Abstract
Gonadotropin-releasing hormone (GnRH) acts via G-protein-coupled receptors on gonadotrophs to stimulate synthesis and secretion of luteinizing hormone and follicle-stimulating hormone. It is secreted in pulses, and its effects depend on pulse frequency, but decoding mechanisms are unknown. Here we have used an extracellular signal regulated kinase-green fluorescent protein (ERK2-GFP) reporter to monitor GnRH signaling. GnRH caused dose-dependent ERK2-GFP translocation to the nucleus, providing a live-cell readout for activation. Pulsatile GnRH caused dose- and frequency-dependent ERK2-GFP translocation. These responses were rapid and transient, showed only digital tracking, and did not desensitize under any condition tested (dose, frequency, and receptor number varied). We also tested for the effects of cycloheximide (to prevent induction of nuclear-inducible MAPK phosphatases) and used GFP fusions containing ERK mutations (D319N, which prevents docking domain-dependent binding to MAPK phosphatases, and K52R, which prevents catalytic activity). These manipulations had little or no effect on the translocation responses, arguing against a role for MAPK phosphatases or ERK-mediated feedback in shaping ERK activation during pulsatile stimulation. GnRH also caused dose- and frequency-dependent activation of the α-gonadotropin subunit-, luteinizing hormone β-, and follicle-stimulating hormone β- luciferase reporters, and the latter response was inhibited by ERK1/2 knockdown. Moreover, GnRH caused frequency-dependent activation of an Egr1-luciferase reporter, but the response was proportional to cumulative pulse duration. Our data suggest that frequency decoding is not due to negative feedback shaping ERK signaling in this model.
Keywords: Dual Specificity Phosphoprotein Phosphatase, ERK, G-protein-coupled Receptors (GPCR), Hormone Receptors, MAP Kinases (MAPKs), Signal Transduction, GnRH, GnRH Receptor
Introduction
Gonadotropin-releasing hormone (GnRH)2 stimulates the synthesis and secretion of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) and thereby mediates central control of reproduction (1–3). It is secreted in brief pulses from the hypothalamus and acts via seven transmembrane receptors on pituitary gonadotrophs to stimulate phospholipase C, mobilize Ca2+, and activate protein kinase C isozymes. This leads to activation of mitogen-activated protein kinase (MAPK) pathways and Ca2+ effectors, such as calmodulin, and these in turn mediate the effects of GnRH on exocytotic gonadotropin secretion as well as its effects on expression of many genes including those for the gonadotropin subunits (1–3). GnRH pulse frequency of GnRH pulses varies under different physiological conditions. For example, pubertal increases in gonadotropin secretion and the pre-ovulatory gonadotropin surge are both driven by increases in GnRH pulse frequency (4). GnRH effects are pulse frequency-dependent; constant GnRH suppresses pituitary LH and FSH secretion, whereas GnRH pulses restore pulsatile gonadotropin secretion in vivo (5). Desensitization of GnRH-stimulated gonadotropin secretion is exploited therapeutically, as pulsatile administration of GnRH agonists can increase circulating gonadotropins and gonadal steroids and thereby increase fertility (e.g. in ovulation induction during assisted reproduction), whereas sustained stimulation ultimately reduces steroid secretion, which underlies agonist efficacy against steroid hormone-dependent cancers (6, 7).
Although frequency decoding is fundamental to the physiology and pharmacology of this system, the mechanisms are poorly understood. Most recent work has focused on transcription and effects of GnRH on expression of gonadotropin subunit genes, which (in both gonadotrophs and LβT2 cells) was found to be sensitive to GnRH pulse frequency. Increasing GnRH pulse frequency to physiological level increases its effects on LHβ, FSHβ, and GnRH receptor (GnRHR) expression but as frequency is further increased to super-physiological levels, transcription is reduced (4, 8–14). Computational models suggest that such bell-shaped frequency-response relationships require feedback mechanisms (15, 16), and these could include GnRHR down-regulation, induction of RGS-2 (regulator of G-protein signaling-2), inhibition of Ca2+ channels by the calmodulin-dependent G-protein Kir/Gem, or induction of MAPK phosphatases (MKPs) (4, 15, 16). Rapid homologous receptor desensitization can be excluded as a mechanism because type I mammalian GnRHR does not show this behavior. Uniquely, they lack the C-terminal tails that mediate phosphorylation, arrestin binding, and desensitization of numerous other seven-transmembrane receptors (17–20). Alternative mechanisms of frequency decoding involve interplay between Egr-1 and a co-regulator (Nab-2) at the LHβ promoter. In this model, low pulse frequency causes transient Egr-1 expression, causing expression of Nab-2, which inhibits LHβ expression. However, at high pulse frequencies, more sustained increases in Egr-1 quench Nab-2 and increase LHβ transcription (21), although it is not clear whether this occurs in vivo (22). For the FSHβ promoter, similar interplay between c-Fos and the co-regulator TGIF has been proposed to underlie preferential activation at low GnRH pulse frequency (23). Ciccone et al. (24) suggest CREB (cyclic AMP response element-binding protein) and inducible cAMP early repressor may be responsible (24). Here, high pulse frequencies preferentially induce inducible cAMP early repressor, causing transcriptional repression by competing for a CRE (cyclic AMP response element) site in the FSHβ promoter.
The data above highlight two distinct possibilities; that GnRH frequency decoding reflects feedback effects on signal generation in the cytoplasm or that frequency decoding occurs at the level of the transcriptome. Using conventional techniques it has been difficult to test the first of these possibilities. We have been developing live cell readouts for GnRH effectors implicated in frequency decoding in other systems. In a recent study we used a cytoplasm-to-nucleus translocation assay with a fusion protein consisting of the nuclear factor of activated T-cells (NFAT) and emerald fluorescent protein (EFP) and found no evidence for GnRH frequency coding in the Ca2+/calmodulin/calineurin/NFAT pathway (25). Here, we focus on an alternative possibility that frequency decoding occurs within the ERK cascade. Like many other seven-transmembrane receptors, GnRHR activate the Raf/MEK/ERK cassette (26–30). On stimulation, ERKs translocate to the nucleus where they phosphorylate transcription factors to control gene expression. GnRH activates ERK1/2, and ERKs can mediate GnRHR-stimulated transcription of the common αGSU subunit as well as the LHβ and FSHβ (1–3). ERKs can mediate responses to pulsatile GnRH stimulation (31–33), and the ERK cascade functions as a frequency decoder in other systems (34–36). GnRH also induces expression of MKP family dual-specificity phosphatases (DUSPs) in vitro and in vivo (30, 37–39), and a recent computational model illustrated the potential for pulse frequency decoding by MAPK pathways and inducible phosphatases (16). To explore the dynamics of ERK signaling, we have developed a knock-down/add-back model in which siRNAs are used to prevent expression of endogenous ERK1/2 and recombinant adenovirus (Ad) is used to add back an ERK2-GFP reporter at a physiological expression level. Using this model and siRNA targeting of DUSPs, we found that 12 of the 16 DUSPs expressed in HeLa cells influenced ERK responses to sustained stimulation with GnRH or phorbol 12-,13-dibutyrate (used to activate protein kinase C) (37, 40).
Effects of GnRH on intracellular signaling pathways have been defined in detail with sustained stimulation, but much less is known for physiologically relevant pulsatile stimuli. Here, mathematical modeling may be helpful, and a recent model describing kinetics of GnRH effects on Ca2+ mobilization can incorporate receptor occupancy and effector activation with GnRH pulses (41). This model predicts desensitization with both sustained and pulsatile GnRH stimulation and that desensitization will become more pronounced with increasing pulse frequency, pulse magnitude, or receptor number (25, 41). Here we have tested for such adaptation using nuclear translocation of ERK2-GFP as a live cell readout for ERK activation. We have also explored the effects of GnRH pulses on MKP expression and have used pharmacological and molecular genetic approaches to test for possible MKP effects on ERK signaling with pulsatile GnRH. We find that the ERK responses are resistant to desensitization and appear independent of nuclear-inducible MKPs or ERK-mediated feedback.
EXPERIMENTAL PROCEDURES
Engineering of Plasmids and Viruses
Ad-expressing wild-type (WT) and D319N ERK2-GFP, mGnRHR, and Egr-1 promoter luciferase reporter were prepared, grown to high titer, and purified as described (17, 40, 42). Ad K52R ERK2-GFP was prepared using 5′-CAA AGT TCG AGT TGC TAT CAG GAA AAT CAG TCC TTT TGA GC-3′ forward and complementary reverse primers to mutate WT ERK2-GFP template with a Stratagene QuikChange mutagenesis kit before Ad manufacture. Plasmid- and Ad-expressing luciferase reporters with promoters from human αGSU, rat βFSH, rat βLH, and murine Egr-1 have been described (40, 43, 44). Blue fluorescent protein (BFP) nuclear marker (designated BFP-NLS) containing the SV40 large T-antigen nuclear localization sequence (NLS) was constructed as described (25). Viruses were made from shuttle vectors as described (45). Briefly, 4.5 μg of shuttle vectors were digested alongside 1.5 μg of pacAd5 9.2–100 sub360 backbone vector (donated by Prof. Beverly Davidson, University of Iowa, IA) with PacI or NheI. Cut shuttle and backbone vectors were then mixed and transfected into low passage HEK293 cells using Superfect (Qiagen, Crawley, UK). Cells were left for 7–10 days to allow recombination between shuttle and backbone vectors, and after cytopathic effects, lysates were collected for further bulking. Ad vectors were grown to high titer and purified according to standard protocols (17).
Cell Culture, Transfection, and Transduction
HeLa cells were cultured in Costar black-wall 96-well plates (Corning, Arlington, UK) or Nunc 6-well plates (supplied by Fisher) using 10% FCS-supplemented Dulbecco's modified Eagle's medium (DMEM). For ERK1/2 knockdown, they were transfected with 1 nm nontargeting control siRNAs or siRNAs targeting non-coding regions of ERK1/2 as described (65–67). Cells were transduced in DMEM with 2% FCS 24 h after siRNA knockdown. Ad WT, D319N, or K52R ERK2-GFP were used at 2 pfu/nl, Ad-NLS-BFP was used at 75 pfu/nl, and Ad mGnRHR was used at 3 pfu/nl. For luciferase assays, Ad vectors were used at 1 pfu/nl. The Ad-containing medium was removed after 4–6 h and replaced with DMEM containing 0.1% FCS. The cells were then cultured for 16–24 h before GnRH stimulation. For plasmid transfections, cells were treated with Superfect (Qiagen, Crawley, UK) using 0.5 μg DNA/well for 2 h before Ad transduction.
Western Blotting
HeLa cells were plated in 6-well plates at 2.5 × 105 cells per well, transfected with siRNA, and transduced with Ad as above. Cells lysis and Western blotting was performed as described (37).
Semi-automated Image Acquisition and Analysis
Imaging experiments were performed using an IN Cell Analyzer 1000 (GE Healthcare) high content imaging platform. Images were acquired with a single field of view (0.6 mm2) and a 10× objective. Experiments were performed in duplicate or triplicate wells and each field typically contained 300–500 cells.
For live cell imaging, cells were plated in 96-well plates at 5 × 103 cells per well and cultured as above. Medium was replaced 25 min before imaging with phenol-red free DMEM:F-12 (with 100 μg/ml BSA and 10 μg/ml apotransferrin) and, if Ad-NLS-BFP was not included, contained 400 nm Hoechst nuclear stain (GE Healthcare). Cells were imaged in an environmental control chamber at 37 °C in a 5% CO2 humidified atmosphere. Stimuli were staggered by 5–10 s to compensate for image acquisition delays. Cells were stimulated with GnRH continuously or for 5 min (followed by washing ×5 in DMEM:F-12). In some experiments cells were subjected to repeat stimulation with GnRH (5 min stimulation, removal by washing as above) at the indicated frequency. For fixed cell imaging, cells were plated in 96-well plates and cultured as above. After treatment, they were washed in ice-cold phosphate-buffered saline, fixed with 4% paraformaldehyde, stained for ppERK1/2, and imaged as described (37, 40, 42).
Image analysis and quantification of fluorescence intensity and localization was performed using IN Cell Analyzer Work station 3.5 software (IN Cell Investigator, GE Healthcare). Green channel (GFP) and blue channel (BFP, Hoechst, and 4′-6-diamidino-2-phenylindole) images were used to define whole-cell and nuclear regions, respectively. Data reported are population-averaged fluorescence intensities (typically, with background subtracted) and ratios of nuclear to cytoplasmic fluorescence intensity (N:C).
Luciferase Assays
Cells were plated and transfected or transduced with luciferase reporters as above. After treatment as detailed in the figure legends, cells were washed in ice-cold phosphate-buffered saline and lysed, and luciferase activity was determined as described (37, 40, 46). Data are reported as relative light units normalized as -fold change over control except where indicated.
Quantitative PCR
HeLa cells were cultured in 6-well plates (2.5 × 105 cells/well) in 10% FCS DMEM, then 16–24 h before GnRH stimulation media was replaced with 0.1% FCS DMEM. Extraction of total RNA was performed using an RNeasy kit according to the manufacturer's instructions (Qiagen). Contaminating genomic DNA was removed from columns using an additional DNase (Qiagen) digestion step. Complementary DNA was then prepared for 1 μg of each total RNA sample using a cloned avian myeloblastosis virus first-strand synthesis kit according to the manufacturer's instructions (Invitrogen). cDNAs were then quantified relative to expression of human GTPase-activating protein using the following primers: human GTPase-activating protein, 5′-GGG AAG GTG AAG GTC GGA GT-3′ and 5′-GAG TTA AAA GCA GCC CTG GTG A-3′; DUSP1, 5′-CAA CGA GGC CAT TGA CTT CAT AG-3′ and 5′-CAA ACA CCC TTC CTC CAG CA-3′; DUSP2, 5′-CTG TCT ACG ACC AGG GTG GC-3′ and 5′-GGT CTG ACG AGT GAC TGC AGC-3′; DUSP4, 5′-CTG GTT CAT GGA AGC CAT AGA GT-3′ and 5′-CGC CCA CGG CAG TCC-3′; DUSP5, 5′-CCG CGG GTC TAC TTC CTC A-3′ and 5′-GGG TTT TAC ATC CAC GCA ACA-3′. PCR primers were mixed with 50 ng of reverse transcription-PCR template and SYBR Green PCR master mix (Applied Biosystems, Warrington, UK), and the comparative cycle threshold method was used to detect relative expression curves on an ABI PRISM 7500 detection system (Applied Biosystems).
Statistical Analysis and Data Presentation
The figures show the mean ± S.E. of data pooled from at least three experiments and normalized as described in the figure legends. Statistical analysis was typically by one- or two-way ANOVA and post-hoc tests (as detailed in figure legends), accepting p < 0.05 as statistically significant. Statistical analysis, curve fitting, and regression were performed using GraphPad Prism 4.0 (GraphPad Software Inc, CA).
RESULTS
GnRH Signaling to ERK in HeLa Cells
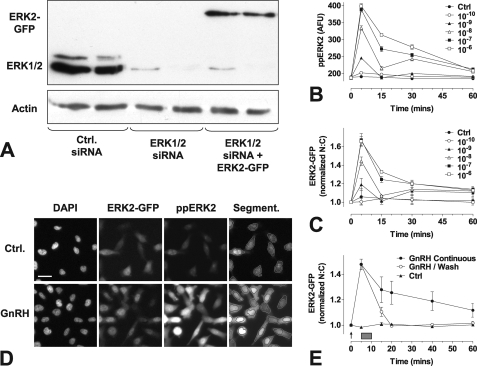
GnRH activates MAPKs in many models, and we have shown that GnRH mediates protein kinase C-dependent activation of ERK in GnRHR-expressing HeLa cells, increasing dual phosphorylated ERK (ppERK) levels as well as ERK nuclear accumulation (17, 37). To monitor this we used a previously characterized knock-down/add-back protocol in which siRNAs are used to knock-down endogenous ERKs 1 and 2 and recombinant Ad is used to add back an ERK2-GFP reporter at a physiological expression level (37, 40, 42). We used Western blotting to confirm that the ERK1/2 siRNAs were effective at knocking down endogenous ERK1/2 expression and that Ad transduction recovered ERK expression to levels seen in control cells (Fig. 1A). These results are in agreement with previous Western blotting and imaging data used to validate ERK expression and phosphorylation in this model (37, 40, 42). Again, consistent with our earlier data (17, 37, 40, 42), we found that GnRH caused a dose-dependent increase in whole cell ppERK2 levels and N:C ERK2-GFP ratio (pEC50 values 8.4 ± 0.1 and 8.3 ± 0.2, respectively) with both effects maximal at 5 min (Fig. 1, B and C). To test the dependence on ongoing GnRHR activation, we adapted the ERK2-GFP translocation assay for use in live cells. As shown (Fig. 1E), GnRH again caused a rapid increase in the ERK2-GFP N:C ratio with a maximum response within 5 min. In contrast, when cells were washed to remove GnRH, ERK2-GFP N:C declined to near basal values by 20 min. Accordingly, the wash was effective at removing GnRH, and the sustained effect of GnRH on ERK2-GFP N:C ratio was dependent on continued receptor activation.
FIGURE 1.
Image-based assays of GnRH-mediated ERK signaling. Panel A, HeLa cells were grown in 6-well plates, transfected with control siRNAs (Ctrl) or ERK1/2 siRNAs, and transduced with Ad mGnRHR with or without Ad ERK2-GFP as indicated. Cells were processed for Western blotting with antibodies targeting total ERK1/2 or β-actin, as described under “Experimental Procedures.” The position of bands showing ERK1/2 and ERK2-GFP were determined by comparison with molecular weight markers. Results are representative of three similar experiments. Panels B and C, cells were plated in 96-well plates and subject to siRNA knockdown of ERK1/2 then transduced with Ad-mGnRHR and Ad-ERK2-GFP before serum starvation overnight. Cells were stimulated with GnRH at the indicated times and molar concentrations before fixation, immunocytochemical staining, and image analysis as described under “Experimental Procedures.” Panel D, cells were treated as above, and representative images are shown before and after treatment with GnRH (10−6 m, peak response shown) for the ppERK2 and ERK2-GFP image channels, with an example of the automated image segmentation. Scale bar, 30 μm. Panel E, cells were plated as above, then stained with Hoechst dye and treated with 10−7 m GnRH at the indicated time points either continuously or for 5 min followed by repeated wash (gray rectangle). Live cell image acquisition and analysis was performed as described under “Experimental Procedures.” Data shown are ppERK fluorescence intensity in arbitrary fluorescence units (AFU) or N:C ratio of ERK2-GFP fluorescence intensity (background subtracted) normalized as the -fold change over control. Results shown are the mean ± S.E. of 3–8 independent experiments. For panel E, statistical analysis by two-way ANOVA indicates that treatment type (brief versus sustained) is a significant source of variation (p < 0.001, F1,68 = 22.49), as is time (p < 0.001, F5,68 = 30.32), and the interaction (p < 0.05, F5,68 = 2.92).
Pulsatile GnRH Signaling to ERK
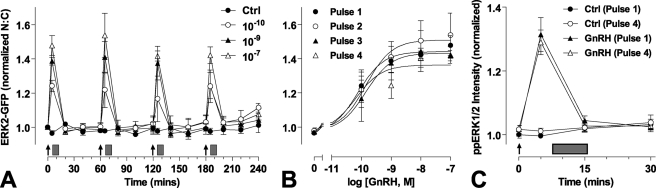
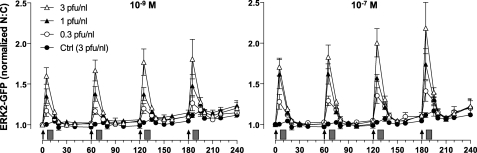
Because brief stimulation with GnRH causes transient translocation of ERK2-GFP to the nucleus (above), we tested the reproducibility of this response with a series of brief GnRH pulses. To do so, cells were stimulated with a train of 4 GnRH stimuli of 5-min duration and a varied dose at 1-h intervals. Brief stimulation with GnRH caused dose-dependent translocation of ERK2-GFP to the nucleus (Fig. 2), and similar dose-dependent responses were seen with each of 4 repeated stimuli (pEC50 values 10.2 ± 0.4 to 9.9 ± 0.4). The responses had comparable kinetics at all doses and pulses, increasing to maxima at 5 min and reducing toward basal values over the next 15 min. No desensitization was seen, as ANOVA revealed the GnRH dose (p < 0.001), but not pulse number or pulse-dose interaction, as a significant source of variation. This live cell assay simplifies monitoring of ERK response kinetics but raises the concern of whether ERK2-GFP translocation faithfully mirrors ERK phosphorylation. Accordingly, we performed similar experiments (repeated 5 min stimulation with 10−7 m GnRH at 1-h intervals) and fixed cells so that ppERK2 could be stained and quantified during the first and fourth pulse. As expected, GnRH increased ppERK2 (Fig. 2C), with a maximal response at 5 min, returning to basal by 15 min. For both pulses kinetics and response magnitudes were identical with no desensitization observed. With this assay GnRH also increased the ERK2-GFP N:C ratio equally at both pulses, and responses were similar to those observed with live cell imaging (supplemental Fig. 3). We also used the live cell ERK2-GFP imaging assay to explore frequency dependence, applying 5-min pulses of 10−7 or 10−9 m GnRH at 0.5-, 1-, or 2- intervals (Fig. 3). As before, repeat stimulation resulted in recurrent ERK2-GFP translocation. ERK2-GFP responses displayed comparable kinetics and maximal responses with no evidence for desensitization at any pulse frequency or concentration. Thus, ERK2-GFP translocation shows digital tracking in response to pulsatile GnRH under these conditions; this is in sharp contrast to the integrative tracking seen with NFAT-EFP translocation at a 30-min pulse frequency (Ref. 25 and supplemental Fig. 1).
FIGURE 2.
ERK2 responses during pulsatile GnRH treatment. Cells were transduced with Ad-mGnRHR and Ad-ERK2-GFP after knockdown of endogenous ERK1/2 then serum-starved overnight. Panels A and B, cells were stained with Hoechst dye and treated with the indicated concentrations of GnRH at 0, 60, 120, and 180 min (arrows) for 5 min followed by repeated wash steps (gray rectangles). Live cell image acquisition and analysis was performed as described under “Experimental Procedures.” Panel C, cells were pretreated with 10−7 m GnRH (for 5 min, followed by wash) or with control (Ctrl) at hourly intervals for 3 h and then stimulated with a final 5-min pulse of GnRH. During the last pulse cells were fixed at the indicated time points, and immunocytochemical staining for ppERK1/2 was performed. Data shown are the N:C ratio of ERK2-GFP fluorescence intensity (background subtracted) or ppERK2 fluorescence intensity, normalized as the -fold change over control. Results shown are the mean ± S.E. of 3–4 independent experiments, performed in duplicate wells. For panel B, curve fitting reveals log EC50 values in the range of −10.2 to −9.8, and two-way ANOVA reveals that GnRH concentration is a significant source of variation (p < 0.01, F4,48 = 32.82), whereas pulse number is not (p = 0.44, F3,48 = 0.91).
FIGURE 3.
Live cell imaging with varied GnRH pulse frequency. Cells were subject to siRNA knockdown of ERK1/2 and transduced with Ad-NLS-BFP, Ad-mGnRHR, and Ad-ERK2-GFP before serum starvation overnight. Cells were treated (for 5 min) with 10−7 or 10−9 m GnRH at 30-min, 1 h, or every 2 h as indicated. As a control (Ctrl), all wells were subject to half-hourly washes (gray rectangles) 5 min after the GnRH addition. Live cell image acquisition and analysis was performed as described under “Experimental Procedures.” Data shown are the N:C ratio of ERK2-GFP fluorescence intensity (background subtracted), normalized as the -fold change over control (at 0 min) and offset by +1, 2, or 3 (for 0.5-, 1-h, and 2-h intervals, respectively). Results shown are the mean ± S.E. of three independent experiments.
ERK Responses to GnRH at the Single Cell Level
The data above were derived from average responses in large populations of cells, but the assays used also provide measures on each individual cell facilitating single cell analysis. The nature of ERK signaling is controversial with all-or-nothing responses seen in some models and graded responses in others (47–51). A mixed digital/analog response was seen for GnRH effects on ppERK1/2 levels in LβT2 cells, but it was not clear whether this is characteristic of the receptor or the cell type (50); to answer this we constructed frequency-distribution plots for GnRH effects on ERK2-GFP N:C ratio or whole cell ppERK2 levels. As shown (supplemental Fig. 2), GnRH reduced the number of non-responsive cells and increased the average response with either of these experimental end-points. Thus, for example, >99% of control cells had ppERK2 levels of <250 arbitrary fluorescence units, whereas >91% had >250 arbitrary fluorescence units after 5 min with 10−6 m GnRH. Using 250 arbitrary fluorescence units as a cut-off to define activated cells, we found that GnRH caused dose-dependent increases in the proportion of cells in which ERK2 was activated and also in the ppERK2 intensity within these activated cells. Thus, GnRH increases ERK phosphorylation in cell populations by increasing the proportion of cells in which ERK is activated and the extent of its activation in these cells (supplemental Fig. 2C). The frequency distribution analysis also revealed a wide range of responses to GnRH (i.e. N:C varying from <1 to >5 in GnRH-stimulated cells; with background subtracted data) raising the question of whether response magnitude is characteristic for a single cell. To test this we plotted the effects of GnRH on N:C ERK2-GFP ratio for individual cells receiving repeated pulses of GnRH, and this revealed a strong linear correlation between response amplitude in the first and second stimulus with GnRH (supplemental Fig. 2D). Thus, despite high cell to cell variability, there is considerable reproducibility over time within individual cells. Similar variability from cell to cell, but reproducibility from one stimulus to the next within an individual cell, has also been seen for GnRH effects on cytoplasmic [Ca2+] and when NFAT2-EFP was used to monitor GnRH signaling in HeLa cells and in gonadotroph cell lines (data not shown and Ref. 25).
Effects of Sustained and Pulsatile GnRH on Transcription Reporters
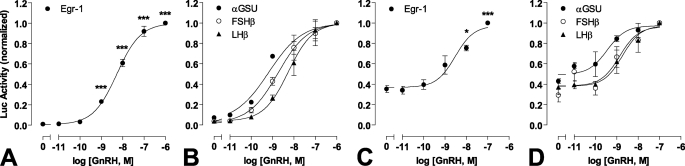
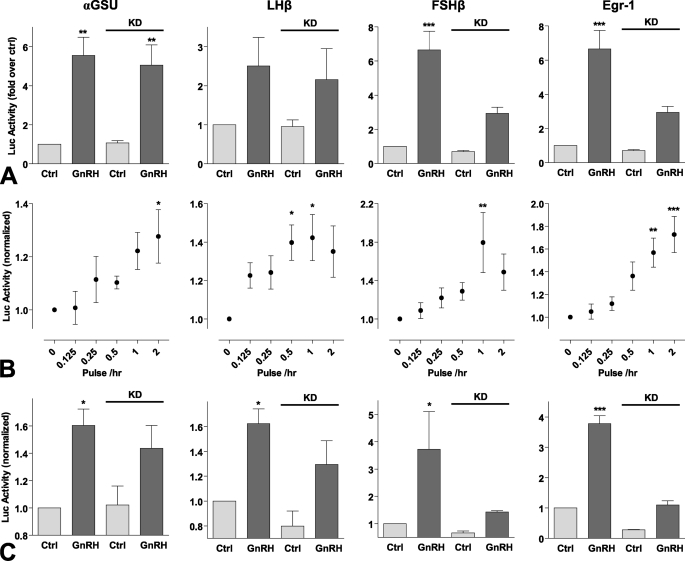
In many models ERK activation increases expression of the zinc finger-containing transcription factor Egr-1. GnRH rapidly increases Egr-1 expression, and this can mediate GnRH effects on gonadotropin β-subunit expression (1–3). Gonadotropin gene expression is differentially regulated by GnRH pulse frequency and amplitude (4), so we sought to compare transcriptional effects of GnRH using luciferase (luc) reporters based on promoter regions of αGSU, FSHβ, LHβ, and Egr-1. Continuous treatment with GnRH for 8 h caused a dose-dependent increase in Egr-1-luc activity with a pEC50 value of 8.3 ± 0.1 (Fig. 4A), which is comparable with the values previously reported for GnRH effects on αGSU-, FSHβ-, and LHβ-luc activity in this model (pEC50 values of 8.2–9.3, Fig. 4B, see also Ref. 25). GnRH also caused a dose-dependent increase in Egr-1 luc activity with pulsatile treatment (5-min pulses at hourly intervals for 8 h), and again the pEC50 value (8.5 ± 0.2, Fig. 4C) was comparable with values for pulsatile GnRH effects on αGSU-, FSHβ- and LHβ-luc activity (pEC50 values of 8.7- 9.4, Fig. 4D, see also Ref. 25). When GnRH pulse frequency was varied (5-min pulses with 10−9 m GnRH at 30 min to 4-h intervals for 8 h) this caused the expected bell-shaped frequency response curves (Fig. 5B; see also Ref. 25). GnRH effects on LHβ-luc and FSHβ-luc activity were maximal at 1–2-h intervals, but in contrast, maximal αGSU-luc and Egr-1-luc responses occurred at the highest pulse frequency (Fig. 5B). In these experiments we observed that basal luc activity was considerably higher in cells undergoing pulsatile treatment than continuous (∼40% of the maximum response compared with <10%, Fig. 4). This may well reflect mechanical stimulation during the repeated wash steps so this was controlled for in the experiments where pulse frequency was varied (Fig. 5A) by changing medium on all cells at 30-min intervals. We also tested for ERK dependence of these responses using siRNA to inhibit ERK1/2 expression. ERK knockdown markedly inhibited FSHβ-luc and Egr-1-luc transcriptional responses to sustained GnRH stimulation (Fig. 5A) and also inhibited these responses to pulsatile treatment (Fig. 5C), although it had no measurable effect on the LHβ-luc or αGSU-luc responses to GnRH with either sustained or pulsatile stimulation (Fig. 5, A and C).
FIGURE 4.
Dose response relationships of transcriptional reporters with sustained and pulsatile GnRH treatment. Cells were transfected with αGSU-Luc, LHβ-Luc, FSHβ-Luc, or Egr1-Luc plasmids and transduced with Ad-mGnRHR. Cells were treated with indicated concentration of GnRH for 8 h either continuously (panels A and B) or briefly for 5 min at hourly intervals (panels C and D). The data shown are luciferase activity in relative luminescence units (RLU) normalized to the maximum response. Results shown are the mean ± S.E. of at 3 or 4 independent experiments, performed in triplicate wells. Panels A and C, significant differences are indicated using one-way ANOVA and Bonferroni's post-hoc test, comparing untreated control versus agonist treated; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 5.
ERK dependence and frequency response relationships of transcriptional reporters with pulsatile GnRH treatment. Cells were transduced with Ad-αGSU-Luc, Ad-LHβ-Luc, Ad-FSHβ-Luc, or Ad-Egr1-Luc and Ad-mGnRHR. Where indicated (panels A and C) cells were subject to siRNA knockdown (KD) of ERK1/2 48 h before stimulation. Panel A, cells were treated with 10−7 m GnRH or control (Ctrl) for 8 h continuously. Panel B, cells were briefly treated (for 5 min followed by repeated wash steps) with 10−9 m GnRH at the indicated frequency (with half-hourly washes as a control) for 8 h. Panel C, cells were briefly treated with 10−7 m GnRH or control at hourly intervals. The data shown are luciferase activity (in RLU) normalized to the control. Results shown are the mean ± S.E. of 3–7 independent experiments, performed in triplicate wells. Panel A, two-way ANOVA reveals that GnRH treatment is a significant source of variation (p < 0.05) for all reporters, whereas siRNA knockdown (and interaction; p < 0.05) is a significant variable for both FSHβ-Luc and Egr1-Luc (both p < 0.01). Panel B, significant differences are indicated using one-way ANOVA and Bonferroni's post-hoc test, comparing untreated control versus agonist treated. Panel C, two-way ANOVA reveals that GnRH treatment is a significant source of variation (p < 0.05) for all reporters, whereas siRNA knockdown and the interaction is a significant variable for Egr1-Luc (both p < 0.001). Significant differences are indicated using Bonferroni's post-hoc test; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Receptor Number
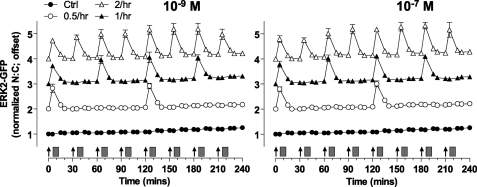
GnRH effects on gonadotropin subunit expression and cell proliferation are dependent on receptor number in pituitary derived cell lines and in other models (43, 52, 53). Accordingly, we tested the effects of GnRHR number on ERK2-GFP and NFAT-EFP responses. Varied Ad titers were used to control mGnRHR expression, and cells were transduced with Ad mGnRHR at 0.3, 1, or 3 pfu/nl, which corresponds to subphysiological (40,000/cell), physiological (80,000/cell), and overexpression (160,000/cell) of GnRH receptors, as determined previously (53, 54). As shown (Fig. 6), ERK2-GFP response magnitude was dependent upon GnRHR number with the greatest response at the highest titer. Again, repeat stimulation caused recurrent ERK2-GFP translocation, but no evidence was seen for response integration or for desensitization at any receptor number.
FIGURE 6.
Influence of GnRH receptor number on ERK2-GFP translocation responses to pulsatile stimulation. Cells were subject to siRNA knockdown of ERK1/2 and transduced with Ad-NLS-BFP, Ad-ERK2-GFP, and 0.3, 1, or 3 pfu/nl Ad-mGnRHR before serum starvation overnight. Cells were treated with the indicated concentrations of GnRH at 0, 60, 120, and 180 min for 5 min followed by repeated wash as indicated (gray rectangles). Live cell image acquisition and analysis was performed as described under “Experimental Procedures.” Data shown are the N:C ratio of ERK2-GFP fluorescence intensity (background subtracted), normalized as the -fold change over control (at 0 min). Results shown are the mean ± S.E. of three independent experiments, performed in duplicate wells.
GnRHR-mediated DUSP Expression in HeLa Cells
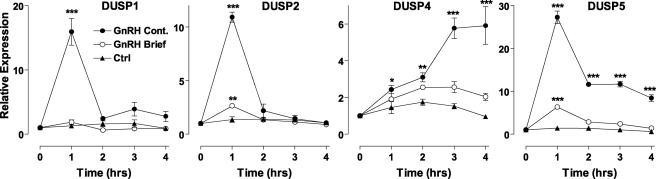
Dual-specificity phosphatases play a major role in shaping ERK responses in many systems (42, 55), and we have found that 5 DUSPs expressed in HeLa cells can influence effects of sustained (2 h) GnRH on ERK signaling in HeLa cells (37). Moreover, sustained exposure to GnRH increases expression of nuclear-inducible MKPs (DUSPs 1, 2, and 4 in HeLa cells and DUSPs 1 and 4 in gonadotroph cell lines (30, 37–39)), and negative feedback effects of DUSPs have been implicated in GnRH pulse-frequency decoding (15, 16). Accordingly, we tested for effects of pulsatile GnRH on expression of nuclear-inducible MKPs and also explored the possible effects of these proteins on ERK2-GFP translocation responses to pulsatile GnRH. In the first experiments we quantified mRNA expression after 5 min of stimulation or continuous exposure to 10−7 m GnRH. As shown (Fig. 7), sustained GnRH treatment caused a marked increase in expression of DUSP1, -2, and -5 mRNAs with maximal expression by 1 h and reduction thereafter. Sustained GnRH also increased expression of DUSP4 mRNA, although this response was slower with a maximal response at 4 h. Interestingly, brief GnRH treatment failed to increase transcription of DUSP1 mRNA and caused much more modest increases in DUSP2, -4, and -5 mRNAs (maximally 10–20% of the response to sustained stimulation).
FIGURE 7.
Nuclear-inducible DUSP mRNA expression after brief or sustained GnRH treatment. Cells were plated in 6-well plates, serum-starved overnight, then treated for 4 h with 10−7 m GnRH either continuously or with GnRH or control for 5 min. Total RNA was isolated and analyzed for relative levels of DUSP1, -2, -4, or -5 mRNA using quantitative PCR as described under “Experimental Procedures.” Data shown are normalized values (mean ± S.E.) obtained from three independent experiments, each with duplicate readings. Significant differences are indicated, comparing untreated control versus treated using one-way ANOVA and Bonferroni's post-hoc test; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Do Nuclear-inducible MKPs Shape ERK Responses to Pulsatile GnRH?
Because MKPs can influence ERK activation with sustained GnRH exposure (37), we tested whether they could also influence responses to pulsatile GnRH. To do so, we first used siRNA to knock-down endogenous ERKs and recombinant Ad to add back either WT ERK2-GFP or D319N ERK2-GFP. This mutation perturbs binding to D-domains, which are required for ERK binding to many partners, including MKPs (56). We have previously shown that this mutation prolongs GnRH effects on ERK signaling with a sustained stimulation protocol (25), but no such effect was seen with pulsatile GnRH stimulation (supplemental Fig. 3). Pulsatile stimulation with GnRH caused the expected rapid, transient, and reproducible translocation of ERK2-GFP from the cytoplasm to the nucleus, and the D319N mutation did not alter the kinetics of the responses that were maximal at 5 min and returned to basal within 20 min for each of 4 GnRH pulses in ERK2-GFP and D319N-ERK2-GFP expressing cells. Similar results were obtained with half-hour stimulation (not shown). Interestingly, the D319N mutation did cause a small reduction (∼20%) in the amplitude of the translocation response with all four GnRH pulses in this live cell imaging assay (supplemental Fig. 3), and a similar effect was seen when cells were fixed 5, 15, or 30 min after stimulation during pulse 1 or 4 (supplemental Fig. 3). However, the fixed cell assay enables ppERK2 to be stained, and this revealed no effect of the D319N mutation on whole cell ppERK2 levels (supplemental Fig. 3). We also tested the effect of this mutation using an Egr-1 luciferase reporter assay (supplemental Fig. 4), and this confirmed our earlier findings (37) that siRNA-mediated knockdown of ERKs 1 and 2 inhibits phorbol 12-,13-dibutyrate- or GnRH-stimulated Egr-1-luc activation, that the responses are rescued by Ad-mediated ERK2-GFP expression, and that the response is increased by D319N-ERK2-GFP expressed at comparable levels. Together, these data confirm that D-domain-dependent binding can have a pronounced effect on ERK signaling with sustained stimulation and does not influence the kinetics or amplitude of ERK activation with pulsatile stimulation, although it can have a modest effect on the proportion of ppERK2 in the nucleus in this model.
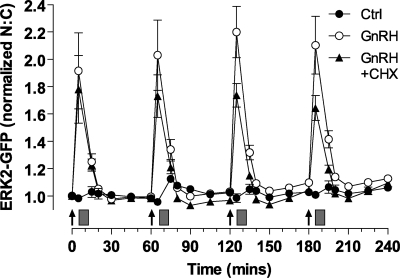
To test for a possible role of nuclear-inducible MKPs in shaping ERK responses, we used cycloheximide (CHX) to inhibit new protein synthesis. As shown (Fig. 8), the rapid and transient translocation of ERK2-GFP caused by GnRH (5 min, 10−7 m) was unaltered by CHX. There was also no measurable desensitization in the absence or presence of CHX (as judged by comparison of maximal responses in pulses 1–4). There was, nevertheless, a tendency for maximal responses to reduce over time with CHX so that the maximal responses in control and CHX treated cells were indistinguishable in pulse 1 but significantly different in pulse 4. Similar data were obtained with cells fixed 5, 15, or 30 min after stimulation during pulse 1 or 4 (supplemental Fig. 3). Interestingly, CHX reduced the GnRH effect on ERK2-GFP N:C in pulse 4 (p < 0.05) without influencing its effect on whole cell ppERK2. Thus, like the D319N mutation, CHX appears not to alter the kinetics or amplitude of ERK activation but may influence the proportion of ERK translocated to the nucleus, implying that protein synthesis influences this parameter.
FIGURE 8.
Effect of mutated D319N-ERK2 and CHX on ERK2-GFP translocation. Cells were subject to siRNA knockdown of ERK1/2 and transduced with Ad-mGnRHR, Ad-NLS-BFP, and Ad-ERK2-GFP. They were treated with 10−7 m GnRH at 0, 60, 120, and 180 min for 5 min followed by repeated washes (gray rectangles); where indicated the cells were pretreated with 30 μm CHX for 20 min, and CHX remained present during wash steps. Live cell image acquisition and analysis was performed as described under “Experimental Procedures.” Data shown are the N:C ratio of ERK2-GFP fluorescence intensity (background subtracted), normalized as the -fold change over control (Ctrl, at 0 min). Results shown are the mean ± S.E. of 3–6 independent experiments, performed in duplicate wells. The relevant controls (without GnRH treatment) in each experiment were pooled as no significant differences were observed. Statistical analysis is by repeated-measure ANOVA and Bonferroni's Multiple Comparison Test. CHX-treated peak responses at 125 and 185 min are significantly different from the corresponding peaks with GnRH treatment alone (p < 0.01).
Does ERK-mediated Feedback Influence ERK Responses to Pulsatile GnRH?
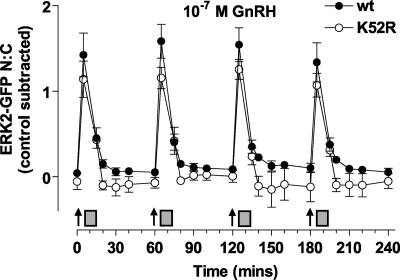
Positive or negative feedback loops can underlie frequency decoding, and ERK activity (i.e. ERK-dependent induction of MKP expression) could mediate such feedback. To test for this, we knocked down endogenous ERKs and added back either WT ERK2-GFP or K52R ERK2-GFP. This mutation prevents ERK catalytic activity, thereby negating any kinase-mediated feedback. This mutation increased the basal K52R-ERK2-GFP N:C ratio (by 0.6 ± 0.1, compared with cells expressing wild-type ERK2-GFP, p < 0.01) as expected because of the loss of negative feedback to MEK and Raf (supplemental Fig. 4 and Refs. 57 and 58). To simplify comparison we have, therefore, subtracted basal values and show the increment in N:C caused by GnRH in Fig. 9. GnRH pulses caused the expected rapid, transient, and reproducible translocation of ERK2-GFP, and the responses were indistinguishable (in terms of kinetics and amplitude) in cells expressing K52R ERK2-GFP. We also used the Egr-1 luciferase reporter assay to test for the effect of this mutation (supplemental Fig. 4), and this confirmed that when added back at a levels comparable with that for ERK2-GFP or D319N-ERK2-GFP, the K52R-ERK2-GFP construct was unable to stimulate Egr-1 luc activity. Accordingly, ERK-mediated feedback apparently influences basal ERK activity and localization but not responses to pulsatile GnRH in this model.
FIGURE 9.
Effect of K52R mutant on ERK2-GFP translocation after pulsatile GnRH treatment. Cells were subject to siRNA knockdown of ERK1/2 and transduced with Ad-mGnRHR, Ad-NLS-BFP, and Ad-ERK2-GFP (wt) or mutated K52R-ERK2-GFP as indicated. Cells were treated with 10−7 m GnRH at 0, 60, 120, and 180 min for 5 min followed by repeated wash as indicated (gray rectangles). Live cell image acquisition and analysis was performed as described under “Experimental Procedures.” Data shown is the N:C ratio of ERK2-GFP fluorescence intensity with the relevant time-matched control values subtracted. Results shown are the mean ± S.E. of three independent experiments, performed in duplicate wells.
DISCUSSION
GnRH is secreted from hypothalamic neurons to control synthesis and secretion of LH and FSH. It is released in brief pulses, with pulse frequency varying under different physiological conditions. GnRH effects on gonadotropin synthesis and secretion are dependent upon pulse frequency. Although this has obvious physiological and pharmacological relevance, the molecular mechanism of frequency decoding by gonadotrophs is unclear. Where increasing pulse frequency increases a response, this could reflect dependence on cumulative pulse duration or on pulse interval. The latter is characteristic of genuine frequency decoders (15) and is well illustrated by GnRH effects on LHβ and FSHβ expression that are maximal at intermediate (physiological) GnRH pulse frequency but reduce as frequency is further increased, generating characteristic bell-shaped frequency-response relationships. Such frequency-decoding behavior requires feedback mechanisms (15), but their nature remains unknown.
Here we have explored the possibility that GnRHR frequency decoding occurs within the Raf/MEK/ERK pathway using an ERK2-GFP reporter to monitor ERK activation in live cells. ERKs are mainly cytoplasmic in unstimulated cells but on stimulation they move to the nucleus to activate targets including transcription factors. However, cytoplasmic scaffolds are readily saturated so that when ERKs are overexpressed they are largely nuclear even without stimulation. Accordingly, we have developed an assay using siRNAs to knock down endogenous ERKs and Ad to add back ERK2-GFP at a physiological level. This recapitulates normal N:C distribution of ERKs and enables translocation to the nucleus to be used as a readout for ERK activation. With this assay, GnRH caused an increase in ERK2-GFP N:C ratio that was rapid (maximal at 5–15 min) and dose-dependent (EC50 ∼ 5 nm). The response was also transient (returning to 50% maximal within 30 min of sustained stimulation with 10−7 m GnRH) but was even more rapidly reversed (returned to base line within 20 min) when GnRH was removed after 5 min (Fig. 1). Thus, most of the translocation response seen with sustained GnRH required ongoing GnRHR activation (after the first 5 min), demonstrating the reversibility that is prerequisite for frequency decoding. Using sustained stimulation, we have previously shown that GnRH-stimulated ERK2-GFP translocation data are similar to those obtained by Western blotting or immunocytochemistry (37, 40, 42) and show here that the live cell ERK2-GFP responses also have similar kinetics to the fixed cell ppERK2 measures with brief (5 min) exposure to GnRH (Figs. 1 and 2). These data show that ERK2-GFP translocation can be used as a live cell readout for ERK activation with brief stimulation and also highlight the paucity of information on ERK inactivation; we know that GnRH causes a protein kinase C-dependent (but EGF receptor and Src-independent) activation of ERK1/2 in this model (17)3 but know nothing about the phosphatases causing the rapid inactivation of ERK when GnRH is removed (Figs. 1 and 2).
Using a published mathematical model, we predicted that pulsatile GnRH would cause desensitization of effector activation and that such desensitization would increase with pulse frequency and GnRHR dose (25, 41). However, when live cell imaging was used to monitor GnRH effects (5-min pulses at 60-min intervals) on ERK2-GFP in GnRHR-expressing HeLa cells, there was clear dose-dependent translocation without desensitization (i.e. response amplitude and pEC50 were comparable during the first and fourth pulse with GnRH) (Fig. 2). There was also no evidence for desensitization of ERK1/2 phosphorylation during the 1st and 4th pulse after hourly GnRH stimulation (Fig. 2, supplemental Fig. 2). Moreover, when frequency was varied to include 0.5-h pulses, the ERK2-GFP translocation responses did not desensitize (Fig. 3). We also explored the possible relationship between receptor number and ERK2-GFP translocation because our modeling predicts greater desensitization to pulsatile GnRH at higher receptor number (25, 41) and because the effects of GnRH on gonadotropin subunit promoter activity are dependent upon receptor number (43). When the Ad titer was varied to give low, high, or super-physiological GnRHR levels (53, 54), GnRH-stimulated ERK2-GFP translocation increased as the receptor number increased, but the responses to hourly GnRH pulses were qualitatively similar at all receptor densities. Importantly, no desensitization was observed (comparing pulses 1 and 4) with either dose (10−9 or 10−7 m) or at any receptor expression level (Fig. 6). We have previously shown that sustained GnRH treatment down-regulates cell surface GnRHR in this model, whereas brief exposure has no measurable effect. Accordingly, the lack of desensitization of pulsatile GnRH effects on ERK2-GFP translocation may reflect the lack of GnRHR down-regulation with intermittent stimulation.
The lack of desensitization seen with 5-min pulses of GnRH (Fig. 1) extends earlier work showing no hysteresis when whole cell ppERK1/2 levels were measured in LβT2 cells stimulated 10 min with 0.1 or 100 nm GnRH and then stimulated for 40 min with varied doses of GnRH (50). This group also used immunocytochemistry to quantify responses in individual cells, enabling frequency distribution analysis. Here, a key question is whether responses are switch-like or graded in individual cells because the dose dependences seen with cell population measures (i.e. Western blotting) could reflect either a dose-dependent increase in the proportion of cells responding or a dose-dependent increase in response within the activated cells. Early work on Xenopus oocytes suggested that ERK activation is switch-like (47), but analog, digital, and mixed responses have been reported in different systems (47–51). Ruf et al. (50) demonstrated that GnRH caused a mixed analogue/digital response and argued that the digital component of this could reflect activation of other signaling pathways effectively providing a gating mechanism for the graded response. To our knowledge this possibility has not been tested in other systems, so it was not clear whether this feature of GnRHR signaling is gonadotroph-specific. The data shown herein (i.e. Figs. 1–3) are population averages derived from thousands of cells (for each condition), but the automated segmentation also provides information on each individual cell so we also constructed frequency-distribution curves for cells treated with various concentrations of GnRH. This revealed that GnRH causes a dose-dependent increase in ppERK2 in cell populations by increasing the proportion of cells responding as well as the average response in the responsive cells. Thus, the mixed digital/analog signal seen with GnRH effects on ppERK1/2 in LβT2 cells is not restricted to gonadotrophs.
Sustained GnRH treatment can increase expression of nuclear-inducible MKPs (30, 37–39), and these protein phosphatases may mediate pulse frequency decoding (16), so we tested for effects of pulsatile and sustained GnRH on expression of DUSPs 1, 2, 4, and 5 (the nuclear-inducible MKPs). A brief pulse of GnRH did indeed increase DUSP2, -4, and -5 mRNA levels, but the effect was only 10–20% that seen with sustained stimulation (Fig. 7). We also tested for possible functional effects of these proteins using image-based assays. MKP phosphatase binding to ERK is dependent upon a common docking domain that binds D-domain-containing proteins, and this interaction can be prevented by introducing a D319N mutation into ERK (56). When we compared effects of pulsatile GnRH in cells transduced with either ERK2-GFP or D319N-ERK2-GFP, the mutation had no measurable effect on the kinetics of the ERK2-GFP translocation response or on whole cell ppERK2 responses (supplemental Fig. 3). As an alternative means of probing possible nuclear-inducible MKP involvement, we used CHX to prevent their synthesis, but again, this treatment had no measurable effect on response kinetics with pulsatile stimulation (Fig. 8 and supplemental Fig. 3). These data are in stark contrast to our previous work with sustained GnRH stimulation in which the D319N mutation and CHX both caused more prolonged responses to GnRH (37). Thus, it is likely that sustained GnRH stimulation causes a more pronounced increase in expression of nuclear-inducible MKPs that may enable them to influence ERK signaling with sustained, but not pulsatile GnRH. To address the related question of whether ERK-mediated feedback influences ERK responses to pulsatile GnRH, we used a catalytically inactive K52R ERK mutant. This mutant prevents all kinase-mediated feedback, and accordingly, we saw an increase in ERK nuclear localization, likely due to the loss of negative-feedback to Raf and MEK (57, 58). We assessed K52R ERK2-GFP responses to pulsatile GnRH, and when increased basal levels were compensated for, we saw no change in ERK response kinetics. The clear implication is that phosphatases other than the nuclear-inducible MKPs (i.e. pre-existing phosphatases) shape ERK responses to pulsatile GnRH and that MKP-mediated feedback does not underlie frequency decoding through the ERK pathway in this model. Indeed, ERK kinase-mediated auto-regulatory feedback (that could include ERK-regulated MKP induction) does not decode GnRH pulse frequency in this model.
In addition to cell imaging, we used luciferase reporters to compare GnRH effects on Egr-1-luc (used as a downstream readout for nuclear ERK signaling) with its effects on gonadotropin subunit transcription. As expected, we found bell-shaped frequency response relationships for GnRH effects on LHβ-luc and FSHβ-luc activities with maximal effects at 1–2-h pulse intervals. In contrast, the GnRH effect on Egr-1-luc simply increased as with pulse frequency and was, therefore, maximal at the highest frequency (30 min intervals). We also used siRNA to knock-down endogenous ERK1/2, and this revealed that GnRH effects on Egr-1-luc and FSHβ-luc are ERK-dependent with both sustained and pulsatile stimulation. Accordingly our data reveal that ERKs may mediate the effects of pulsatile GnRH on gonadotropin expression but that the Ras/Raf/MEK pathway appears not to function as a genuine frequency decoder in this model.
When considering pulse frequency-dependent cellular responses, the simplest scenario is one in which a train of brief stimuli elicits a series of corresponding responses in a process known as digital tracking (59). However, downstream responses characteristically have slower activation and inactivation kinetics than upstream signals, so responses may not have returned to the basal level before repeat stimulation. This can yield cumulative responses (59) in a process termed integrative tracking. In a recent study (25) we used the cellular localization of an NFAT-EFP as a downstream readout for Ca2+ mobilization (activation of the Ca2+/calmodulin/calineurin/NFAT cascade). Pulsatile stimulation with GnRH caused rapid translocation of this reporter from the cytoplasm to the nucleus and interestingly; GnRH effects on NFAT2-EFP N:C ratio were slower in onset and more slowly reversed than the ERK2-GFP translocation responses reported here. Consequently, at high pulse frequency there was insufficient time for return to basal conditions before repeat stimulation, and (at 30-min pulse frequency) marked cumulative or “saw-toothed” NFAT-EFP responses occurred. Thus, at 30-min pulse intervals we see digital tracking with an ERK2-GFP translocation reporter and integrative tracking with an NFAT-EFP translocation reporter (25), providing an illustration of how the relative activation and inactivation kinetics of effectors can lead to signal specificity with a common pulsatile input (supplemental Fig. 1).
Integrative tracking can amplify signaling and provide signal specificity (59) but cannot alone explain the bell-shaped frequency-response relationships seen in many systems. These require positive or negative feedback or feed-forward loops (15, 25). The data described here clearly argue against feedback loops shaping ERK signal from the cytoplasm to the nucleus with pulsatile GnRH, and a similar conclusion was reached for the Ca2+/calmodulin/calcineurin/NFAT signaling pathway. In both cases we found that these pathways could mediate transcriptional effects of pulsatile GnRH, but the adaptive changes suspected to underlie frequency decoding were not seen, raising the obvious possibility that other pathways mediate frequency decoding. These could include other MAPK pathways as GnRHR mediate activation of JNK and p38 in several models (1–3), although response kinetics with GnRH pulses are largely unknown. Alternatively, G-proteins other than Gq/11 could be involved as GnRHR-mediated G-protein activation is context-dependent. Thus, although GnRHR are thought to couple faithfully to Gq/11 in αT3–1 gonadotrophs, they may also activate Gs, Gi, or G11/12 in other cell types (1–3, 23). This possibility has not been explored in HeLa cells, but recent work suggests that intermittent GnRH exposure can cause pulsatile increases in cAMP and that this pathway mediates transcriptional effects of GnRH pulses in LβT2 cells (23).
This possibility of cell context-dependent GnRHR signaling raises an obvious caveat to the current work that it was performed with a HeLa cell model rather than in normal or immortalized gonadotrophs. However, we have found that genuine frequency decoding does occur in this model (i.e. the bell-shaped frequency-response relationships seen for pulsatile GnRH effects on LHβ-luc and FSHβ-luc in HeLa cells demonstrate that GnRH frequency decoding is not restricted to gonadotrophs) and also found that the effects of pulsatile GnRH on NFAT-EFP location are very similar in HeLa and LβT2 cells. The LβT2 cell line was derived by targeted expression of SV40 T antigens in murine gonadotrophs and has proved invaluable for work on GnRHR signaling. However, T-antigens interact with a wide range of target proteins including protein phosphatase 2A, which accounts for a large proportion of protein phosphatase activity in many cells and is markedly inhibited by the small T antigen (25). Suspecting that this might influence the kinetics of ERK inactivation, we elected not to use LβT2 cells for the experiments herein.
In summary, we have found that ERK2-GFP translocation to the nucleus provides a robust live-cell readout for ERK activation and that pulsatile GnRH causes reversible, dose- and frequency-dependent ERK2-GFP translocation, which effectively tracks GnRHR occupancy. Known feedback mechanisms that could underlie GnRH frequency decoding include down-regulation of cell surface GnRHR or intracellular inositol 1,4,5-trisphosphate receptors and induction of RGS-2 or MKPs (4, 15). However, any of these would be expected to influence GnRH effects on ERK phosphorylation and translocation. The fact that we have not seen desensitization of ERK responses under any of the experimental conditions argues against a role for ERK in GnRH frequency decoding despite the fact that the ERK pathway can act as a frequency decoder in other systems (155–157) and despite the fact that genuine frequency decoding does occur in our GnRHR-expressing HeLa cell model. We also found that effects of pulsatile GnRH on FSHβ-luc and Egr-1-luc reporters were ERK-dependent but that Egr-1 luc activity was directly proportional to cumulative pulse duration, consistent with a role for ERKs in mediating gonadotropin expression but not frequency decoding. A similar conclusion was reached for the Ca2+/calmodulin/calcineurin/NFAT pathway that may mediate the effects of pulsatile GnRH on LHβ, FSHβ, and αGSU expression but appeared not to act as a genuine frequency decoder (25). Alternative possibilities are that frequency decoding occurs within other signaling pathways and/or that frequency decoding occurs further downstream. The latter possibility is implicit in models where differential regulation of FSHβ and LHβ expression is attributed to the interplay of transcription factors and co-activators (21, 23, 24).
Supplementary Material
This work was supported by Wellcome Trust Grants 084588 and 078407 (to C. A. M.) and a Biotechnology and Biological Sciences Research Council, Swindon, United Kingdom Doctoral Training Grant Award to the University of Bristol Laboratories for Integrative Neuroscience and Endocrinology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
S. P. Armstrong, C. J. Caunt, and C. A. McArdle, unpublished observations.
- GnRH
- gonadotropin-releasing hormone
- GnRHR
- GnRH receptor
- mGnRHR
- mouse GnRHR
- LH
- luteinizing hormone
- FSH
- follicle-stimulating hormone
- GSU
- gonadotropin subunit
- ERK
- extracellular signal-regulated kinase
- ppERK
- dual phosphorylated ERK
- MAPK
- mitogen-activated protein kinase
- MKP
- MAPK phosphatase
- DUSP
- dual specificity phosphatase
- RGS
- regulator of G-protein-coupled receptor signaling
- MEK
- MAPK/ERK kinase
- NFAT
- nuclear factor of activated T-cells
- GFP
- (enhanced) green fluorescent protein
- EFP
- emerald fluorescent protein
- BFP
- blue fluorescent protein
- NLS
- nuclear localization sequence
- Ad
- adenovirus
- WT
- wild type
- FCS
- fetal calf serum
- DMEM
- Dulbecco's modified Eagle's medium
- N:C
- nuclear to cytoplasmic ratio
- CHX
- cycloheximide
- luc
- luciferase
- siRNA
- small interfering RNA
- pfu
- plaque-forming units
- ANOVA
- analysis of variance
- pEC50
- negative log of effective concentration producing 50% maximal response.
REFERENCES
- 1.Millar R. P., Lu Z. L., Pawson A. J., Flanagan C. A., Morgan K., Maudsley S. R. (2004) Endocr. Rev. 25, 235–275 [DOI] [PubMed] [Google Scholar]
- 2.Burger L. L., Haisenleder D. J., Dalkin A. C., Marshall J. C. (2004) J. Mol. Endocrinol. 33, 559–584 [DOI] [PubMed] [Google Scholar]
- 3.Naor Z. (2009) Front. Neuroendocrinol. 30, 10–29 [DOI] [PubMed] [Google Scholar]
- 4.Ferris H. A., Shupnik M. A. (2006) Biol. Reprod. 74, 993–998 [DOI] [PubMed] [Google Scholar]
- 5.Belchetz P. E., Plant T. M., Nakai Y., Keogh E. J., Knobil E. (1978) Science 202, 631–633 [DOI] [PubMed] [Google Scholar]
- 6.Schally A. V. (1999) Peptides 20, 1247–1262 [DOI] [PubMed] [Google Scholar]
- 7.Conn P. M., Crowley W. F., Jr. (1994) Annu. Rev. Med. 45, 391–405 [DOI] [PubMed] [Google Scholar]
- 8.Bédécarrats G. Y., Kaiser U. B. (2003) Endocrinology 144, 1802–1811 [DOI] [PubMed] [Google Scholar]
- 9.Dalkin A. C., Haisenleder D. J., Ortolano G. A., Ellis T. R., Marshall J. C. (1989) Endocrinology 125, 917–924 [DOI] [PubMed] [Google Scholar]
- 10.Haisenleder D. J., Dalkin A. C., Ortolano G. A., Marshall J. C., Shupnik M. A. (1991) Endocrinology 128, 509–517 [DOI] [PubMed] [Google Scholar]
- 11.Shupnik M. A. (1990) Mol. Endocrinol. 4, 1444–1450 [DOI] [PubMed] [Google Scholar]
- 12.Weiss J., Jameson J. L., Burrin J. M., Crowley W. F., Jr. (1990) Mol. Endocrinol. 4, 557–564 [DOI] [PubMed] [Google Scholar]
- 13.Yasin M., Dalkin A. C., Haisenleder D. J., Kerrigan J. R., Marshall J. C. (1995) Endocrinology 136, 1559–1564 [DOI] [PubMed] [Google Scholar]
- 14.Kaiser U. B., Jakubowiak A., Steinberger A., Chin W. W. (1993) Endocrinology 133, 931–934 [DOI] [PubMed] [Google Scholar]
- 15.Krakauer D. C., Page K. M., Sealfon S. (2002) J. Theor. Biol. 218, 457–470 [PubMed] [Google Scholar]
- 16.Lim S., Pnueli L., Tan J. H., Naor Z., Rajagopal G., Melamed P. (2009) PLoS. ONE 4, e7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caunt C. J., Finch A. R., Sedgley K. R., Oakley L., Luttrell L. M., McArdle C. A. (2006) J. Biol. Chem. 281, 2701–2710 [DOI] [PubMed] [Google Scholar]
- 18.Heding A., Vrecl M., Hanyaloglu A. C., Sellar R., Taylor P. L., Eidne K. A. (2000) Endocrinology 141, 299–306 [DOI] [PubMed] [Google Scholar]
- 19.Hislop J. N., Caunt C. J., Sedgley K. R., Kelly E., Mundell S., Green L. D., McArdle C. A. (2005) J. Mol. Endocrinol. 35, 177–189 [DOI] [PubMed] [Google Scholar]
- 20.Willars G. B., Heding A., Vrecl M., Sellar R., Blomenröhr M., Nahorski S. R., Eidne K. A. (1999) J. Biol. Chem. 274, 30146–30153 [DOI] [PubMed] [Google Scholar]
- 21.Lawson M. A., Tsutsumi R., Zhang H., Talukdar I., Butler B. K., Santos S. J., Mellon P. L., Webster N. J. G. (2007) Mol. Endocrinol. 21, 1175–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burger L. L., Haisenleder D. J., Aylor K. W., Marshall J. C. (2009) Biol. Reprod. 81, 1206–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsutsumi R., Webster N. J. (2009) Endocr. J. 56, 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciccone N. A., Xu S., Lacza C. T., Carroll R. S., Kaiser U. B. (2010) Mol. Cell. Biol. 30, 1028–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armstrong S. P., Caunt C. J., Fowkes R. C., Tsaneva-Atanasova K., McArdle C. A. (2009) J. Biol. Chem. 284, 35746–35757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caunt C. J., Finch A. R., Sedgley K. R., McArdle C. A. (2006) Trends Endocrinol. Metab. 17, 276–283 [DOI] [PubMed] [Google Scholar]
- 27.Caunt C. J., Finch A. R., Sedgley K. R., McArdle C. A. (2006) Trends Endocrinol. Metab. 17, 308–313 [DOI] [PubMed] [Google Scholar]
- 28.Mitchell R., Sim P. J., Leslie T., Johnson M. S., Thomson F. J. (1994) J. Endocrinol. 140, R15–R18 [DOI] [PubMed] [Google Scholar]
- 29.Sim P., Mitchell R., Thorfinn L. (1993) Biochem. Soc. Trans. 21, 357S. [DOI] [PubMed] [Google Scholar]
- 30.Roberson M. S., Misra-Press A., Laurance M. E., Stork P. J., Maurer R. A. (1995) Mol. Cell. Biol. 15, 3531–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burger L. L., Haisenleder D. J., Aylor K. W., Marshall J. C. (2008) Biol. Reprod. 79, 947–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haisenleder D. J., Cox M. E., Parsons S. J., Marshall J. C. (1998) Endocrinology 139, 3104–3111 [DOI] [PubMed] [Google Scholar]
- 33.Kanasaki H., Bedecarrats G. Y., Kam K. Y., Xu S., Kaiser U. B. (2005) Endocrinology 146, 5503–5513 [DOI] [PubMed] [Google Scholar]
- 34.Kupzig S., Walker S. A., Cullen P. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cullen P. J., Lockyer P. J. (2002) Nat. Rev. Mol. Cell Biol. 3, 339–348 [DOI] [PubMed] [Google Scholar]
- 36.Durham P. L., Russo A. F. (2000) Mol. Endocrinol. 14, 1570–1582 [DOI] [PubMed] [Google Scholar]
- 37.Armstrong S. P., Caunt C. J., McArdle C. A. (2009) Mol. Endocrinol. 23, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang T., Roberson M. S. (2006) J. Mol. Endocrinol. 36, 41–50 [DOI] [PubMed] [Google Scholar]
- 39.Zhang T., Mulvaney J. M., Roberson M. S. (2001) Mol. Cell. Endocrinol. 172, 79–89 [DOI] [PubMed] [Google Scholar]
- 40.Caunt C. J., Armstrong S. P., Rivers C. A., Norman M. R., McArdle C. A. (2008) J. Biol. Chem. 283, 26612–26623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Washington T. M., Blum J. J., Reed M. C., Conn P. M. (2004) Theor. Biol. Med. Model. 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caunt C. J., Rivers C. A., Conway-Campbell B. L., Norman M. R., McArdle C. A. (2008) J. Biol. Chem. 283, 6241–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaiser U. B., Sabbagh E., Katzenellenbogen R. A., Conn P. M., Chin W. W. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 12280–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holdstock J. G., Aylwin S. J., Burrin J. M. (1996) Mol. Endocrinol. 10, 1308–1317 [DOI] [PubMed] [Google Scholar]
- 45.Anderson R. D., Haskell R. E., Xia H., Roessler B. J., Davidson B. L. (2000) Gene Ther. 7, 1034–1038 [DOI] [PubMed] [Google Scholar]
- 46.Maru B. S., Tobias J. H., Rivers C., Caunt C. J., Norman M. R., McArdle C. A. (2009) Bone 44, 102–112 [DOI] [PubMed] [Google Scholar]
- 47.Ferrell J. E., Jr., Machleder E. M. (1998) Science 280, 895–898 [DOI] [PubMed] [Google Scholar]
- 48.Harding A., Tian T., Westbury E., Frische E., Hancock J. F. (2005) Curr. Biol. 15, 869–873 [DOI] [PubMed] [Google Scholar]
- 49.Mackeigan J. P., Murphy L. O., Dimitri C. A., Blenis J. (2005) Mol. Cell. Biol. 25, 4676–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruf F., Park M. J., Hayot F., Lin G., Roysam B., Ge Y., Sealfon S. C. (2006) J. Biol. Chem. 281, 30967–30978 [DOI] [PubMed] [Google Scholar]
- 51.Whitehurst A., Cobb M. H., White M. A. (2004) Mol. Cell. Biol. 24, 10145–10150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morgan K., Stewart A. J., Miller N., Mullen P., Muir M., Dodds M., Medda F., Harrison D., Langdon S., Millar R. P. (2008) Cancer Res. 68, 6331–6340 [DOI] [PubMed] [Google Scholar]
- 53.Finch A. R., Green L., Hislop J. N., Kelly E., McArdle C. A. (2004) J Clin. Endocrinol. Metab. 89, 1823–1832 [DOI] [PubMed] [Google Scholar]
- 54.Hislop J. N., Madziva M. T., Everest H. M., Harding T., Uney J. B., Willars G. B., Millar R. P., Troskie B. E., Davidson J. S., McArdle C. A. (2000) Endocrinology 141, 4564–4575 [DOI] [PubMed] [Google Scholar]
- 55.Patterson K. I., Brummer T., O'Brien P. M., Daly R. J. (2009) Biochem. J. 418, 475–489 [DOI] [PubMed] [Google Scholar]
- 56.Tanoue T., Adachi M., Moriguchi T., Nishida E. (2000) Nat. Cell Biol. 2, 110–116 [DOI] [PubMed] [Google Scholar]
- 57.Dougherty M. K., Müller J., Ritt D. A., Zhou M., Zhou X. Z., Copeland T. D., Conrads T. P., Veenstra T. D., Lu K. P., Morrison D. K. (2005) Mol. Cell 17, 215–224 [DOI] [PubMed] [Google Scholar]
- 58.Ramos J. W. (2008) Int. J. Biochem. Cell Biol. 40, 2707–2719 [DOI] [PubMed] [Google Scholar]
- 59.Berridge M. J. (2008) Cell Signalling Biology. Module 6, Spatial and Temporal Aspects of Signalling. Portland Press Ltd., London, www.biochemj.org/csb [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.