-acyl/acetyltransferases were shown to be differentially expressed and suggested to provide complementary systems for exporting β-oxidation products from peroxisomes (33). Whereas S. cerevisiae has a single peroxisomal thioesterase (Tes1) (34), the C. albicans genome encodes five putative thioesterases, of which four have a likely PTS1 (-AKLCOOH or -SRLCOOH). Thioesterases hydrolyze acyl-CoAs to the corresponding free fatty acids. Several studies have suggested, however, that peroxisomes may also contain acetyl-CoA thioesterase activity (35–37), and an acetyl-CoA thioesterase referred to as ACOT12 has been detected in isolated peroxisomes of rats and mice (38, 39). It remains to be investigated if one of the C. albicans isoenzymes is localized to peroxisomes and has acetyl-CoA thioesterase activity. Another possibility to produce acetate from acetyl-CoA is posed by the enzyme acetyl-CoA hydrolase (Ach1); however, this protein is predicted to be cytosolic in C. albicans because it lacks obvious (peroxisomal) targeting signals. The produced acetate might be able to cross the peroxisomal membrane by active transport or diffusion. Recent data show that the peroxisomal membrane contains pore-forming proteins that enable transfer of small molecules across the membrane (40, 41). It remains to be investigated whether these channel-forming proteins also mediate the export of acetate from the peroxisome. In plants, transport of acetate in the other direction (from cytosol to peroxisomes) was shown to be dependent on the Comatose transporter (Cts) that is also the acyl-CoA transporter in plants (42). Pxa1 and Pxa2 are the yeast orthologs of Cts; therefore, these transporters might also play a role in acetate transport (43). Upon arrival in the cytosol, acetate can be converted to acetyl-CoA by one of the cytosolic acetyl-CoA synthases (Acs1 or Asc2), linked to carnitine by a cytosolic carnitine acetyltransferase (most likely Yat1), and transported to mitochondria. The fact that oxidation of oleate by the mitCAT2 strain is dependent on cytosolic Yat1/(Yat2) supports the hypothesis that it is indeed acetate that is being transported to the cytosol. If true, it is remarkable that this carnitine-independent export of C2 units appears to be very efficient, as can be deduced form the high CO2 production measured in the β-oxidation assay (Fig. 5A). Despite the high β-oxidation activity, biomass formation from fatty acids is strongly reduced in the mitCAT2 strain both in minimal and rich oleate medium (Fig. 4, A and B, and supplemental Fig. S1A), an unexpected finding that is discussed below.
Uncoupling of β-Oxidation and Glyoxylate Cycle in the mitCAT2 Strain
One of our most striking observations is that the mitCAT2 strain is still able to β-oxidize fatty acids but cannot efficiently use these fatty acids for energy conservation and biomass formation and thus for growth. Analysis of the mitCAT2 strain suggests that the tricarboxylic acid cycle and the key enzymes of the glyoxylate cycle, Icl1 and Mls1, are functional (data not shown). However, the levels of (iso)citrate were found to be strongly elevated when cells were incubated in rich oleate medium (data not shown). Because (iso)citrate produced in the mitochondrial tricarboxylic acid cycle must be transported to peroxisomes to feed into the glyoxylate cycle, the accumulation of (iso)citrate suggests a transport defect of this metabolite, either its export out of the mitochondria or its import into peroxisomes. We also found that peroxisomes in the mitCAT2 strain grown on rich oleate medium have a very irregular shape and a change in size compared with wild type cells (supplemental Fig. S1, B–F). An increase in size and a changed peroxisomal morphology have been previously connected to transport deficiencies of glyoxylate cycle intermediates in C. albicans (7), and we speculate that the aberrant peroxisomal morphology of the mitCAT2 strain might cause a similar transport defect. In this scheme, (iso)citrate would not be able to enter the peroxisomal glyoxylate cycle due to changed peroxisomal morphology, resulting in a block in synthesis of C4 units for gluconeogenesis and thus growth (Fig. 6, marked 3). This hypothesis is strengthened by our observation that although the icl1 null mutant is unable to grow on fatty acids (31, 44), it does show wild type levels of β-oxidation activity (Fig. 5).
Transport of Acetyl Units from Cytosol to Peroxisomes
During growth of C. albicans on non-fermentable carbon sources other than fatty acids, such as ethanol and acetate, acetyl-CoA is produced in the cytosol and needs to be imported into peroxisomes where the key enzymes of the glyoxylate cycle, Mls1 and Icl1, are localized (7). It is conceivable that under these conditions, Yat1 is involved in acetylcarnitine synthesis in the cytosol (see above), which is subsequently imported into peroxisomes where perCAT2 catalyzes the reverse reaction, generating acetyl-CoA in the peroxisomal matrix (Fig. 6, dashed box 2). However, our results suggest that under certain conditions, acetate is transported over the peroxisomal membrane and converted to acetyl-CoA in the peroxisomal matrix (Fig. 6, dashed box 1). In the presence of acetate or ethanol as a carbon source, lack of peroxisomal Cat2 leads to a growth delay that can be solved by the addition of carnitine (Fig. 4, C and D). We have shown previously that the substrate of the carnitine biosynthesis pathway, trimethyllysine, is the limiting factor during growth of C. albicans on non-fermentable carbon sources (3). The carnitine-dependent growth phenotype of the mitCAT2 strain on acetate and ethanol suggests that intracellular carnitine is most likely depleted by trapping of carnitine in acetylcarnitine in the peroxisomal compartment. The nearly wild type growth rates on acetate/ethanol of the mitCAT2 strain in the presence of sufficient carnitine suggest that transport of acetyl units from the cytosol to the mitochondria is carnitine-mediated but that transport to the peroxisomes occurs independent of carnitine and perCAT2, most likely in the form of acetate. In this model, acetate is converted to acetyl-CoA by a peroxisomal acetyl-CoA synthase and can subsequently enter the glyoxylate cycle. Whether acetyl-CoA synthase is peroxisomal in C. albicans remains to be established. Therefore, we conclude that in C. albicans, bidirectional acetate transport over the peroxisomal, but not the mitochondrial, membrane is likely to occur in addition to the carnitine-dependent transport of acetyl units between compartments.
Acknowledgments
We thank the mass spectrometry section of the Department of Genetic Metabolic Diseases, especially Arno van Cruchten, for technical assistance.
This work was supported by a grant from the Academic Medical Center, Amsterdam.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Fig. S1.
I. Smaczynska-de Rooij, unpublished results.
- Cat
- carnitine acetyltransferase
- MTS
- mitochondrial targeting signal
- YNB
- yeast nitrogen base
- MCS
- multiple cloning site
- MES
- 4-morpholineethanesulfonic acid
- HA
- hemagglutinin.
REFERENCES
- 1.Kunau W. H., Dommes V., Schulz H. (1995) Prog. Lipid. Res. 34, 267–342 [DOI] [PubMed] [Google Scholar]
- 2.van Roermund C. W., Elgersma Y., Singh N., Wanders R. J., Tabak H. F. (1995) EMBO J. 14, 3480–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strijbis K., van Roermund C. W., Hardy G. P., van den Burg J., Bloem K., de Haan J., van Vlies N., Wanders R. J., Vaz F. M., Distel B. (2009) FASEB J. 23, 2349–2359 [DOI] [PubMed] [Google Scholar]
- 4.Zhou H., Lorenz M. C. (2008) Microbiology 154, 500–509 [DOI] [PubMed] [Google Scholar]
- 5.Strijbis K., van Roermund C. W., Visser W. F., Mol E. C., van den Burg J., MacCallum D. M., Odds F. C., Paramonova E., Krom B. P., Distel B. (2008) Eukaryot. Cell 7, 610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elgersma Y., van Roermund C. W., Wanders R. J., Tabak H. F. (1995) EMBO J. 14, 3472–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piekarska K., Hardy G., Mol E., van den Burg J., Strijbis K., van Roermund C., van den Berg M., Distel B. (2008) Microbiology 154, 3061–3072 [DOI] [PubMed] [Google Scholar]
- 8.Prigneau O., Porta A., Maresca B. (2004) Fungal Genet. Biol. 41, 783–793 [DOI] [PubMed] [Google Scholar]
- 9.Swiegers J. H., Dippenaar N., Pretorius I. S., Bauer F. F. (2001) Yeast 18, 585–595 [DOI] [PubMed] [Google Scholar]
- 10.Kispal G., Sumegi B., Dietmeier K., Bock I., Gajdos G., Tomcsanyi T., Sandor A. (1993) J. Biol. Chem. 268, 1824–1829 [PubMed] [Google Scholar]
- 11.Schmalix W., Bandlow W. (1993) J. Biol. Chem. 268, 27428–27439 [PubMed] [Google Scholar]
- 12.Franken J., Kroppenstedt S., Swiegers J. H., Bauer F. F. (2008) Curr. Genet. 53, 347–360 [DOI] [PubMed] [Google Scholar]
- 13.Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. (1987) Methods Enzymol. 154, 164–175 [DOI] [PubMed] [Google Scholar]
- 14.Noble S. M., Johnson A. D. (2005) Eukaryot. Cell 4, 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chevray P. M., Nathans D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5789–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elgersma Y., Kwast L., Klein A., Voorn-Brouwer T., van den Berg M., Metzig B., America T., Tabak H. F., Distel B. (1996) J. Cell Biol. 135, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walther A., Wendland J. (2003) Curr. Genet. 42, 339–343 [DOI] [PubMed] [Google Scholar]
- 18.Klein A. T., van den Berg M., Bottger G., Tabak H. F., Distel B. (2002) J. Biol. Chem. 277, 25011–25019 [DOI] [PubMed] [Google Scholar]
- 19.Wanders R. J., IJlst L., van Gennip A. H., Jakobs C., de Jager J. P., Dorland L., van Sprang F. J., Duran M. (1990) J. Inherit. Metab. Dis. 13, 311–314 [DOI] [PubMed] [Google Scholar]
- 20.van Roermund C. W., Drissen R., van Den Berg M., Ijlst L., Hettema E. H., Tabak H. F., Waterham H. R., Wanders R. J. (2001) Mol. Cell. Biol. 21, 4321–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond L. E., Neschen S., Romanelli A. J., Cline G. W., Ilkayeva O. R., Shulman G. I., Muoio D. M., Coleman R. A. (2005) J. Biol. Chem. 280, 25629–25636 [DOI] [PubMed] [Google Scholar]
- 22.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 23.van Vlies N., Tian L., Overmars H., Bootsma A. H., Kulik W., Wanders R. J., Wood P. A., Vaz F. M. (2005) Biochem. J. 387, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Roermund C. W., Hettema E. H., Kal A. J., van den Berg M., Tabak H. F., Wanders R. J. (1998) EMBO J. 17, 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hettema E. H., Girzalsky W., van Den Berg M., Erdmann R., Distel B. (2000) EMBO J. 19, 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jong-Gubbels P., Vanrolleghem P., Heijnen S., van Dijken J. P., Pronk J. T. (1995) Yeast 11, 407–418 [DOI] [PubMed] [Google Scholar]
- 27.Kunze M., Kragler F., Binder M., Hartig A., Gurvitz A. (2002) Eur. J. Biochem. 269, 915–922 [DOI] [PubMed] [Google Scholar]
- 28.Visser W. F., van Roermund C. W., Waterham H. R., Wanders R. J. (2002) Biochem. Biophys. Res. Commun. 299, 494–497 [DOI] [PubMed] [Google Scholar]
- 29.Vélot C., Lebreton S., Morgunov I., Usher K. C., Srere P. A. (1999) Biochemistry 38, 16195–16204 [DOI] [PubMed] [Google Scholar]
- 30.Maeting I., Schmidt G., Sahm H., Revuelta J. L., Stierhof Y. D., Stahmann K. P. (1999) FEBS Lett. 444, 15–21 [DOI] [PubMed] [Google Scholar]
- 31.Piekarska K., Mol E., van den Berg M., Hardy G., van den Burg J., van Roermund C., MacCallum D., Odds F., Distel B. (2006) Eukaryot. Cell 5, 1847–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein A. T., Barnett P., Bottger G., Konings D., Tabak H. F., Distel B. (2001) J. Biol. Chem. 276, 15034–15041 [DOI] [PubMed] [Google Scholar]
- 33.Westin M. A., Hunt M. C., Alexson S. E. (2008) Cell Mol. Life Sci. 65, 982–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leighton F., Bergseth S., Rørtveit T., Christiansen E. N., Bremer J. (1989) J. Biol. Chem. 264, 10347–10350 [PubMed] [Google Scholar]
- 35.Wilcke M., Alexson S. E. (1994) Eur. J. Biochem. 222, 803–811 [DOI] [PubMed] [Google Scholar]
- 36.Osmundsen H., Neat C. E., Borrebaek B. (1980) Int. J. Biochem. 12, 625–630 [DOI] [PubMed] [Google Scholar]
- 37.Hovik R., Brodal B., Bartlett K., Osmundsen H. (1991) J. Lipid Res. 32, 993–999 [PubMed] [Google Scholar]
- 38.Nakanishi Y., Okamoto K., Isohashi F. (1994) J. Biochem. 115, 328–332 [DOI] [PubMed] [Google Scholar]
- 39.Wiese S., Gronemeyer T., Ofman R., Kunze M., Grou C. P., Almeida J. A., Eisenacher M., Stephan C., Hayen H., Schollenberger L., Korosec T., Waterham H. R., Schliebs W., Erdmann R., Berger J., Meyer H. E., Just W., Azevedo J. E., Wanders R. J., Warscheid B. (2007) Mol. Cell Proteomics 6, 2045–2057 [DOI] [PubMed] [Google Scholar]
- 40.Grunau S., Mindthoff S., Rottensteiner H., Sormunen R. T., Hiltunen J. K., Erdmann R., Antonenkov V. D. (2009) FEBS J. 276, 1698–1708 [DOI] [PubMed] [Google Scholar]
- 41.Rokka A., Antonenkov V. D., Soininen R., Immonen H. L., Pirilä P. L., Bergmann U., Sormunen R. T., Weckström M., Benz R., Hiltunen J. K. (2009) PLoS ONE 4, e5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hooks M. A., Turner J. E., Murphy E. C., Johnston K. A., Burr S., Jarosławski S. (2007) Biochem. J. 406, 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hettema E. H., van Roermund C. W., Distel B., van den Berg M., Vilela C., Rodrigues-Pousada C., Wanders R. J., Tabak H. F. (1996) EMBO J. 15, 3813–3822 [PMC free article] [PubMed] [Google Scholar]
- 44.Lorenz M. C., Fink G. R. (2001) Nature 412, 83–86 [DOI] [PubMed] [Google Scholar]