Abstract
Foot-and-mouth disease virus (FMDV), a positive sense, single-stranded RNA virus, causes a highly contagious disease in cloven-hoofed livestock. Like other picornaviruses, FMDV has a conserved 2C protein assigned to the superfamily 3 helicases a group of AAA+ ATPases that has a predicted N-terminal membrane-binding amphipathic helix attached to the main ATPase domain. In infected cells, 2C is involved in the formation of membrane vesicles, where it co-localizes with viral RNA replication complexes, but its precise role in virus replication has not been elucidated. We show here that deletion of the predicted N-terminal amphipathic helix enables overexpression in Escherichia coli of a highly soluble truncated protein, 2C(34–318), that has ATPase and RNA binding activity. ATPase activity was abrogated by point mutations in the Walker A (K116A) and B (D160A) motifs and Motif C (N207A) in the active site. Unliganded 2C(34–318) exhibits concentration-dependent self-association to yield oligomeric forms, the largest of which is tetrameric. Strikingly, in the presence of ATP and RNA, FMDV 2C(34–318) containing the N207A mutation, which binds but does not hydrolyze ATP, was found to oligomerize specifically into hexamers. Visualization of FMDV 2C-ATP-RNA complexes by negative stain electron microscopy revealed hexameric ring structures with 6-fold symmetry that are characteristic of AAA+ ATPases. ATPase assays performed by mixing purified active and inactive 2C(34–318) subunits revealed a coordinated mechanism of ATP hydrolysis. Our results provide new insights into the structure and mechanism of picornavirus 2C proteins that will facilitate new investigations of their roles in infection.
Keywords: ATPases, Enzyme Mechanisms, Positive Strand RNA Viruses, Protein Assembly, RNA-binding Protein, Viral Protein, Viral Replication, AAA+ Protein, Foot-and-mouth Disease Virus, Protein Oligomerization
Introduction
Foot-and-mouth disease virus (FMDV)5 causes a highly contagious disease of cloven-hoofed animals, such as cattle, sheep, and pigs (1, 2). As a member of the picornavirus family that also includes human pathogens, such as poliovirus (PV), hepatitis A virus (HAV), and human rhinovirus, FMDV possesses a single-stranded, positive sense RNA genome of some 8500 nucleotides, containing one large open reading frame. This is translated as a single polyprotein that is co- and post-translationally cleaved by viral proteases to release the mature proteins needed for virus replication (1).
Following the initial rounds of translation, the RNA genome is used as the template for the synthesis of a complementary negative strand that, in turn, becomes the template for the synthesis of many positive strands. This asymmetric replication process generates genomic copies that are packaged to make new virions (1). Picornavirus infection causes rearrangement of intracellular membranes into characteristic vesicles that provide platforms for assembly of the viral RNA replication complex, which largely comprises mature and precursor proteins derived from the P2 and P3 regions of the polyprotein (3–5). Targeting of replication complexes to vesicular membranes has a number of potential benefits, such as increasing the local concentration of the viral and cellular proteins co-opted for RNA synthesis or shielding the viral RNA from cellular sensors of infection (6). Rearrangement of intracellular membranes during FMDV and PV infection is accompanied by the disruption of normal cellular protein trafficking and cell surface presentation of viral peptides or release of cytokines; both of these effects may dampen the antiviral response of the host (7–10).
Several of the picornavirus proteins involved in RNA replication (2B, 2C, and 3A) have membrane binding properties, although their precise roles in replication have yet to be fully worked out. Although they are implicated in disturbing the structure and/or function of membranes in host cells, their effects show some variations between different picornaviruses (3, 9, 11, 12). For PV, transient expression of 2C or 2BC in mammalian cells induces vesicular clustering, similar to the membrane rearrangements observed during infection (11); however, PV 2B, 2BC, or 3A (but not 2C) are each capable of disrupting protein trafficking and cell surface protein expression, with 3A being the most efficient blocker (9, 13). In contrast, the vesicles observed during FMDV infection do not cluster (5, 12). The particular FMDV proteins responsible for vesicularization have not been identified, although it has been shown that transient expression of 2B and 2C or of the 2BC precursor (but not 3A) is sufficient to impede trafficking of proteins between the endoplasmic reticulum and Golgi and their expression on the cell surface (7, 12).
2C is the largest and arguably the most complex of the membrane-binding components of the virus RNA replication complex. FMDV 2C, a 318-amino acid polypeptide, exemplifies the common architecture of picornavirus 2C proteins. It has a predicted amphipathic helix in its N terminus (residues 17–34) (14), a feature that has been shown for other picornaviruses to confer the ability to bind to cell membranes (15–18) and is thought to be required for both membrane rearrangement and formation of the viral replication complex (3). This predicted helix lies upstream of a well conserved ATPase domain (19) formed by residues 60–270. However, FMDV 2C is unusual in lacking a Cys-rich motif at its C terminus that, although conserved in several other 2C proteins, is of unknown function (20).
The ATPase domains of picornavirus 2C proteins contain Walker A and Walker B motifs characteristic of many enzymes that hydrolyze NTPs (19, 21, 22) (Fig. 1a). The detection of a third motif, Motif C, resulted in 2C being classified as an AAA+ (ATPases associated with various cellular activities) protein subgrouped with superfamily 3 (SF3) helicases, such as the SV40 large T-antigen (LTag) and bovine papillomavirus E1 (E1) DNA helicases (19). This designation implies that 2C may act as an RNA helicase, but, although the enzyme is capable of hydrolyzing nucleotides (23–26) and binds RNA (see below), no helicase activity has yet been reported for the 2C protein of any picornavirus. In the case of PV, the ATPase activity appears not to be necessary for the induction of membrane rearrangements by 2C (22); nevertheless, this activity is clearly important for virus infection because mutations of the Walker A or Walker B motifs that block ATP hydrolysis by PV 2C (23) also prevent virus replication (21, 22).
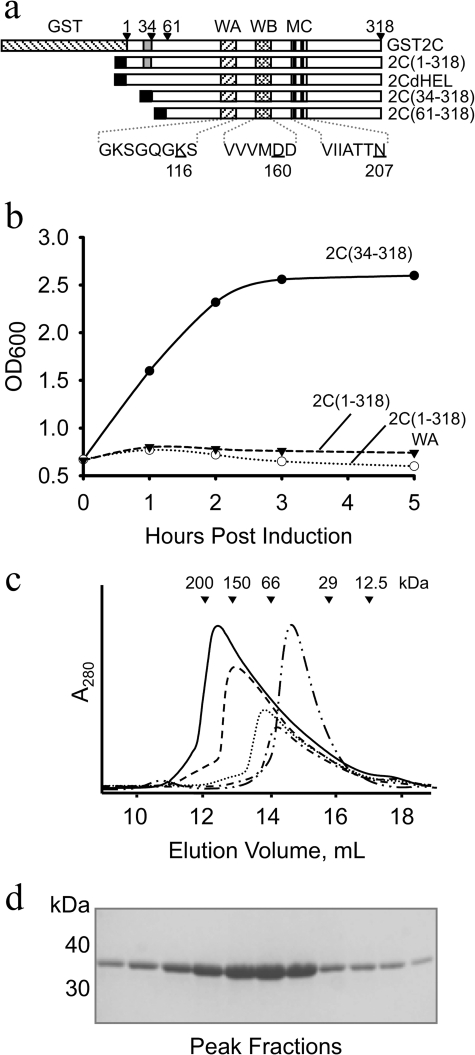
FIGURE 1.
Modifications of the N terminus of FMDV 2C to enhance soluble overexpression in E. coli. a, schematic representation of the modified FMDV 2C proteins produced for this study. The Walker A (WA), Walker B (WB), and C (MC) motifs are indicated. Residues mutated in these motifs are underlined. The predicted N-terminal amphipathic region is shaded gray; the black box indicates the N-terminal His6 tag. b, E. coli growth curves following induction of His6-tagged wild type 2C(1–318) (broken line), the Walker A mutant (K116A) 2C(1–318)WA (dotted line), or the deletion mutant 2C(34–318) (solid line); cell density was monitored by OD measurements at 600 nm. c, size exclusion chromatography of purified FMDV 2C(34–318) and 2C(61–318). In five separate runs, 10 mg (solid line), 7.5 mg (dashed line), 5 mg (dotted line), and 2.5 mg (dashed-single dotted line) of 2C(34–318) or 10 mg of 2C(61–318) (dashed-double dotted line) was loaded onto the column. The elution positions of molecular mass standards are indicated: 1) β-amylase (200 kDa), 2) alcohol dehydrogenase (150 kDa); 3) BSA (66 kDa), 4) carbonic anhydrase (29 kDa), and 5) cytochrome c (12.5 kDa). d, 12% SDS-PAGE of the peak fractions from the 10-mg run shown in c.
The replication of several picornaviruses is also inhibited by low (2–3 mm) concentrations of guanidine hydrochloride (GdnHCl), an effect that is generally attributable to drug interactions with the 2C protein because mutations in 2C confer resistance on the virus (27–31). Such mutations include an M159L substitution in the Walker B motif of FMDV 2C (29) and a N179A mutation within the ATPase active site of PV 2C (27). In vitro studies confirm that 2C is the target of GdnHCl for PV and suggest that the drug works by interfering with the ability of the protein to hydrolyze ATP. The ATPase activity of purified recombinant PV 2C containing the N179A mutation was 100-fold higher than the wild-type protein in the presence of 1 mm drug (23). However, not all picornavirus 2C proteins are so sensitive to the drug; for example, 2C from echovirus-9 (EV9) was only very weakly inhibited by GdnHCl even at concentrations as high as 20 mm (28).
Consistent with the predicted helicase role of 2C, the protein also binds RNA. PV 2C was readily cross-linked to replicating viral RNA by exposure of infected cells to UV light (3). However, the specificity of RNA binding by 2C is still unclear. A significant problem in the investigation of the RNA binding, which also impacts the study of other 2C activities, is the generally poor solubility of the recombinant protein. His6-tagged 2C proteins from PV, HAV, and human rhinovirus, which had to be denatured and refolded during purification from inclusion bodies, were reported to bind a specific sequence in the 3′ negative strand viral RNA (32, 33). In contrast, following purification under non-denaturing conditions, PV 2C fused to maltose-binding protein (MBP-2C) and His6-tagged versions of 2C from HAV and EV9 were all found to interact nonspecifically with RNA (17, 24, 34). It is notable that in experiments done with His-tagged proteins from HAV and EV9, the 2C-RNA complexes formed aggregates that were unable to enter polyacrylamide gels during mobility shift assays (17, 34).
In common with other AAA+ proteins, SF3 helicases appear to oligomerize into hexamers to form a functional unit (35–37), although the ability of these proteins to assemble into heptamers in the absence of substrates has also been reported (38). Crystallographic analysis of the LTag and E1 helicases, for example, revealed a ring structure formed from six subunits (39–41). The ATPase active sites of these hexamers are formed at the interfaces between pairs of subunits. In the active sites, one subunit provides the Walker A and B motifs (42) and Motif C (19) (sensor 1 in AAA+ nomenclature (35)), whereas an adjacent subunit contributes an arginine residue (the “R finger”) that interacts with the triphosphate tail during catalysis. Consistent with this architecture, the ATPase activity of these proteins is dependent on oligomerization (36). The structure of BPV E1 co-crystallized with ATP, and a short length of single-stranded DNA showed that the oligonucleotide was threaded through the pore formed at the center of the hexameric protein and makes differential contacts with each subunit that suggest a plausible helicase mechanism (39).
Is hexamerization also necessary to activate ATPase hydrolysis and other functions attributed to picornavirus 2C proteins? An interesting recent study showed that the MBP-2C fusion protein prepared from PV 2C formed homo-oligomers that were necessary for the ATPase activity of the protein (43), consistent with some earlier work (27, 44, 45). Moreover, the study found that deletion of the N-terminal 38 residues of the 2C portion of the fusion protein, which removes the predicted amphipathic region involved in membrane binding, abrogated both the oligomerization and ATPase activity of this MBP-2C construct (43). Curiously, analysis by size exclusion chromatography and electron microscopy revealed that the MBP-2C protein formed oligomeric species that ranged from tetramers to octamers. It is currently not clear why this range of sizes is much broader than found with other AAA+ and related proteins, which are typically hexameric or heptameric (46–48).
To further the investigation of the assembly and enzymatic properties of picornavirus 2C proteins, we aimed to engineer recombinant FMDV 2C that could be produced as a soluble protein in Escherichia coli without the encumbrance of a large protein tag. We now show that overexpression of soluble FMDV 2C with an N-terminal His tag can be achieved by deleting the N terminus of the protein up to and including the predicted amphipathic helix. The FMDV 2C(34–318) construct has been used in in vitro studies that complement ongoing investigations of the effects of 2C in transfected mammalian cells (7, 12). Following proteolytic removal of the His tag, the resulting protein can be concentrated to at least 20 mg/ml, and it exhibits specific ATPase activity and nonspecific RNA binding activity. In the absence of ligands, FMDV 2C(34–318) exhibits concentration-dependent oligomerization, the largest form of which is tetrameric. However, in the presence of ATP and RNA, a mutant of 2C(34–318) that can bind but not hydrolyze the nucleotide specifically assembles, even at low concentrations, into a hexameric ring structure, demonstrating that the N terminus of the protein is not required for oligomerization. ATPase assays performed with mixtures of active and inactive 2C subunits support the hypothesis that hexamerization is necessary for a coordinated mechanism of nucleotide hydrolysis. We propose that assembly into hexameric rings is a general requirement for the proper functioning of picornavirus 2C proteins.
EXPERIMENTAL PROCEDURES
Cloning
All PCR and mutagenesis primers used in this study are listed in supplemental Table S1. A pCR-XL-TOPO plasmid containing the type O1K FMDV 2C sequence (GenBankTM accession number X00871) (derived from a full-length infectious cDNA) was used as template for the generation of FMDV 2C constructs. cDNAs for 2C(1–318), 2C(38–318), 2C(52–318), and 2C(61–318) were amplified by PCR using primers designed to incorporate an upstream BamHI site and a downstream HindIII site to facilitate ligation into a modified pETM-11 plasmid (EMBL) to express the 2C product with a TEVpro cleavable N-terminal His6 tag (49); because of an internal NcoI site in the FMDV 2C sequence, the NcoI site in pETM-11 was first changed to a BamHI site using the QuikChange method (Stratagene). The pET-h2C(38–318) construct was used as template in QuikChange reactions to generate pET-h2C(34–318), which, in turn, served as the template for introduction of point mutations by the same method. Cloning and expression of GST-FMDV 2C is described in the supplemental material. All expression plasmids were sequenced by MWG Eurofins Ltd.
Expression and Purification
Proteins were produced in E. coli BL21 (DE3) pLysS cells. Cells for expression of His6-tagged 2C constructs were grown at 37 °C to an A600 of 0.7. Isopropyl β-d-thiogalactopyranoside (0.5 mm) was added, and cells were incubated with shaking for a further 4 h at 37 °C before being harvested by centrifugation and stored at −80 °C. Pellets were resuspended in lysis buffer, containing 50 mm HEPES, pH 7.1, 400 mm NaCl, 1 mm β-mercaptoethanol 0.1% (v/v) Triton X-100; cells were lysed by sonication. His6-tagged protein was batch-purified on TALON (Clontech). The tag was removed with His6-tagged TEVpro before the protein was reverse purified over a second bed of TALON. The 2C protein was further purified by gel filtration on a Superdex S200 10/30 column (GE Healthcare) in 50 mm HEPES, pH 7.1, 200 mm NaCl, and 1 mm β-mercaptoethanol at a flow rate of 0.4 ml/min. Except where stated otherwise, the soluble 2C proteins obtained were concentrated in the gel filtration buffer to 20 mg/ml and stored at −80 °C.
ATPase Assays
ATP hydrolysis was measured with a colorimetric assay based on the malachite green dye method described previously (50, 51). ATPase assays were performed in 96-well plates in 100-μl reaction volumes containing 50 mm HEPES, pH 7.3, 1 mm dithiothreitol, 2 mm MgCl2 in the presence of 2C (typically 1.25 μm) and ATP (or another NTP) at the concentrations indicated in the relevant figure legends. The reactions were terminated by the addition of 200 μl of the color reagent described by Mahuren et al. (51). The production of phosphate was measured by the increase in absorbance at 590 nm read on a Sunrise 96 well microplate reader (Tecan).
ATP Binding Assays
UV cross-linking of ATP to FMDV 2C was performed as described by Schumacher et al. (52). FMDV 2C (20 μm) was incubated with 40 μCi of [α-32P]ATP (3000 Ci/mmol) (PerkinElmer Life Sciences), in 25 mm HEPES, pH 7.1, 100 mm NaCl, 2 mm MgCl2, 0.5 mm β-mercaptoethanol. Samples (10 μl), spotted onto glass slides, were irradiated for 20 min over ice at 254 nm using a 6-watt UVG-54 lamp (UVP Inc.) before analysis by SDS-PAGE. Protein bands were visualized with Instant Blue protein stain (Expedeon) prior to gel drying. Radiolabel was detected by exposure to a phosphor screen, which was read on a Fuji FLA-5000 phosphor imager with AIDA software (Raytest).
RNA Binding Assays
Synthetic RNA oligomers used in this study were purchased from Dharmacon Ltd. RNA was radiolabeled at the 5′ terminus using [γ-32P]ATP and T4 polynucleotide kinase (Promega). RNA binding assays were performed essentially as described previously (53) using 0.5 nm RNA and final binding conditions of 50 mm HEPES, pH 7.1, 40 mm NaCl, 2 mm MgCl2, 1 mm dithiothreitol, and 2C(34–318) at 0–60 μm. Samples were analyzed in 10% acrylamide gels using 50 mm Tris, 50 mm boric acid running buffer and autoradiographed on a Fuji FLA-5000 phosphor imager.
Size Exclusion Chromatography-Multiangle Light Scattering (SEC-MALS)
FMDV 2C samples were applied to a Superdex 200 10/30 column (GE Healthcare) pre-equilibrated in buffer containing 50 mm HEPES, pH 7.3, 200 mm NaCl, 2 mm β-mercaptoethanol, 0.02% sodium azide. MgCl2 and ATP were included in the running buffer as indicated throughout. The buffer and sample were pumped through the column using a Varian HPLC system (Varian, Inc.) coupled in line to a DAWN Helios II light scatter detector (18 detectors) (Wyatt) and Optilab rEX Differential Refractive Index Detector (Wyatt) at a flow rate of 0.5 ml/min. Peak analysis was performed using the ASTRA software package (Wyatt).
Electron Microscopy
2 μl of the 2C-ATP-RNA complex at 0.1 mg/ml was taken directly from a gel filtration experiment, applied within 30 min to glow-discharged continuous carbon grids (TAAB), and then stained with 2% uranyl acetate. Electron micrographs were taken on a Philips CM200 electron microscope operating at 200 kV under low dose conditions of ∼10 e−/Å2. Images were captured directly at ×50,000 magnification on a 4096 × 4096 pixel F415 CCD camera (Tietz Video and Image Processing Systems), giving a pixel size of 1.76 Å/pixel. Image processing was performed using IMAGIC-5 software (54). Particles were picked manually, and the data were coarsened by a factor of 2, giving a working pixel size of 3.52 Å/pixel. The final data set consisted of 1600 particles, which were normalized and band pass-filtered between 10 and 150 Å. Individual images were translationally aligned to the rotationally averaged total sum and analyzed by multivariate statistical analysis (MSA), followed by classification to form class averages containing ∼20 particles/class. The eigenimages resulting from MSA can be used to assess the main symmetry components present in the particle data set (55).
RESULTS
Expression of Soluble FMDV 2C
Previous studies of picornaviral 2C proteins have frequently involved the use of large N-terminal fusion tags to overcome the poor solubility of native 2C (23, 24, 43, 56). In anticipation of similar problems with FMDV 2C, we tried a number of different approaches to obtain a soluble, active form of the protein that would facilitate structural and functional characterization. The various constructs designed for production of FMDV 2C in E. coli are summarized in Fig. 1a. First, following previous work on other picornavirus 2C proteins, we found that GST-FMDV 2C was strongly expressed in E. coli (supplemental Fig. S1a). However, only a very small fraction of the protein was recovered from the soluble fraction in cell lysates, and that had suffered significant proteolysis (supplemental Fig. S1b). The low molecular weight contaminants present in the partially purified sample were removed by gel filtration (supplemental Fig. S1c), but the addition of thrombin to remove the GST tag resulted in cleavage of GST-FMDV 2C at a site internal to 2C, as shown by the generation of a 50-kDa GST-2C′ cleavage product (supplemental Fig S1d).
The inability to process the GST-FMDV 2C fusion and the poor yield of purified protein (0.5 mg from 8 liters of E. coli culture) prompted us to seek alternative methods for obtaining soluble protein. We next created pET-h2C(1–318), which was designed to produce full-length FMDV 2C with an N-terminal His6 tag that could be removed by TEVpro cleavage. However, expression of this construct was toxic to E. coli because the growth of the culture was arrested upon induction with isopropyl β-d-thiogalactopyranoside (Fig. 1b, dotted line). Modification of this construct to introduce a K116A mutation that abrogates ATP hydrolysis by FMDV 2C (see below) did not relieve the growth arrest due to the induction of protein expression in E. coli (Fig. 1b, broken line), indicating that the observed toxicity is not due to the ATPase activity of the protein.
To examine if the predicted N-terminal amphipathic region of FMDV 2C was responsible for the growth arrest of E. coli cultures, the hydrophobic residues of the putative amphipathic helix (Trp18, Leu19, Val20, Leu22, Ile23, Leu24, Ile26, and Trp29) were mutated to Ala. This mutation strategy successfully eliminated the toxicity of the protein and resulted in good expression of FMDV 2C (data not shown). Unfortunately, this helix mutant was poorly soluble and precipitated at concentrations over 0.5 mg/ml; it was therefore not used in any experiments. Significantly, however, this observation suggested that the predicted amphipathic region of FMDV 2C is responsible for the toxic effects of the protein in E. coli. We suspect that expression of the GST-2C protein is permissible in E. coli because the large GST tag may interfere with the membrane interactions of the N-terminal amphipathic helix in FMDV 2C. Our findings are also consistent with work on HAV 2C, which showed that partial deletion of the predicted amphipathic region relieved the toxicity of the protein in E. coli (17).
We therefore generated FMDV 2C deletion mutants lacking the putative amphipathic helix (residues 17–34) (14). The design of constructs was guided by sequence alignments with homologous AAA+ proteins of known structure, such as p97 (57) and BPV E1 (39), which indicated that the body of the ATPase domain in FMDV 2C starts at around residue 60. These constructs included pET-h2C(34–318), pET-h2C(38–318), pET-h2C(52–318), and pET-h2C(61–318) that produce proteins with an N-terminal TEVpro-cleavable His6 tag, which lack the first 33, 37, 51, and 60 residues of 2C, respectively. Expression from each of these plasmids proved to be non-toxic to E. coli; a typical growth curve following induction of expression of 2C(34–318) is shown in Fig. 1b. What is more, these 2C proteins were all expressed at a high level in the bacteria (typically 10 mg/liter culture) and could be purified from the soluble fraction of cell lysates (see “Experimental Procedures”). After processing with TEVpro, these 2C proteins could be concentrated to >20 mg/ml (see “Experimental Procedures”).
Interestingly, analyses by size exclusion chromatography showed that the apparent Mr of the larger soluble 2C N-terminal deletion mutants increased with the amount of protein loaded, suggesting that concentration-dependent subunit association was occurring. The chromatographic profiles for each sample contained a single asymmetric peak, with a lagging slope on the low Mr side of the peak, similar to the elution profile shown for 2C(34–318) in Fig. 1c. They were very different from the gel filtration chromatograms obtained with MBP-2C from PV; in particular, they did not contain any high Mr aggregates in the void peak (43). 2C(34–318) and 2C(38–318) were found to form oligomers with apparent Mr values of up to about 180 kDa. The oligomerization of 2C(52–318) was less efficient, requiring higher concentrations to give the same yield of the largest species with an apparent Mr of ∼180 kDa (data not shown). In contrast, the shortest construct, 2C(61–318), appeared to elute at a maximum apparent Mr of about 60 kDa (Fig. 1c) (see below). These observations suggested that the N-terminal amphipathic helix is not required for oligomerization of FMDV but nevertheless provided an initial indication that residues 34–60 may contribute to oligomer stability. Because 20 mg/ml solutions of 2C(38–318) displayed a weak tendency to aggregate when warmed to room temperature, we opted to use the slightly longer 2C(34–318) deletion mutant (which did not exhibit this behavior) for further analysis of the enzymatic properties of FMDV 2C.
FMDV 2C Is an ATPase
FMDV 2C contains Walker A, Walker B, and C (or Sensor 1) motifs typical of SF3 helicases and other AAA+ ATPases (Fig. 1a) (36) and, unsurprisingly, was found to have good ATPase activity (Fig. 2a). To characterize this activity in greater detail, we examined the ability of FMDV 2C(34–318) to hydrolyze ATP in a variety of different conditions (Fig. 2 and supplemental Fig. S2). The ATPase activity was optimal in the presence of 1–2 mm Mg2+ (supplemental Fig. S2a, closed circles) but was about 5 times less efficient when the cation was replaced by Mn2+ (supplemental Fig. S2a, open circles), similar to results obtained with PV 2C (23). The activity was also maximal when the hydrolysis reaction was performed at 32 °C (supplemental Fig. S2b) and pH 7.4 (supplemental Fig. S2c), although the enzyme did not display a very strong dependence on temperature or pH. In contrast, the hydrolysis of ATP by FMDV 2C was very sensitive to ionic strength; the catalytic reaction rate was reduced by 50% in the presence of only 20 mm NaCl (Fig. 2b). The FMDV enzyme is therefore more strongly affected by salt than PV 2C, which was inhibited by 50% at 50 mm NaCl (23).
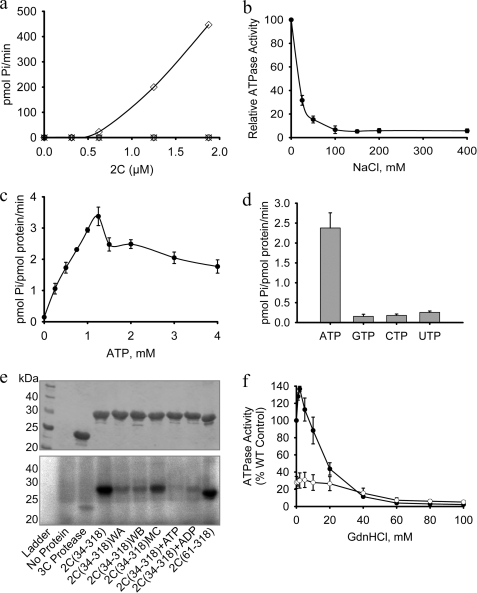
FIGURE 2.
FMDV 2C is a specific ATPase that is sensitive to NaCl and GdnHCl. The ATPase activity of FMDV 2C(34–318) was measured in 100-μl reactions containing 4 μg of protein (1.25 μm), 1 mm ATP, and 2 mm MgCl2, unless otherwise stated. a, ATP hydrolysis was measured in the presence of varying concentrations of protein (open diamonds). There was no detectable ATPase activity for the Walker A, K116A (WA) (open circles), Walker B, D160A (WB) (open triangles), and Motif C, N207A (MC) (crosses). The data plotted are an average of two experiments. b, the ATPase activity is very sensitive to NaCl and decreases monotonically with an ED50 of about 10 mm. c, the ATPase activity of 2C(34–318) varies in a complex manner with increases in ATP concentration, exhibiting a sharp decline between 1.25 and 1.5 mm substrate. d, comparative NTPase activity of 2C(34–318) with 1 mm each ATP, GTP, CTP, and UTP. e, UV cross-linking of [α-32P]ATP to FMDV 2C(34–318) and various mutants (identified below each lane). After cross-linking, the protein sample was run on a 12% SDS-polyacrylamide gel. Top, the positions of protein bands were confirmed by staining with Coomassie Blue before gel drying; bottom, cross-linked ATP was detected by exposure to a phosphor screen (bottom). 5 mm unlabeled ADP or ATP was included as indicated. f, effect of GdnHCl on the ATPase activity of wild-type FMDV 2C(34–318) (WT) (closed circles) and the 2C(34–318) M159L mutant (open circles). Error bars, S.E. determined from three independent measurements. WA, Walker A; WB, Walker B; MC, motif C.
To examine the roles of each of the AAA+ ATPase motifs, key residues in each were mutated. No ATP hydrolysis was detected for 2C(34–318) proteins containing amino acid substitutions in the Walker A motif (K116A), in the Walker B motif (D160A), or in Motif C (N207A) (Fig. 2a). These results suggest that that each of the conserved AAA+ ATPase motifs are involved in this activity. They also confirm that the ATP hydrolysis detected in our assays is a result of 2C(34–318) activity and not due to an E. coli contaminant.
Interestingly, we observed that FMDV GST-2C had ∼7-fold higher ATPase activity than 2C(34–318) (Table 1). The ATP hydrolysis rate of FMDV GST-2C was comparable with that reported for similar fusions of PV 2C and for the homologous murine norovirus protein, MNV p41 (Table 1) (23, 43, 58). The precise reason for the increased activity of the GST-2C fusion over the 2C(34–318) construct is not yet known. However, it is possible that the retention of the full-length N terminus, even in the context of a GST fusion, may stimulate the ATPase activity of 2C.
TABLE 1.
ATPase activity of FMDV deletion and active site mutants
| Construct | ATPase activity | Source/Reference |
|---|---|---|
| pmol Pi/pmol protein/min | ||
| GST-FMDV 2C | 17 | This report |
| GST-PV 2C | 13.3a | Ref. 23 |
| GST-MNV p41 | 17.3a | Ref. 58 |
| FMDV 2C(34–318) | 2.3 | This report |
| FMDV 2C(34–318) K116A (Walker A) | 0.017 | This report |
| FMDV 2C(34–318) D160A (Walker B) | 0.017 | This report |
| FMDV 2C(34–318) N207A (Motif C) | 0.042 | This report |
| FMDV 2C(61–318) | 0.46 | This report |
a The values reported in the associated references were converted to these units for comparison.
FMDV 2C ATPase activity increased linearly from 0 to 1.25 mm ATP but then dropped by ∼30% at 1.5 mm and declined slowly as the ATP concentration was increased further (Fig. 2c). The observed enzyme kinetics therefore differ significantly from classical Michaelis-Menten behavior. For PV 2C, it was necessary to maintain Mg2+ concentrations at 1 mm above that of the ATP concentration to obtain a hyperbolic substrate response curve from which Michaelis-Menten kinetic parameters could be derived (23). In the case of FMDV 2C, maintaining this elevated level of Mg2+ failed to rescue the apparent inhibition at higher ATP concentrations. Nevertheless, based on the half-maximal activity, an apparent value for the Km is estimated to be ∼0.5 mm (similar to the Km of 0.7 mm found for PV 2C (23)). FMDV 2C is quite specific for ATP because the hydrolysis rates measured with GTP, CTP, and UTP substrates were at least 10-fold lower than with ATP (Fig. 2d).
The binding of ATP to wild type and mutant 2C proteins was analyzed by UV cross-linking. We observed efficient cross-linking of ATP to wild type 2C(34–318), consistent with good binding of the substrate (Fig. 2e, lane 3). The addition of 5 mm unlabeled ATP or ADP reduced the cross-linking of radiolabeled ATP to 2C(34–318) to background levels (lanes 8 and 9), confirming the specificity of the interaction detected in this experiment. Both the Walker A (K116A) (lane 5) and the Walker B mutant (D160A) (lane 6) mutants of 2C(34–318) displayed very low levels of cross-linking, comparable with the signal obtained with a control protein (FMDV 3C protease), which does not bind ATP (lane 3). In marked contrast, introduction of an N207A mutation within Motif C caused only a very modest decrease in UV cross-linking to FMDV 2C(34–318) (lane 7), suggesting that the ability to bind ATP is retained in this mutant.
GdnHCl Has Stimulatory and Inhibitory Effects on FMDV 2C ATP Hydrolysis
FMDV infections are sensitive to millimolar concentrations of GdnHCl (29, 59). For example, the addition of 3 mm GdnHCl led to a greater than 16-fold reduction in FMDV RNA synthesis in infected cells (29). In common with other picornaviruses, the drug seems to target 2C because mutations that confer resistance to GdnHCl have been mapped to this protein (29, 60). We found that purified FMDV 2C(34–318) was inhibited by GdnHCl but only at much higher concentrations than affect virus replication (Fig. 2f, closed circles). Surprisingly, at low millimolar concentrations of GdnHCl (1–5 mm) that are inhibitory to virus infection, there is an increase in 2C(34–318) ATPase activity of up to 30%. Above 5 mm, the drug starts to inhibit the enzyme; by 10 mm GdnHCl, the rate of ATP hydrolysis has returned to that observed in the absence of the drug, and at 20 mm GdnHCl, the activity is reduced by about 55%. Complete inhibition is observed at drug concentrations above 60 mm.
The M159L mutation in FMDV 2C that renders the virus resistant to GdnHCl (29) was inserted into 2C(34–318). Despite the conservative nature of the amino acid substitution, the mutant protein was significantly less soluble than wild type 2C and could only be concentrated to 1.5 mg/ml. In the absence of GdnHCl, the ATPase activity of the 2C(34–318) M159L mutant was only 25% of wild type (Fig. 2f, open circles). The response of this mutant enzyme to increasing concentrations of GdnHCl was quite different from the behavior exhibited by wild-type 2C. There was no significant stimulation of ATPase activity at low concentrations of GdnHCl, and the enzyme was less sensitive to inhibition by the drug, requiring ∼35 mm to be inhibited 50% compared with an ED50 of 18 mm observed with the wild-type protein (supplemental Fig. S2d). These results are consistent with the observed effect of the M159L mutation in infected cells (29), although there are obvious discrepancies in the sensitivities of 2C and the virus to the presence of the drug.
ATP binding in the presence of varying concentrations of GdnHCl was also analyzed by UV cross-linking. As shown in supplemental Fig. S3a (solid line), there is a slight increase in ATP cross-linking in the presence of low concentrations of GdnHCl, suggesting that low levels of the drug may act by enhancing affinity for the ATP substrate. In the presence of 40 mm GdnHCl, a concentration of the drug that inhibits ATPase activity by about 50% (Fig. 2f), UV cross-linking of ATP is as strong as that observed in the absence of the drug (supplemental Fig. S3a). The differential effects of GdnHCl, at low and high concentrations, on the binding and hydrolysis of ATP by 2C(34–318) appear to indicate dual modes of interaction between the protein and the drug.
We also examined whether increases in ionic strength produce their marked inhibitory effect on ATPase activity by reducing binding of ATP to FMDV 2C. Increasing the NaCl concentration clearly decreased UV cross-linking of ATP (supplemental Fig. S3b). In fact, the dose dependence of the decrease in UV cross-linking in the presence of NaCl follows a similar trend to the observed reduction in ATPase activity (supplemental Fig. S3b). These results suggest that, in contrast to GdnHCl, NaCl inhibits the enzyme by reducing binding of the ATP substrate.
FMDV 2C Binds ssRNA
As mentioned in the Introduction, previous experiments to examine the RNA binding properties of picornavirus 2C proteins may have been hampered by the poor solubility and nonspecific aggregation of the recombinant proteins used (17, 32, 34, 61). In our experiments, FMDV 2C(34–318) was purified as a soluble protein without denaturation, and electrophoretic mobility shift assay experiments were performed under native conditions. We observed that increasing concentrations of 2C(34–318) formed a single, well defined slower migrating complex with a randomly selected 20-base ssRNA oligomer (5′-AAUUCUAAGGGCCAGCGAGA) (Fig. 3a). 2C(34–318) bound the 20-base-long ssRNA with an apparent Kd of ∼5 μm under our experimental conditions (estimated as the concentration needed to bind 50% of the RNA and taking no account of possible oligomerization of 2C). The interaction between RNA and FMDV 2C was dependent on the length of the RNA. 2C(34–318) bound 12 and 20 nucleotide RNAs (Fig. 3a and supplemental Fig. S4b) with similar affinity, but binding to RNAs that were 7 or 10 nucleotides long was significantly weaker (apparent KD > 40 μm) (Fig. 3b and supplemental Fig. S4a). Notably, 2C(34–318) had a much higher affinity for ssRNA over blunt-ended dsRNA of the same length (20 bp), which was bound with an apparent KD of over 60 μm (Fig. 3c).
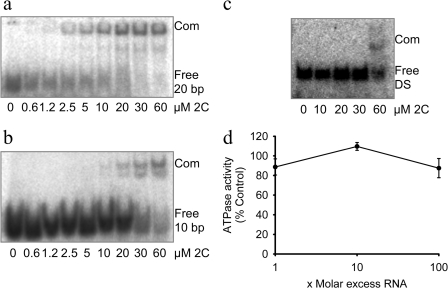
FIGURE 3.
FMDV 2C binds to ssRNA with higher affinity than to dsRNA. The binding of 2C(34–318) to 32P-5′-end-labeled RNA was measured in electrophoretic mobility shift assays (see “Experimental Procedures”). Protein-RNA complexes were resolved on a native 10% polyacrylamide gel, which was then exposed to a phosphor screen. The positions of the free unbound probe (Free) and protein-RNA complex (Com) are indicated. Varying concentrations of 2C(34–318) (2C) were added to labeled 20-base ssRNA (5′-AAUUCUAAGGGCCAGCGAGA-3′) (a) or labeled 10-base ssRNA (5′-CCUUAGAAUU-3′) (b). c, a dsRNA probe (DS) was prepared by mixing the 20-base-long probe from a with an unlabeled complementary RNA of exactly the same length, at a molar ratio of 1:1.2, heating to 70 °C for 5 min, and cooling to 22 °C over 2 h. d, the ATPase activity of 2C(34–318) was measured in the presence of various amounts of single-stranded RNA. Error bars, S.E. determined from three independent measurements.
Although the biological function of 2C is unknown, its similarity to the SF3 helicase family of proteins has prompted suggestions that it may have a role in unwinding dsRNA (19). The ATPase activity of other hexameric helicases, such as BPV E1 and gene 4 protein of T7 bacteriophage, is observed to be stimulated in the presence of a DNA substrate (46, 62). However, the addition of as much as a 100-fold molar excess of the 20-nucleotide ssRNA oligomer used in the electrophoretic mobility shift assays had no effect on the ATPase activity of 2C(34–318) (Fig. 3d).
RNA and ATP Binding Stabilize Hexameric Form of FMDV 2C
In most cases, AAA+ proteins form hexameric rings, with the ATPase active site at the interface between neighboring subunits (39, 57). An MBP fusion of PV 2C was recently reported to form homo-oligomeric structures required for the ATPase activity of the protein (43); in this case, a mixed population of oligomeric states ranging from tetramers to octamers was observed, but it was not clear if all of these represent functional assemblies.
As noted above, FMDV 2C(34–318), which is soluble to at least 20 mg/ml, was found to exhibit concentration-dependent oligomerization during SEC (Fig. 1c). Increasing the loading of purified protein samples ranging from 5 to 20 mg/ml onto the SEC column progressively reduced the elution volume, indicating the formation of higher molecular mass species (Fig. 1c) that appeared to attain a maximum value of about 180 kDa when compared with the elution position of molecular mass standards. Since the calculated molecular mass for 2C(34–318) is 32 kDa, these data were initially interpreted to indicate the formation of hexamers of 2C, consistent with the oligomerization behavior of members of the SF3 helicase/AAA+ ATPase superfamily.
To provide a more precise examination of 2C(34–318) oligomerization, we used SEC-MALS, which permits absolute Mr determination of macromolecules (63). For a sample loading of 0.5 ml at 20 mg/ml (equal to the highest concentration used in the SEC experiment shown in Fig. 1c), fitting of the light scattering data revealed a distribution of molar mass indicative of concentration-dependent oligomerization of 2C(34–318) with the maximum observed mass corresponding to a tetrameric species (Fig. 4a, solid line). There was no evidence to indicate the presence of hexamers under these conditions. This result shows that the initial SEC analysis had significantly overestimated the Mr of 2C(34–318) oligomers, perhaps due to aberrant retention of the protein on the column. In contrast, the largest oligomeric species identified by SEC-MALS for 2C(61–318) was ∼60 kDa (Fig. 4b, solid line), consistent with previous SEC measurements that suggested that this form of the protein cannot assemble into oligomeric states larger than dimers. These results suggest that residues in the region 34–61 of FMDV 2C are involved in stabilization of higher order oligomers of 2C, probably by mediating interactions between subunits.
FIGURE 4.
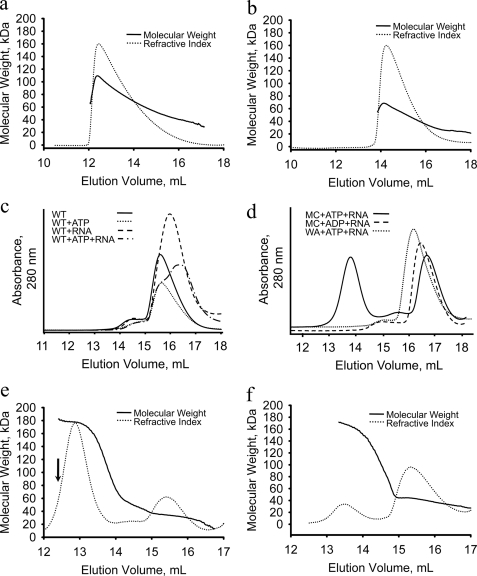
ATP and RNA stimulate the hexamerization of FMDV 2C. Oligomerization of 2C(34–318) and 2C(61–318) was analyzed by SEC and SEC-MALS. 10 mg of 2C(34–318) (a) or 2C(61–318) (b) was loaded onto a Superdex S200 10/30 column coupled to in-line light scattering and refractive index detectors. The refractive index measurement (dotted line) indicates the concentration of protein in the eluate. The molecular weight of the protein species in the eluate (determined from the refractive index and light scattering measurements) (63)) is indicated by the solid line. c–f, 100 μl of 5 mg/ml wild type or mutant 2C(34–318) or 2C(61–318) protein were mixed with 2 mm ATP or ADP, 5 mm MgCl2, and a 5-fold molar excess of 20-nucleotide ssRNA (as indicated) and incubated for 10 min at 22 °C prior to injection onto a Superdex S200 10/30 column for analysis by standard SEC (c and d) or SEC-MALS (e and f). The arrow in c indicates the sample used for electron microscopy work (see Fig. 5).
Because 2C(34–318) was found to bind both ATP and ssRNA (Figs. 2 and 3), the effects of these ligands on oligomerization were investigated, first with SEC and subsequently with SEC-MALS. In these experiments, a 0.5-mg loading (100 μl at 5 mg/ml) was used to ensure that unliganded 2C(34–318) was in the monomeric form so that the assay would provide a sensitive measure of any enhancement of oligomerization due to ligand binding. When added, ATP was maintained at 2 mm in the running buffer in these experiments to counteract dissociation during the chromatographic run. As shown in Fig. 4c (solid line), at this low protein concentration, 2C(34–318) eluted from the SEC column with an estimated Mr of ∼30 kDa, which is consistent with it being monomeric. The position of the peak was not affected by the addition of ATP (dotted line). It is shifted slightly in the presence of RNA (dashed line). This shift may be due to the effect of RNA on the interaction of 2C(34–314) with the column matrix. In the presence of both ATP and RNA, the peak profile appeared as a mixture of the ATP-bound and RNA-bound forms, but there was no indication of hexamer formation (dashed and dotted line).
The effect of ATP and ssRNA on the 2C(34–318) Motif C mutant, N207A, was then tested by SEC because this mutant binds but does not hydrolyze ATP (Fig. 4d). This gave elution profiles that were broadly similar to wild-type 2C(34–318) in the presence of either ATP or ssRNA (data not shown). However, when both ATP and ssRNA were mixed with 2C(34–318), N207A, a well defined species with high molecular weight was detected (Fig. 4d, solid line). Notably, this peak was not observed upon the addition of ADP and ssRNA, indicating that the intact nucleotide triphosphate is necessary for the observed oligomerization (Fig. 4d, dashed line). Consistent with this interpretation, the 2C(34–318) K116A Walker A mutant, which does not hydrolyze or bind ATP (Fig. 2, a and e), was unable to form high molecular weight complexes in the presence of ssRNA and ATP (Fig. 4d, dotted line).
Determination of Mr using SEC-MALS revealed that the Motif C N207A mutant of 2C(34–318) forms hexamers upon the addition of ligands. As shown in Fig. 4e (solid line), the molecular mass of the 2C(34–318) N207A oligomer obtained in the presence of ATP and ssRNA is 182 kDa, consistent with hexamerization of the protein.
Strikingly, although the largest oligomeric species of 2C(61–318) detected in the absence of ligand was dimeric (Fig. 4b), SEC-MALS analysis showed that ssRNA and ATP also stabilized hexamers of 2C(61–318) N207A (Fig. 4f, solid line). The stabilization of hexamers was less efficient for the 2C(61–318) mutant than for the 2C(34–318) mutant, as shown by the relative areas of the peaks corresponding to the hexameric species and monomer/dimer species in each case (Fig. 4, e and f). The decreased propensity of 2C(61–318) to oligomerize is accompanied by a proportional impairment of ATPase activity; this enzyme hydrolyzes ATP 5 times less efficiently than the longer 2C(34–318) (Table 1). 2C(61–318) nevertheless appears to bind ATP as strongly as 2C(34–318), as shown in UV cross-linking experiments (Fig. 2e). These results are consistent with the idea that the reduced ATPase activity results from decreased oligomerization (although a more direct effect of residues 34–61 on ATPase activity cannot be ruled out).
FMDV 2C Forms Hexameric Ring Structures Characteristic of AAA+ ATPases
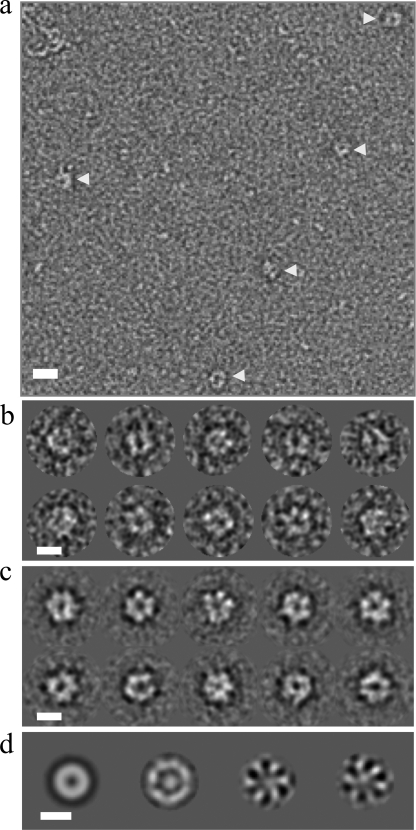
The detection of stable oligomerization of the 2C(34–318) N207A Motif C mutant in the presence of ATP and RNA allowed us to analyze the complex by negative stain electron microscopy. A sample of the stabilized hexameric species, at a concentration of 1 μm, was taken directly from the hexameric peak in an SEC experiment and negatively stained with uranyl acetate within 30 min (see “Experimental Procedures”). Examination of micrographs revealed the presence of small ringlike structures with diameter of about 140 Å (Fig. 5a). The majority of the particles appeared to be aligned so that the plane of the ring of the 2C oligomers was parallel to the plane of the micrograph, suggesting that the 2C oligomers adopt a preferred orientation on the grid. Because of the relatively small size of FMDV 2C, it is possible that only this view of the oligomeric form of the protein is distinct enough to be clearly observed using electron microscopy. Alternatively, the fixing of the protein on the grid before staining may also have led to preferential views of the complex. A total of 1600 particles were picked to assemble a data set for further analysis of the population of 2C complexes (Fig. 5b).
FIGURE 5.
The Motif C mutant of FMDV 2C(34–318) forms hexameric rings in the presence of ATP and RNA. A sample of the 2C(34–318) Motif C mutant (N207A) hexamers obtained in the presence of ATP and RNA (indicated by the arrow in Fig. 4c) was applied to glow-discharged carbon grids and negatively stained with uranyl acetate. Electron micrographs were taken on a Philips CM200 electron microscope operating at 200,000 V under low dose conditions of 10 e−/Å2. Images were captured directly at 50,000 times magnification over a range of nominal defocus (0.5–2 μm) on a CCD camera. a, a representative micrograph of FMDV 2C particles (indicated by arrows). Scale bar, 200 Å. b, a selection of FMDV 2C particles following initial processing by IMAGIC (54). Scale bar, 100 Å. c, a range of hexameric FMDV 2C class averages (∼20 particles/class). Scale bar, 100 Å. d, left to right, first, second, third, and fourth eigenimages reveal the 6-fold symmetry in the FMDV 2C data set.
To identify symmetry components in the data set, the images of the 2C(34–318)-ATP-RNA complexes were translationally aligned to a rotational average of the entire data set using the method of Dube et al. (55). MSA followed by classification was performed on the final data set, and class averages were generated, many of which displayed clear hexamerization of FMDV 2C (Fig. 5c). The eigenimages from the initial MSA, which describe the interimage variance, also indicate a clear 6-fold symmetry (Fig. 5d). The diameter of the FMDV 2C rings observed in the class averages is ∼140 Å, consistent with the dimensions of E1 (97 Å) and LTag (128 Å) SF3 helicase hexamers (39, 40), which have similarly sized ATPase domains. Our electron microscopy and SEC-MALS data demonstrate for the first time that FMDV 2C can form stable hexamers upon binding of ATP and RNA.
Consistent with the absence of a hexameric peak for wild-type 2C(34–318) in the presence of ATP and RNA in gel filtration experiments, no ringlike structures of FMDV 2C were observed in micrographs taken from negatively stained samples prepared by mixing the protein with these two ligands (data not shown).
FMDV 2C Hydrolyzes ATP through a Coordinated Mechanism
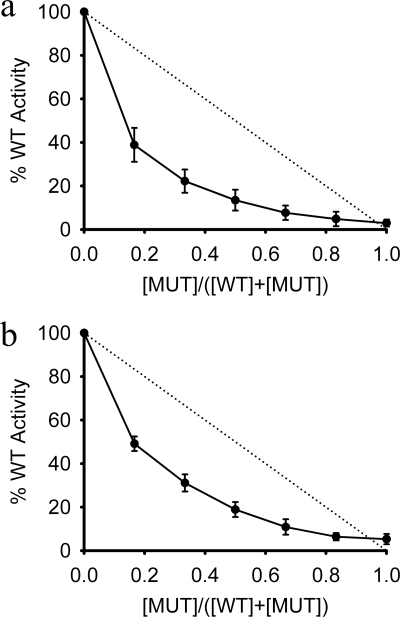
The mechanism of ATP hydrolysis in AAA+ hexameric rings has been studied in a number of systems. For example, ClpX, a protein-unfolding AAA+ ATPase, was reported to hydrolyze ATP in an uncoordinated manner (64) (hydrolysis occurs randomly at any site), whereas SV40 LTag (65) and BPV E1 (39) helicases were both found to hydrolyze ATP through coordinated mechanisms (hydrolysis occurs at multiple sites simultaneously or with rotational synchronization around the ring). To probe the mechanism of ATP hydrolysis by FMDV 2C, subunit-mixing experiments were performed. Wild type 2C(34–318) subunits were mixed with the K116A Walker A mutant in molar ratios ranging from 6:0 to 0:6 and assayed for ATPase activity. For an uncoordinated mechanism, the ATPase activity would be expected to decrease linearly as the ratio of inactive to active 2C subunits was increased. Instead, the ATPase activity decreased non-linearly (Fig. 6a); a sharp decline in activity of 50–60% was observed when only one-sixth of the subunits were catalytically inactive. This result clearly indicates that hydrolysis by 2C is coordinated around the subunits in the ring. A similar experiment performed with the 2C(61–318) protein showed the same non-linear decrease in ATPase activity upon mixing with 2C(61–318) K116A (Fig. 6b), suggesting that the ATPase activity detected from 2C(61–318) also results from the transient formation of hexamers that are needed to coordinate ATP hydrolysis. This is consistent with the detection by SEC-MALS of hexamers of 2C(61–318) in the presence of ATP and ssRNA (Fig. 4e).
FIGURE 6.
ATP hydrolysis by FMDV 2C is coordinated. a, various ratios of wild type 2C(34–318) (WT) and 2C(34–318) Walker A mutant, K116A (MUT), were mixed before addition to an ATPase assay. b, various ratios of wild type 2C(61–318) and 2C(61–318) Walker A mutant, K116A, were mixed before addition to an ATPase assay. Asynchronous ATP hydrolysis would be expected to decrease linearly as the amount of inactive enzyme is increased (dotted line in both plots). Data are averages of three experiments ± S.E. (error bars).
The results of our subunit-mixing experiments with FMDV 2C are consistent with the dominant negative effects observed on co-expression of active and inactive subunits of PV MBP-2C (43).
DISCUSSION
Expression of FMDV 2C
Most previous characterizations of picornaviral 2C proteins have been performed using recombinant N-terminal fusions to GST or MBP without removing the large protein tag because this generally resulted in loss of the protein through aggregation (23–26). The ability of these particular fusion partners to increase the solubility of different proteins is well documented (66–70). In the case of 2C, such large tags may aid solublization by sterically blocking the hydrophobic interactions made by the conserved N-terminal amphipathic helix. Although these reagents have facilitated analyses of the enzymatic and RNA binding activities of the protein, the use of large proteinaceous tags does not completely resolve the solubility of 2C proteins because the tags cannot be removed without resulting in significant loss of protein (43); in at least one case, proteolytic processing failed to efficiently remove the tag, which is suggestive of misfolding (24). Moreover, even when the tag is retained, there is evidence of residual aggregation (43) and difficulties with purification (23).
In the present study, we adopted a different approach. Although fusion of GST to the N terminus of full-length FMDV 2C facilitated the expression in E. coli, the resulting protein was poorly soluble. Expression of soluble FMDV 2C protein with a short N-terminal His6 tag was only possible after genetic deletion of the predicted N-terminal amphipathic region of the protein was found to eliminate its toxicity in E. coli. This result is consistent with the boost in solubility observed due to deletion of the first 21 amino acids of HAV 2C, which would be expected to remove most of the predicted amphipathic helix (14, 17). However, in that case, no quantitative assessments of yield or protein solubility were made. The largest FMDV 2C protein obtained by this deletion approach in the present study yielded 2C(34–318), which, following proteolytic removal of the His6 tag, remained highly soluble and could be concentrated to at least 20 mg/ml. The reason for the toxic effects of retaining the N-terminal amphipathic helix of 2C are not known, although it seems likely to be related to the membrane-binding properties attributed to this part of the protein in studies of 2C from PV (15, 16) and HAV (17). Although this approach produces an N-terminally truncated 2C protein, it retained ATPase, RNA binding, and oligomerization activities that could be analyzed in detail within in vitro experiments. Given the high level of sequence identity between picornavirus 2C proteins, we believe that similar N-terminal deletions should facilitate the production of soluble 2C proteins in E. coli. However, the method is not foolproof; we were able to clone and express similar soluble deletion mutants of 2C from type C FMDV but not 2C from swine vesicular disease virus.6
FMDV 2C Has ATPase Activity
Unsurprisingly, each of the Walker A and B motifs and Motif C was found to be required for the ATPase activity of FMDV 2C (Fig. 2a). The side chains of Lys116 and Asp160 in the Walker A and B motifs, respectively, appear to be important for ATP binding because mutation to Ala decreased UV cross-linking of the nucleotide to background levels. Interestingly, this pattern of binding differs from that recently reported for PV 2C (43); the D177A Walker B mutation in MBP-PV 2C (equivalent to D160A in FMDV 2C) had no effect on ATP cross-linking, whereas the K135A mutation (equivalent to K116A) was found to greatly reduce ATP cross-linking to the protein. The reason for the different effects of mutating the conserved Asp residue in the Walker B motif of the 2C proteins from the two viruses is not clear. The ATP hydrolysis sites of PV 2C and FMDV 2C would be expected to be quite similar because the regions thought to form the active sites are well conserved (62% identical across A, B, and C) (19). However, it is notable that the ATPase activities of 2C proteins from PV and FMDV respond quite differently to GdnHCl, which may also be due to structural differences at or near the ATP binding site; FMDV 2C is stimulated by low concentrations of the drug and only inhibited at about 20 mm (Fig. 2f), whereas PV 2C exhibits monotonic inhibition with an ED50 of about 2 mm (23). A further indication of possible structural differences at the ATP binding site comes from the observation that the M159L mutation that confers FMDV resistance to GdnHCl (29) is inside the Walker B motif, whereas the N179G/A mutations that confer GdnHCl resistance on the virus and rescue PV 2C ATPase activity are just outside its Walker B motif (Val172–Asp177) (23).
The clustering of mutations that confer resistance to GdnHCl around the ATPase active site of 2C from different picornaviruses suggests that the drug may bind close to the nucleotide binding site (23, 29). Although it was misassigned as “non-competitive,” the detailed kinetic analysis by Pfister and Wimmer revealed that GdnHCl is actually an uncompetitive inhibitor of PV 2C (see Fig. 7b of Ref. 23), which means that the drug only binds to inhibit in the presence of ATP. This observation is consistent with the results of UV cross-linking experiments with FMDV 2C, which showed that GdnHCl does not reduce ATP binding at concentrations known to inhibit its ATPase activity (supplemental Fig. S3). However, although the inhibition of the ATPase activity of PV 2C by submillimolar concentrations of GdnHCl appears to fully account for the block of PV infectivity by this drug (23), the ATPase activity of purified 2C from FMDV is about an order of magnitude less sensitive to GdnHCl than the virus infectivity (Fig. 2) (23, 29), a pattern of response that is similar to that observed with EV9 (28) (although, like PV, EV9 is an enterovirus). This relative insensitivity of 2C from FMDV and EV9 to inhibition by GdnHCl remains unexplained; it may be that 2BC is a more sensitive target for the drug or that it acts on additional targets in these viruses.
RNA Binding by FMDV 2C
We found that FMDV 2C(34–318) binds short ssRNA in a non-sequence-specific manner, with at least a 10-fold higher affinity than it bound to a randomly selected dsRNA (Fig. 3). Similarly, EV9 2C was reported to bind single-stranded RNA nonspecifically but with a preference for unstructured RNAs (34). If 2C indeed functions as an RNA helicase, it would be expected to interact with RNA in a sequence-independent manner, although the possibility that 2C can interact with RNA in other ways (perhaps to fulfill a different role during replication) cannot be discounted. It should be noted that PV 2C has been reported to bind in a sequence-specific manner to the 3′-end of the viral negative strand RNA, which is predicted to adopt a cruciform stem loop structure (33, 71), an observation that has been repeated with 2C from HAV and human rhinovirus (32). FMDV is not predicted to contain a similar structure at the 3′-end of its negative strand RNA, and further work will be necessary to determine if FMDV 2C also possesses specific RNA binding.
ATP and RNA Binding Stimulate Hexamerization of FMDV 2C
An N-terminal MBP fusion of PV 2C was recently shown to form homo-oligomeric structures necessary for ATPase activity (43). Negative stain and scanning electron microscopy were used to examine the structure of PV 2C oligomers and to determine the stoichiometry of their assembly. This latter analysis, which was aided by the retention of the MBP tags that appeared as distinct lobes of density surrounding the core of the oligomer, revealed the presence of mixed populations of oligomeric states of the fusion protein ranging from tetramer to octamer (43).
Oligomerization of recombinant FMDV 2C(34–318) occurred in a concentration-dependent manner; in the absence of ligand, the largest oligomeric state detectable was tetrameric (Figs. 1c and 4). This result is broadly similar to findings with other SF3 proteins; in the presence of DNA but not ATP, the largest assembly of LTag observed is tetrameric (72), whereas intermediate oligomeric assemblies were also identified for E1 (73). However, in contrast to the LTag and E1 helicases, which both form stable hexamers in the presence of ATP (40, 74), the addition of the nucleotide failed to stabilize FMDV 2C(34–318) hexamers (Fig. 4c). Instead, we found that both ATP and RNA are required to observe stable hexamers of FMDV 2C(34–318) and that this can only be achieved if the protein incorporates the Motif C mutation N207A, which abrogates ATP hydrolysis but does not significantly impair binding of the nucleotide. Strikingly, in contrast to MBP-PV 2C, this ternary 2C-ATP-RNA complex forms a single oligomeric species with six protein subunits (Fig. 4). The stabilization due to ATP probably arises because its binding site is predicted to occur at the interface between pairs of subunits in the hexameric ring; moreover, by analogy with the co-crystal structure of the BPV E1-ATP/ADP-DNA complex (39), the RNA may contribute to hexamerization by binding to the pore through the center of the hexamer and making contacts with all six subunits. Although we have not observed hexamerization of wild-type 2C(34–318), due presumably to its ATPase activity, it should be noted that absence of the authentic N terminus may also have impaired subunit interactions (see below). Further work will be needed to examine the oligomerization behavior of full-length FMDV 2C and to determine the structure of the hexameric FMDV 2C-ATP-RNA complex.
It is difficult to precisely compare the results obtained using FMDV 2C(34–318) with those obtained using PV MBP-2C due to gross structural differences between the two proteins and differences in experimental approaches; for example, the effects of ATP and RNA on the oligomerization behavior of PV MBP-2C were not analyzed (43). Nevertheless we believe there is at least suggestive evidence that the two proteins have important properties in common. In the absence of ligands, both proteins formed a range of oligomers of a range of sizes, ranging from one to four subunits for FMDV 2C(34–318) and from four to eight subunits for PV MBP-2C. We suspect that the larger size range of PV MBP-2C particles may be partly due to the residual aggregation of this protein (as indicated by the significant void peak observed by SEC analysis), but there may also be contributions from the Cys-rich motif in the C terminus of PV 2C that is absent in the FMDV protein. It is also notable that hexamers were the most commonly observed form of PV MBP-2C (43). Given the homology between picornavirus 2C proteins and the observation that many other AAA+ proteins are observed to function as hexamers, we propose that the hexameric rings observed for FMDV 2C(34–318) are the most physiologically relevant form of picornavirus 2C proteins. This hypothesis is supported by the finding that FMDV 2C hydrolyzes ATP by a coordinated mechanism (Fig. 6), which is likely to involve subunit-subunit interactions.
We suspect that the observations of the effects of N-terminal modifications on oligomerization of FMDV and PV 2C support a common mode of assembly although they superficially appear to be at odds with one another. When examined within the context of an MBP fusion protein, removal of the first 38 residues from the N terminus of PV 2C was found to completely abrogate the ability of the protein to oligomerize and to hydrolyze ATP (43). The authors therefore concluded that the N terminus of PV 2C was necessary for oligomerization. Strikingly, the MPB-PV 2C(39–329) used in these experiments lacks all residues up to and including the predicted N-terminal amphipathic helix (residues 20–36 (14)) and therefore contains a very similar deletion to the FMDV 2C(34–318) Motif C mutant that was shown to hexamerize in the present study. What is the source of this discrepancy? Significantly, the investigation of PV 2C oligomerization was performed with MBP-2C constructs in which MBP remained attached to the N terminus. We suggest that deletion of the N terminus of 2C within such a construct may have brought the MBP domain close enough to the body of the ATPase domain of PV 2C to interfere sterically with the subunit-subunit contacts needed for oligomerization and ATPase activity. Consistent with this idea, there is no tag in FMDV 2C(34–318) to interfere with the formation of hexamers.
Our results show that the first 33 amino acids of FMDV 2C, including the predicted amphipathic helix, are not required for hexamerization, at least in the presence of ATP and ssRNA. However, it is still possible that the presence of the N terminus of the protein may further stabilize the hexameric form of the enzyme; for example, the predicted amphipathic helix may contribute to interactions between subunits or may help to concentrate 2C subunits on the two-dimensional surfaces on internal cellular membranes.
We did find clear evidence that the N-terminal residues of the FMDV 2C(34–318) truncation mutant are important for higher order oligomerization because 2C(61–318) was significantly less efficient at assembling into hexamers (Fig. 4e). Intriguingly, both the LTag and E1 helicases possess small modules of about 50 amino acids on the N-terminal side of their ATPase domains that are reported to facilitate hexamerization (40, 74). Structural analyses have shown that these N-terminal modules fold into small helical domains (containing 4–5 α-helices) that associate with one another into a second hexameric ring structure above the ring formed by the ATPase domains (39–41). However, there is no amino acid sequence homology between these N-terminal domains and the segment at the N terminus of 2C(34–318) that aids hexamerization (residues 34–60). The structural basis for the ability of residues 34–60 to stabilize hexamers of FMDV 2C therefore remains to be determined.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
T. R. Sweeney and S. Curry, unpublished data.
- FMDV
- foot-and-mouth disease virus
- PV
- poliovirus
- HAV
- hepatitis A virus
- SF3
- superfamily 3
- LTag
- large T-antigen
- E1
- bovine papillomavirus E1
- EV9
- echovirus-9
- SEC
- size exclusion chromatography
- MALS
- multiangle light scattering
- MSA
- multivariate statistical analysis
- GST
- glutathione S-transferase
- ssRNA
- single-stranded RNA
- dsRNA
- double-stranded RNA.
REFERENCES
- 1.Mason P. W., Grubman M. J., Baxt B. (2003) Virus Res. 91, 9–32 [DOI] [PubMed] [Google Scholar]
- 2.Grubman M. J., Baxt B. (2004) Clin. Microbiol. Rev. 17, 465–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bienz K., Egger D., Troxler M., Pasamontes L. (1990) J. Virol. 64, 1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlegel A., Giddings T. H., Jr., Ladinsky M. S., Kirkegaard K. (1996) J. Virol. 70, 6576–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monaghan P., Cook H., Jackson T., Ryan M., Wileman T. (2004) J. Gen. Virol. 85, 933–946 [DOI] [PubMed] [Google Scholar]
- 6.Miller S., Krijnse-Locker J. (2008) Nat. Rev. Microbiol. 6, 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moffat K., Knox C., Howell G., Clark S. J., Yang H., Belsham G. J., Ryan M., Wileman T. (2007) J. Virol. 81, 1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanz-Parra A., Sobrino F., Ley V. (1998) J. Gen. Virol. 79, 433–436 [DOI] [PubMed] [Google Scholar]
- 9.Doedens J. R., Giddings T. H., Jr., Kirkegaard K. (1997) J. Virol. 71, 9054–9064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodd D. A., Giddings T. H., Jr., Kirkegaard K. (2001) J. Virol. 75, 8158–8165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho M. W., Teterina N., Egger D., Bienz K., Ehrenfeld E. (1994) Virology 202, 129–145 [DOI] [PubMed] [Google Scholar]
- 12.Moffat K., Howell G., Knox C., Belsham G. J., Monaghan P., Ryan M. D., Wileman T. (2005) J. Virol. 79, 4382–4395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doedens J. R., Kirkegaard K. (1995) EMBO J. 14, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teterina N. L., Gorbalenya A. E., Egger D., Bienz K., Rinaudo M. S., Ehrenfeld E. (2006) Virology 344, 453–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Echeverri A., Banerjee R., Dasgupta A. (1998) Virus Res. 54, 217–223 [DOI] [PubMed] [Google Scholar]
- 16.Echeverri A. C., Dasgupta A. (1995) Virology 208, 540–553 [DOI] [PubMed] [Google Scholar]
- 17.Kusov Y. Y., Probst C., Jecht M., Jost P. D., Gauss-Müller V. (1998) Arch. Virol. 143, 931–944 [DOI] [PubMed] [Google Scholar]
- 18.Murray L., Luke G. A., Ryan M. D., Wileman T., Knox C. (2009) Virus Res. 144, 74–82 [DOI] [PubMed] [Google Scholar]
- 19.Gorbalenya A. E., Koonin E. V., Wolf Y. I. (1990) FEBS Lett. 262, 145–148 [DOI] [PubMed] [Google Scholar]
- 20.Pfister T., Jones K. W., Wimmer E. (2000) J. Virol. 74, 334–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirzayan C., Wimmer E. (1992) Virology 189, 547–555 [DOI] [PubMed] [Google Scholar]
- 22.Teterina N. L., Kean K. M., Gorbalenya A. E., Agol V. I., Girard M. (1992) J. Gen. Virol. 73, 1977–1986 [DOI] [PubMed] [Google Scholar]
- 23.Pfister T., Wimmer E. (1999) J. Biol. Chem. 274, 6992–7001 [DOI] [PubMed] [Google Scholar]
- 24.Rodríguez P. L., Carrasco L. (1993) J. Biol. Chem. 268, 8105–8110 [PubMed] [Google Scholar]
- 25.Klein M., Eggers H. J., Nelsen-Salz B. (1999) Virus Res. 65, 155–160 [DOI] [PubMed] [Google Scholar]
- 26.Samuilova O., Krogerus C., Fabrichniy I., Hyypiä T. (2006) J. Virol. 80, 1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolskaya E. A., Romanova L. I., Kolesnikova M. S., Gmyl A. P., Gorbalenya A. E., Agol V. I. (1994) J. Mol. Biol. 236, 1310–1323 [DOI] [PubMed] [Google Scholar]
- 28.Klein M., Hadaschik D., Zimmermann H., Eggers H. J., Nelsen-Salz B. (2000) J. Gen. Virol. 81, 895–901 [DOI] [PubMed] [Google Scholar]
- 29.Belsham G. J., Normann P. (2008) J. Gen. Virol. 89, 485–493 [DOI] [PubMed] [Google Scholar]
- 30.Pincus S. E., Diamond D. C., Emini E. A., Wimmer E. (1986) J. Virol. 57, 638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pariente N., Airaksinen A., Domingo E. (2003) J. Virol. 77, 7131–7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee R., Dasgupta A. (2001) J. Gen. Virol. 82, 2621–2627 [DOI] [PubMed] [Google Scholar]
- 33.Banerjee R., Echeverri A., Dasgupta A. (1997) J. Virol. 71, 9570–9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein M., Eggers H. J., Nelsen-Salz B. (2000) J. Gen. Virol. 81, 2481–2484 [DOI] [PubMed] [Google Scholar]
- 35.Singleton M. R., Dillingham M. S., Wigley D. B. (2007) Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 36.Erzberger J. P., Berger J. M. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 [DOI] [PubMed] [Google Scholar]
- 37.Iyer L. M., Leipe D. D., Koonin E. V., Aravind L. (2004) J. Struct. Biol. 146, 11–31 [DOI] [PubMed] [Google Scholar]
- 38.Costa A., Pape T., van Heel M., Brick P., Patwardhan A., Onesti S. (2006) J. Struct. Biol. 156, 210–219 [DOI] [PubMed] [Google Scholar]
- 39.Enemark E. J., Joshua-Tor L. (2006) Nature 442, 270–275 [DOI] [PubMed] [Google Scholar]
- 40.Li D., Zhao R., Lilyestrom W., Gai D., Zhang R., DeCaprio J. A., Fanning E., Jochimiak A., Szakonyi G., Chen X. S. (2003) Nature 423, 512–518 [DOI] [PubMed] [Google Scholar]
- 41.Hickman A. B., Dyda F. (2005) Curr. Opin. Struct. Biol. 15, 77–85 [DOI] [PubMed] [Google Scholar]
- 42.Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams P., Kandiah E., Effantin G., Steven A. C., Ehrenfeld E. (2009) J. Biol. Chem. 284, 22012–22021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirzayan C., Wimmer E. (1994) Virology 199, 176–187 [DOI] [PubMed] [Google Scholar]
- 45.Teterina N. L., Levenson E., Rinaudo M. S., Egger D., Bienz K., Gorbalenya A. E., Ehrenfeld E. (2006) J. Virol. 80, 5327–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crampton D. J., Mukherjee S., Richardson C. C. (2006) Mol. Cell 21, 165–174 [DOI] [PubMed] [Google Scholar]
- 47.Lee S. Y., De La Torre A., Yan D., Kustu S., Nixon B. T., Wemmer D. E. (2003) Genes Dev. 17, 2552–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toth E. A., Li Y., Sawaya M. R., Cheng Y., Ellenberger T. (2003) Mol. Cell 12, 1113–1123 [DOI] [PubMed] [Google Scholar]
- 49.Birtley J. R., Curry S. (2005) Acta Crystallogr. D. 61, 646–650 [DOI] [PubMed] [Google Scholar]
- 50.Hohenwallner W., Wimmer E. (1973) Clin. Chim. Acta 45, 169–175 [DOI] [PubMed] [Google Scholar]
- 51.Mahuren J. D., Coburn S. P., Slominski A., Wortsman J. (2001) Anal. Biochem. 298, 241–245 [DOI] [PubMed] [Google Scholar]
- 52.Schumacher J., Zhang X., Jones S., Bordes P., Buck M. (2004) J. Mol. Biol. 338, 863–875 [DOI] [PubMed] [Google Scholar]
- 53.Simpson P. J., Monie T. P., Szendröi A., Davydova N., Tyzack J. K., Conte M. R., Read C. M., Cary P. D., Svergun D. I., Konarev P. V., Curry S., Matthews S. (2004) Structure 12, 1631–1643 [DOI] [PubMed] [Google Scholar]
- 54.van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 55.Dube P., Tavares P., Lurz R., van Heel M. (1993) EMBO J. 12, 1303–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Palma A. M., Heggermont W., Lanke K., Coutard B., Bergmann M., Monforte A. M., Canard B., De Clercq E., Chimirri A., Pürstinger G., Rohayem J., van Kuppeveld F., Neyts J. (2008) J. Virol. 82, 4720–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X., Shaw A., Bates P. A., Newman R. H., Gowen B., Orlova E., Gorman M. A., Kondo H., Dokurno P., Lally J., Leonard G., Meyer H., van Heel M., Freemont P. S. (2000) Mol. Cell 6, 1473–1484 [DOI] [PubMed] [Google Scholar]
- 58.Pfister T., Wimmer E. (2001) J. Virol. 75, 1611–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saunders K., King A. M. (1982) J. Virol. 42, 389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saunders K., King A. M., McCahon D., Newman J. W., Slade W. R., Forss S. (1985) J. Virol. 56, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodríguez P. L., Carrasco L. (1995) J. Biol. Chem. 270, 10105–10112 [DOI] [PubMed] [Google Scholar]
- 62.Seo Y. S., Müller F., Lusky M., Hurwitz J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 702–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wyatt P. J. (1993) Anal. Chim. Acta 272, 1–40 [Google Scholar]
- 64.Martin A., Baker T. A., Sauer R. T. (2005) Nature 437, 1115–1120 [DOI] [PubMed] [Google Scholar]
- 65.Gai D., Zhao R., Li D., Finkielstein C. V., Chen X. S. (2004) Cell 119, 47–60 [DOI] [PubMed] [Google Scholar]
- 66.Edwards A. M., Arrowsmith C. H., Christendat D., Dharamsi A., Friesen J. D., Greenblatt J. F., Vedadi M. (2000) Nat. Struct. Biol. 7, (suppl.) 970–972 [DOI] [PubMed] [Google Scholar]
- 67.Hammarström M., Hellgren N., van Den Berg S., Berglund H., Härd T. (2002) Protein Sci. 11, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Braun P., Hu Y., Shen B., Halleck A., Koundinya M., Harlow E., LaBaer J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shih Y. P., Kung W. M., Chen J. C., Yeh C. H., Wang A. H., Wang T. F. (2002) Protein Sci. 11, 1714–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smyth D. R., Mrozkiewicz M. K., McGrath W. J., Listwan P., Kobe B. (2003) Protein Sci. 12, 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Banerjee R., Tsai W., Kim W., Dasgupta A. (2001) Virology 280, 41–51 [DOI] [PubMed] [Google Scholar]
- 72.Mastrangelo I. A., Hough P. V., Wall J. S., Dodson M., Dean F. B., Hurwitz J. (1989) Nature 338, 658–662 [DOI] [PubMed] [Google Scholar]
- 73.Schuck S., Stenlund A. (2005) Mol. Cell 20, 377–389 [DOI] [PubMed] [Google Scholar]
- 74.Abbate E. A., Berger J. M., Botchan M. R. (2004) Genes Dev. 18, 1981–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.