Abstract
In Pseudomonas putida, the expression of the pWW0 plasmid genes for the toluene/xylene assimilation pathway (the TOL pathway) is subject to complex regulation in response to environmental and physiological signals. This includes strong inhibition via catabolite repression, elicited by the carbon sources that the cells prefer to hydrocarbons. The Crc protein, a global regulator that controls carbon flow in pseudomonads, has an important role in this inhibition. Crc is a translational repressor that regulates the TOL genes, but how it does this has remained unknown. This study reports that Crc binds to sites located at the translation initiation regions of the mRNAs coding for XylR and XylS, two specific transcription activators of the TOL genes. Unexpectedly, eight additional Crc binding sites were found overlapping the translation initiation sites of genes coding for several enzymes of the pathway, all encoded within two polycistronic mRNAs. Evidence is provided supporting the idea that these sites are functional. This implies that Crc can differentially modulate the expression of particular genes within polycistronic mRNAs. It is proposed that Crc controls TOL genes in two ways. First, Crc inhibits the translation of the XylR and XylS regulators, thereby reducing the transcription of all TOL pathway genes. Second, Crc inhibits the translation of specific structural genes of the pathway, acting mainly on proteins involved in the first steps of toluene assimilation. This ensures a rapid inhibitory response that reduces the expression of the toluene/xylene degradation proteins when preferred carbon sources become available.
Keywords: Bacterial Metabolism, Gene Expression, Metabolic Regulation, RNA-binding Protein, Translation Regulation, Catabolite Repression, Global Regulation
Introduction
Pseudomonas putida is a ubiquitous soil bacterium that shows great metabolic versatility and the ability to live under a wide range of environmental conditions (1). Its biotechnological potential and its ability to degrade many aromatic compounds that generate pollution problems have led to the detailed study of its metabolism. A number of global regulation networks carefully control the numerous catabolic pathways of P. putida. Some of these regulatory systems orchestrate a carbon catabolite repression (CCR)3 process, which allows the sequential and hierarchical assimilation of different carbon sources when several are available at non-growth-limiting concentrations (for reviews, see Refs. 2–4). In Pseudomonas, catabolite repression is not well understood. Available evidence shows that in this bacterial group, certain organic acids and amino acids are the most strongly preferred compounds, whereas hydrocarbons are less preferred; the preference for glucose lies somewhere in between (5–7). For this reason, the potential of P. putida to degrade aromatic hydrocarbons is frequently compromised by CCR when in addition to the aromatic substrates, other more readily degradable compounds are present. Understanding CCR in P. putida may help to improve biodegradation rates and the usefulness of this bacterium in biotechnology and in bioremediation.
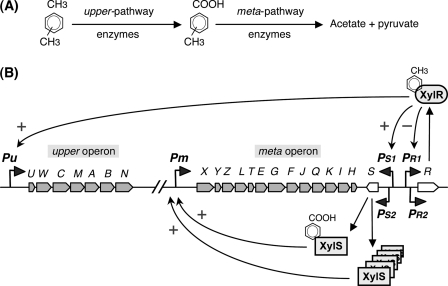
The CCR imposed on the assimilation of hydrocarbons has been studied with particular emphasis on the pathway for toluene and xylene assimilation specified by the genes carried by the P. putida pWW0 plasmid (8, 9). This pathway has become a model system to understand how recalcitrant aromatic compounds are biodegraded in nature. The genes coding for the enzymes that transform toluene into benzoate and xylenes into methylbenzoates are clustered in the “upper operon,” whereas those that convert benzoate or methylbenzoates into Krebs cycle intermediates are grouped in the “meta operon” (Fig. 1) (reviewed in Refs. 9 and 10). The xylR and xylS genes, located downstream of the meta operon, encode the specific transcriptional regulators that activate expression of the pathway genes. Benzoate or methylbenzoates, the substrates of the meta pathway enzymes, induce transcription of the meta operon from promoter Pm by acting as effectors of the XylS transcriptional activator. Toluene and xylenes induce transcription of the upper operon from promoter Pu by interacting with the XylR transcriptional activator. Toluene and xylenes also induce expression of the meta operon without the need of their conversion into benzoate or methylbenzoates (11, 12). This occurs because xylS can be transcribed from two promoters. Promoter PS2 provides for low level constitutive expression, allowing for XylS levels that can activate Pm only in the presence of effectors. However, in the presence of toluene or xylene, XylR activates the expression of xylS from another promoter, PS1, which considerably increases xylS transcription and generates an mRNA translated 10 times more efficiently than that originating at PS2. XylS is then synthesized at levels high enough to activate Pm even in the absence of effectors (13–15). The xylR gene, in turn, is expressed from two contiguous promoters, PR1 and PR2. The XylR protein negatively controls these promoters (16).
FIGURE 1.
Genetic map and transcriptional regulation of the TOL pathway. A, the enzymes encoded in the upper operon sequentially transform toluene or xylenes into benzoate or methylbenzoates, respectively. These are then transformed into acetate and pyruvate by the enzymes encoded in the meta operon. B, transcriptional regulation of the upper and meta operons. Structural genes (in gray), the regulatory genes xylS and xylR (in white), and the Pu, Pm, PS1, PS2, PR1, and PR2 promoters, are indicated. The activating (+) or repressing (−) effects of XylR and XylS are specified. Activation of Pu and PS1 by the XylR regulator requires the presence of an effector such as toluene, xylenes, or 3-methylbenzyl alcohol. To activate Pm, XylS must either bind to an aromatic effector (benzoate or methylbenzoates) or be expressed at high levels.
When in addition to the pathway substrates, the growth medium contains preferred compounds such as succinate or glucose or when a complete medium is used, the induction of the toluene/xylene assimilation (TOL) pathway is inhibited by a CCR phenomenon. Repression is primarily directed to reduce XylR activation of the Pu and PS1 promoters (17–24). Repression of the meta pathway genes also occurs, although this is milder. Two global regulation systems participate in this CCR effect. One involves the PTSNtr proteins, and the other one involves the Crc regulatory protein. The repression exerted on Pu activity by succinate or glucose depends mainly on the PTSNtr system (25). The repression induced by the components of a complete medium on the Pu and PS1 promoters depends on both PTSNtr and Crc (26). The PTSNtr system acts by interfering, directly or indirectly, with the ability of XylR to activate transcription from promoter Pu (26). However, it is unclear how Crc inhibits the induction of the TOL genes. The present work addresses this issue.
The Crc protein is a global regulator of carbon metabolism that inhibits the expression of genes involved in the assimilation of certain sugars (27), aromatic compounds (6), hydrocarbons (28), and amino acids (5, 7, 29) when preferred substrates are present. The final role of this regulator is to optimize metabolism, improving bacterial fitness (7). Crc is an RNA-binding protein that recognizes specific sites located at the translation initiation regions of target mRNAs, inhibiting translation (30–32). In Pseudomonas aeruginosa, the levels of free Crc in the cell are regulated by a small RNA named CrcZ, which binds to and sequesters Crc (33). CrcZ expression in turn varies depending on the medium composition.
Footprinting and mutagenesis techniques have shown that Crc specifically recognizes a short unpaired AA(C/U)AA(C/U)AA motif (32, 33). Several TOL pathway genes show sequences similar to the Crc motif close to their translation start site. The present work analyzes whether Crc can bind these sequences and examines the effect of this global regulator on the expression of the TOL pathway genes. The results show that Crc modulates the expression of these genes by directly interfering with the translation of the transcriptional regulators of the pathway and with that of some structural genes encoding proteins involved in the uptake and assimilation of aromatic substrates. This is a remarkable example of how the global regulation networks of a host cell can interfere with and control the expression of genes carried by a conjugative plasmid, the final aim being the improvement of the metabolic efficiency and fitness of the bacterium.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Media
P. putida KT2440 is derived from P. putida mt-2 by the curing of the pWW0 TOL plasmid (34). P. putida KT2442 is a rifampicin-resistant derivative of P. putida KT2440 (34). The pWW0 plasmid was introduced into strain KT2442 by conjugation using P. putida mt-2 as a donor and selecting for rifampicin-resistant colonies able to assimilate 3-methylbenzoate. The crc gene of P. putida KT2442(pWW0) was inactivated by allele exchange using the suicide donor plasmid pCRC10, as described previously (28). The resulting strain was named KT2442C(pWW0). Cells were grown at 30 °C in aerated flasks containing LB medium (35) and, where indicated, 5 mm 3-methylbenzyl alcohol (3MBA).
Total RNA Purification and Real-time Reverse Transcription-PCR
Strains KT2442(pWW0) and KT2442C(pWW0) were grown at 30 °C in aerated flasks in LB medium, in the absence or presence of 5 mm 3MBA. When cultures reached a turbidity (A600) of 0.6, cells (20 ml) were harvested by centrifugation, and the pellets were frozen at −70 °C. Three independent cultures were grown for each strain under each of the above conditions. RNA was purified from cell pellets using the RNeasy RNA purification kit (Qiagen) following the manufacturer's instructions. Purified RNA was treated with RNase-free DNase I (TURBO DNA-free, Ambion), as specified by the supplier. RNA integrity was analyzed by agarose gel electrophoresis. The absence of DNA was checked by real-time PCR using primers for rpoN.
RNA samples (10 μg) were transformed to cDNA using the High Capacity cDNA archive kit (Applied Biosystems). Real-time PCR was performed using SYBR Green PCR master mix (Applied Biosystems) and 0.2 mm of each primer in a 7300 real-time PCR system (Applied Biosystems), as described previously (36). The extension of PCR products was performed at 60 °C. Results were normalized relative to those obtained for the rpoN gene because its expression in P. putida remains constant throughout the growth phase and under the conditions used (22, 37). The oligonucleotides used for the amplification of each gene are indicated in supplemental Table S1.
Western Blotting
Immunodetection of the XylR protein was performed using a recombinant single-chain antibody expressed in an M13 phage display system (phage antibody), as described previously (38). This antibody was kindly provided by V. de Lorenzo (Centro Nacional de Biotecnología-CSIC, Madrid, Spain). P. putida strains KT2442(pWW0) and KT2442C(pWW0) were grown at 30 °C in aerated flasks in LB medium containing, where indicated, 5 mm 3MBA. To obtain whole-cell protein extracts, 10 ml of culture (turbidity of 0.6 at 600 nm) were centrifuged, and the cell pellets were resuspended in 180 μl of 60 mm Tris-HCl, pH 6.8. Cells were sonicated briefly, and the protein concentration was measured using the BCA protein assay kit (Pierce). The protein concentration of all samples was brought to 4 mg/ml. After the addition of SDS sample buffer (60 mm Tris-HCl, pH 6.8, 1% SDS, 5% glycerol, 0.01% bromphenol blue, 1% 2-mercaptoethanol, final concentration), samples were boiled for 10 min and centrifuged (14,000 × g, 10 min) to eliminate insoluble material. The proteins were separated by SDS-PAGE using the Mini-PROTEAN system (Bio-Rad) using the Precision Plus dual-color protein standards (Bio-Rad) as molecular weight markers. Proteins were transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore), which were then blocked for 2 h at room temperature using MBT buffer (1× phosphate-buffered saline, 5% powdered milk, 1% bovine serum albumin, 0.1% Tween 20). All membranes were incubated with 30 ml of MBT buffer containing 0.1% sodium deoxycholate and a 1:2000 dilution of the phage antibody solution containing 1014 plaque-forming units/ml. They were then washed as described previously (38), incubated with anti-M13-peroxidase conjugate (1:5,000 in MBT buffer), developed using the ECL plus Western blotting detection system (GE Healthcare), and placed in the proximity of x-ray film.
Protein Purification
The Crc protein containing a His6 tag at the C terminus, which is biologically active (30), was overproduced in Escherichia coli BL21(DE3)(pLysS) (39) containing plasmid pCRCH. The latter contains the crc(His6) gene under the control of a T7 promoter (30). Crc protein was purified using a nickel-nitrilotriacetic acid column, as described previously (30).
RNA Fragments and Oligonucleotides for Band-shift Assays
RNA fragments were obtained by in vitro run-off transcription reactions with T7 RNA polymerase, the template being appropriate DNA fragments obtained by PCR amplification performed over total DNA from KT2440(pWW0). The sequence of the amplified fragments was verified in all cases. The forward oligonucleotide contained a promoter sequence for T7 RNA polymerase followed by nucleotides complementary to the transcription start sites of the Pu, Pm, PR1, or PS1 promoters, respectively. The transcription start sites for these promoters have been described earlier (11, 14, 16). The reverse oligonucleotides were designed to hybridize 43 nt downstream of the AUG start site of the first gene of each transcript. The sequence of the oligonucleotides used is described in supplemental Table S1.
Transcription was performed for 1 h at 37 °C in 40 mm Tris-HCl (pH 7.9), 6 mm MgCl2,2 mm spermidine, 10 mm NaCl, 1 μg of DNA template, 10 mm dithiothreitol, 0.5 mm (each) ATP, CTP, GTP, and [α-32P]UTP (3,000 Ci/mmol), and 20 units of T7 RNA polymerase. Reactions were loaded onto a denaturing 6% urea-polyacrylamide gel, and the desired RNA fragments were excised from it and purified. RNA oligonucleotides (synthesized by Sigma) were labeled with T4 polynucleotide kinase and [γ-32P]ATP, as described previously (30), and purified through MicroSpin G-25 columns (GE Healthcare).
RNA Band-shift Assays
Reactions (20-μl volumes) involved 10 mm Hepes-KOH, pH 7.9, 35 mm KCl, 2 mm MgCl2, 0.1 nm radioactively labeled RNA, 1 μg of yeast tRNA and, where indicated, purified Crc(His6). After a 1-h incubation at room temperature, 4 μl of loading buffer (60% glycerol, 0.025% xylene cyanol) were added, and samples were loaded onto a non-denaturing 4% polyacrylamide gel containing TBM buffer (45 mm Tris-HCl, pH 8.3, 43 mm boric acid, 2 mm MgCl2, 5% glycerol). Electrophoresis was performed at 4 °C using TBM as the running buffer.
Enzyme Assays
To prepare crude cell extracts for enzyme reactions, cells were grown at 30 °C in aerated flasks in LB medium containing, where indicated, 5 mm 3MBA. At mid-log phase (turbidity 0.6), 100-ml culture samples were collected, and cells were harvested by centrifugation at 4 °C. Cells were washed twice with 100 mm phosphate buffer, pH 7.5, and disrupted by sonication in 7 ml of the same buffer containing 10% acetone. Cell debris was eliminated by centrifugation (30 min, 30,000 × g, 4 °C). The protein concentration of the extracts was determined using the BCA protein assay kit (Pierce).
The activities of benzyl alcohol dehydrogenase (BADH) and benzaldehyde dehydrogenase (BZDH) were measured essentially as described previously (40), determining the rate of NAD+ reduction following the increase in absorbance at 340 nm. The reaction mixtures (1 ml) consisted of 100 mm Tris-HCl, pH 8.8, 3.3 mm NAD+ (Sigma), 0.5 mm of substrate (benzyl alcohol for BADH and benzaldehyde for BZDH, added to start reactions) and cell extract. The extinction coefficient for NADH is 6,200 m−1cm−1. Catechol-2,3-dioxygenase (C23O) activity was determined as described previously (41) following the increase in absorbance at 375 nm due to the accumulation of 2-hydroxymuconic semialdehyde after the addition of catechol. The reaction mixtures (1 ml) contained 100 mm phosphate buffer, pH 7.5, 0.3 mm catechol (added to start the reaction) and cell extract. The extinction coefficient for 2-hydroxymuconic semialdehyde is 33,000 m−1 cm−1. One unit of enzyme activity is defined as the amount of enzyme required to convert 1 μmol of substrate to product in 1 min at 22 °C; specific activities are given as units/mg of protein.
RESULTS
Effect of Crc on the mRNA Levels of TOL Pathway Genes
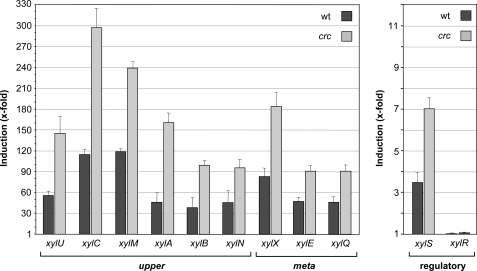
Previous work has shown that the effect of Crc is particularly strong when cells grow exponentially in a complete medium such as LB or in a defined medium containing yeast extract or casamino acids (26, 28). Therefore, to analyze the effect of Crc on the expression of the TOL pathway genes, P. putida KT2442 containing the pWW0 plasmid and its isogenic Crc-deficient derivative KT2442C(pWW0) were grown in LB medium in the absence or presence of 3MBA, an effector of the XylR transcriptional activator (42). At mid-exponential phase, cells were collected, total RNA was purified, and the mRNA levels of selected genes from the upper and meta operons and of the xylR and xylS genes were analyzed by real-time reverse transcription-PCR. As shown in Fig. 2, in the presence of 3MBA, the mRNA levels of the upper pathway genes xylU, xylC, xylM, xylA, xylB, and xylN, and of the meta pathway genes xylX, xylE, and xylQ were 2–3.7 times higher when the crc gene was inactivated. The mRNA levels of the xylS gene were also twice higher in the absence of Crc, an effect that was not observed for xylR.
FIGURE 2.
Effect of Crc on the mRNA levels of the TOL pathway genes. Strain KT2442(pWW0) and its isogenic crc-deficient derivative KT2442C(pWW0) were grown in LB medium in the absence or presence of 3MBA. At mid-exponential phase, cells were collected, and the mRNA levels of the indicated genes were determined by real-time reverse transcription-PCR. The bars show the mRNA levels observed in induced cultures (+3MBA) relative to non-induced cultures (−3MBA) for the wild type (wt) strain (dark bars) and for the crc-deficient derivative (light gray bars). The S.E. is indicated.
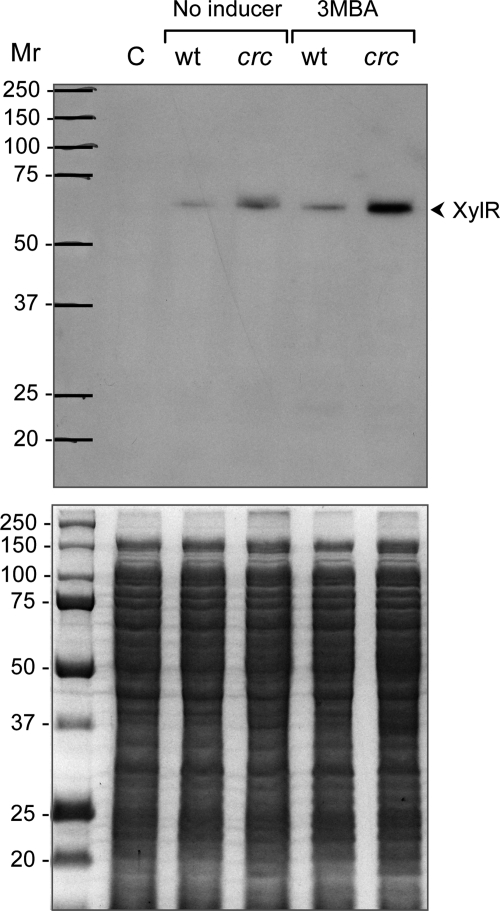
Because Crc reduces the mRNA levels of the XylR-activated genes xylU, xylC, xylM, xylA, xylB, xylN, and xylS, but not those of xylR, and given that Crc is a translational repressor, the results suggest that Crc represses the translation of xylR mRNA. This would reduce the levels of XylR protein available to activate the Pu and PS1 promoters, thus reducing the mRNA levels of the genes transcribed from these promoters. The use of a recombinant antibody directed against XylR (38) showed that the levels of XylR protein were significantly higher in the strain lacking the Crc regulator than in the wild type strain, particularly in the presence of 3MBA (Fig. 3).
FIGURE 3.
Influence of Crc on the levels of the XylR transcriptional activator. Strain KT2442(pWW0) and its isogenic crc-deficient derivative KT2442C(pWW0) were grown in LB medium in the absence or presence of 3MBA. At mid-exponential phase, cells were collected, and the total proteins were resolved by SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane, and the presence of the XylR protein was probed with a recombinant antibody. The lower panel shows the gel stained with Coomassie Blue, and the upper panel shows the Western blot. Equal amounts of proteins were loaded in all wells of the PAGE gel. Lane C corresponds to a protein extract of P. putida strain KT2442 lacking the pWW0 plasmid. The size of the proteins used as markers is indicated on the left (Mr). wt, wild type.
Crc Binds to the Leader Regions of the mRNAs Generated at the Pu, Pm, PS1, and PR1 Promoters
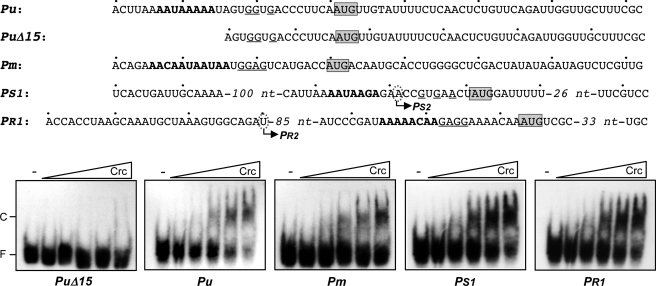
Crc specifically binds to a short unpaired AA(C/U)AA(C/U)AA motif located at, or close to, the translation initiation sites of the genes it regulates, inhibiting translation (32, 33). Similar sequences were found close to the AUG start codons of the first genes of the polycistronic mRNAs originating at the Pu and Pm promoters (the xylU and xylX genes, respectively), as well as at the translation initiation regions of the genes coding for the XylR and XylS regulators (Fig. 4, sequences indicated in boldface). In the case of xylS, the putative Crc binding site is located on the mRNA originating at the XylR-dependent promoter PS1 but is not present in the mRNA transcribed from the constitutive promoter PS2, which initiates immediately downstream of the presumed Crc target just 10 nt upstream from the AUG start site (Fig. 4). Purified Crc can bind in vitro short RNA fragments containing a proper target; the specificity of this binding has been demonstrated by competition and mutagenesis assays (30, 31, 32). Band-shift assays were therefore used to analyze the ability of purified Crc to bind radioactively labeled run-off RNA fragments corresponding to the 5′-end of the mRNAs originating at promoters Pu, Pm, PS1, and PR1. As a control, an RNA was prepared similar to that originating at Pu but lacking the first 15 nt, where the presumed Crc target lies (PuΔ15 RNA; Fig. 4). Crc was able to bind with similar affinity to the RNAs corresponding to promoters Pu, PS1, and PR1 and with a lower affinity to that corresponding to promoter Pm (Fig. 4). No binding was detected to the PuΔ15 RNA, even at the highest Crc concentrations used, supporting the idea that the A-rich sequence in the first 15 nt of Pu mRNA is necessary for Crc binding.
FIGURE 4.
Binding of Crc to RNA fragments including the leader regions of the mRNAs originating at the Pu, Pm, PS1, and PR1 promoters. The ability of Crc (106, 212, 425, 850, or 1,700 nm) to bind to the indicated radioactively labeled RNA fragments was determined by band-shift assays in the presence of 1 μg of tRNA. The free RNA and the retarded band corresponding to the Crc-RNA complex are indicated as F and C, respectively. The presumed Crc target on each RNA is indicated in boldface. The AUG start codons of xylU (Pu and PuΔ15 RNAs), xylX (Pm RNA), xylS (PS1 RNA), and xylR (PR1 RNA) are boxed. The putative Shine-Dalgarno sequences are underlined. The transcription initiation sites corresponding to the PS2 and PR2 promoters are indicated by circled letters.
Crc Binds to the Translation Initiation Region of Genes Located at Internal Positions of the Upper and Meta Polycistronic mRNAs
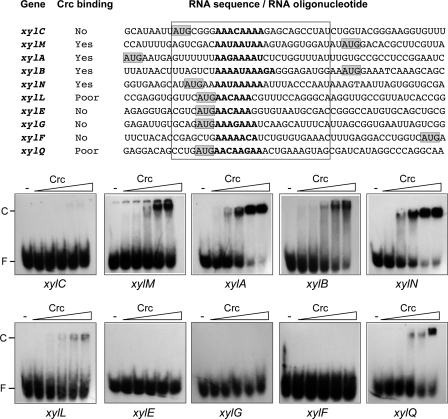
Sequences showing different degrees of similarity to the Crc consensus motif were visualized in the vicinity of the AUG start codons of the xylC, xylM, xylA, xylB, xylN, xylL, xylE, xylG, xylF, and xylQ genes. All these genes map to internal positions in the upper and meta polycistronic mRNAs rather than to their 5′-ends. The ability of Crc to recognize these presumed sites was analyzed through band-shift assays using end-labeled 26-nt RNA oligonucleotides, all of which contained the A-rich region at a similar distance from the RNA 5′-end (Fig. 5). Among the upper operon genes, Crc was able to bind the RNA oligonucleotides corresponding to xylM, xylA, xylB, and xylN, but not that of xylC (Fig. 5). Among the meta operon genes tested, Crc bound to the xylQ RNA oligonucleotide, although only at the highest Crc concentrations tested. Weak binding was also detected for the xylL RNA. No binding was observed for the RNAs corresponding to the xylE, xylG, and xylF genes (Fig. 5).
FIGURE 5.
Binding of Crc to RNA oligonucleotides containing the translation initiation regions of internal genes of the upper and meta polycistronic mRNAs. The RNA sequences shown correspond to the translation initiation regions of the indicated genes. The predicted Crc binding site within each gene is shown in boldface. The AUG start codon is shaded. The sequences boxed by a rectangle correspond to the 26-nt RNA oligonucleotides used to test Crc binding in band-shift assays. The ability of Crc (106, 212, 425, 850, or 1,700 nm) to bind to the radioactively labeled RNA oligonucleotides corresponding to the indicated genes was determined by band-shift assays in the presence of 1 μg of tRNA. The free RNA and the retarded band corresponding to the Crc-RNA complex are indicated as F and C, respectively.
Effect of Crc on the Activity of Key Enzymes of the TOL Pathway
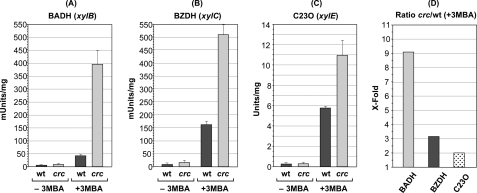
The presence of Crc targets at the translation initiation regions of several structural genes of the pathway suggests that this regulator controls expression of the TOL genes not only by reducing the levels of the XylR transcriptional regulator but also by impacting directly on the translation of selected structural genes. This suggests that the repressing effect of Crc may be stronger on genes having a target for this regulator than on genes lacking such a target. This idea was tested measuring the activity of the BADH encoded by xylB and of the BZDH encoded by xylC in cells growing exponentially in LB medium containing 3MBA. The xylB and xylC genes are both encoded in the upper operon mRNA and therefore depend on the XylR activator, but xylB contains a target for Crc at the translation initiation region, although xylC does not (Figs. 1 and 5). As a control, the expression of the C23O, which is encoded in the meta operon gene xylE but does not contain a target for Crc and depends on XylR indirectly, was measured.
The activity of BADH in the Crc-deficient cells growing exponentially in LB medium containing 3MBA was nine times that seen in the wild type strain. In contrast, BZDH activity was only three times higher in the absence of Crc, whereas that of C23O was twice higher (Fig. 6). It should be recalled that under the same conditions, Crc led to a 3–4 times lower level of the XylR transcriptional activator (Fig. 3), which in turn is reflected in the 2–2.5 times lower levels of the xylB, xylC, and xylE mRNAs (Fig. 2). Therefore, the nine times lower Crc-dependent level of BADH activity, as compared with the 2–3 times lower activity seen in BZDH and C23O activities, suggests that the Crc target present in the xylB (BADH) gene is functional and is responsible for strong Crc-dependent reduction in BADH activity.
FIGURE 6.
Effect of Crc on the activity of key enzymes of the TOL pathway. Strain KT2442(pWW0) and its isogenic crc-deficient derivative KT2442C(pWW0) were grown in LB medium in the presence or absence of 3MBA. At mid-exponential phase, cells were collected and disrupted by sonication. The specific activity of the enzymes BADH (A), BZDH (B), and C23O (C) was determined in the cell extracts as indicated under “Experimental Procedures.” D shows the repression exerted by Crc on the induction of BADH, BZDH, and C23O, expressed as the ratio of enzyme activity observed under induced conditions (+3MBA) for the crc-deficient strain versus that observed for the wild type (wt) strain. mUnits, milliunits; −3MBA, non-induced conditions. The S.E. is indicated.
DISCUSSION
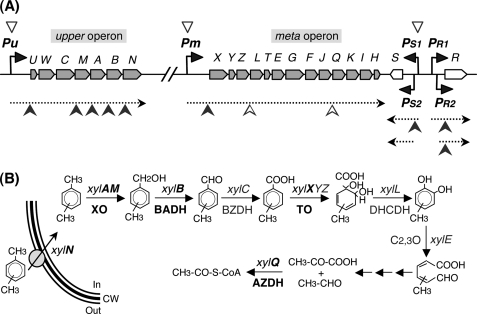
The present results indicate that Crc regulates the expression of the TOL genes by acting at two different levels. First, it inhibits the expression of the two transcriptional activators of the pathway, which is reflected in a 2–3 times lower level of the upper and meta polycistronic mRNAs. Second, Crc inhibits the translation of some key structural genes of the pathway. Most notably, Crc targets were found not only at the 5′-end of the regulated mRNAs, as observed for other Crc-regulated genes (31, 32, 33), but also at internal positions in the upper and meta polycistronic mRNAs. Fig. 7 provides a diagram showing the global effect of Crc on the TOL pathway genes.
FIGURE 7.
Crc-mediated regulation of the TOL pathway genes. A, the structural genes (in gray), the regulatory genes xylS and xylR (in white), and the Pu, Pm, PS1, PS2, PR1, and PR2 promoters of the TOL pathway are shown at the top of the figure. Black arrowheads indicate the direction of transcription. The mRNAs corresponding to the upper and meta operons and to the xylS and xylR genes are indicated by dotted arrows. The Crc binding sites at these mRNAs are indicated by filled arrowheads (strong sites) or open arrowheads (weak sites). The open triangles over promoters Pu, Pm, and PS1 indicate the indirect repressing effect of Crc on these promoters, derived from the direct effect of Crc on translation of the XylR activator. B, proteins involved in toluene uptake and assimilation. Proteins or genes directly regulated by Crc are shown in boldface, whereas those not directly affected by Crc are indicated in Roman text. XO, xylene monooxygenase; TO, toluate/benzoate dioxygenase; DHCDH, dihydroxycyclohexadiene carboxylate dehydrogenase; AZDH, acetaldehyde dehydrogenase; CW, cell wall.
It was earlier reported that Crc leads to the reduced transcription of the Pu and PS1 promoters, although no mechanistic explanation could be advanced at that time (26). The results show that the effect of Crc on the activity of the Pu and PS1 promoters is indirect, the direct target being XylR, which activates transcription from these two promoters. Although Crc had no measurable influence on the levels of xylR mRNA, it bound to a target that overlaps the xylR translation initiation region, leading to a clear reduction in the levels of the XylR protein. The Crc-dependent reduction of XylR levels was paralleled by a 2–3-fold decrease in the levels of the upper and meta pathway mRNAs. This can be explained if it is assumed that Crc reduces XylR levels below those needed for the full induction of the Pu promoter. In addition, transcription of the meta pathway genes depends on the action of the XylS transcriptional activator, which can be expressed from two promoters, PS1 and PS2. In the presence of an upper pathway inducer such as 3MBA, xylS is transcribed mainly from promoter PS1, a XylR-dependent promoter that leads to a transcript translated much more efficiently than that originating at PS2 (14). The transcript originating at PS1, but not that originating at PS2, contains a target for Crc. The present results indicate that Crc reduces xylS mRNA levels by half, which can be explained if it is assumed that the XylR levels in the presence of Crc are insufficient to attain the full induction of the PS1 promoter. In addition, the binding of Crc to the target present at the xylS mRNA originating at PS1 should reduce the translation of XylS. Therefore, it is here proposed that in the presence of an upper pathway inducer such as 3MBA, the effect of Crc on the mRNA levels of the genes that belong to the meta operon is due to the combined action of Crc on the translation of XylR (which determines xylS mRNA levels and which of the two xylS mRNAs will be more abundant) and on the translation of the xylS mRNA originating at promoter PS1.
This work provides evidence showing that the effect of Crc on the expression of the TOL genes is not limited to reducing the levels of the XylR and XylS regulatory proteins. The translation initiation regions of eight structural genes contained sequences similar to the previously characterized Crc targets. The ability of Crc to recognize these sequences was confirmed by band-shift assays with purified protein. Two of these Crc targets were located at the 5′-ends of the upper and meta transcripts, overlapping the translation initiation regions of the xylU and xylX genes, respectively. The other six Crc targets, however, mapped to internal positions within these polycistronic mRNAs, overlapping the translation initiation regions of the xylM, xylA, xylB, xylN, xylL, and xylQ genes. The functionality of these internal sites is supported by the observation that Crc reduced the activity of the BADH enzyme nine times, whereas the effect on the BZDH enzyme was only to reduce its activity by three times. These two enzymes are encoded by xylB and xylC, respectively, two genes located on the same polycistronic mRNA but that differ in that xylB contains a target for Crc, although xylC does not. The reduction of BZDH (xylC) activity can be explained by the Crc-dependent reduction of XylR levels, which in turn reduces Pu activity by 2–3 times. The higher reduction of BADH (xylB) levels probably derives from the additive effect of Crc on the translation of xylR and xylB. Post-transcriptional regulation of BADH has earlier been suggested in cells growing under phosphorous, sulfur, or nitrogen limitation but containing succinate in excess (23). The regulator responsible for this control was not identified at the time.
The presence of internal sites for Crc at polycistronic mRNAs, reported here for the first time, implies that this translational regulator can differentially modulate the expression of certain genes within polycistronic mRNAs, presumably optimizing the levels of each component of a given catabolic pathway. Some other translational regulators follow a similar strategy, selectively inhibiting the translation of internal genes carried by a polycistronic mRNA. This is the case of some E. coli ribosomal proteins (43, 44) and of several small antisense RNAs (45, 46).
It is worth noting that five of the seven genes of the upper operon contained a strong binding site for Crc, whereas among the 13 meta operon genes, only three contained a target for Crc, two of which were weak. This suggests a tighter regulation of the genes involved in the first steps of the pathway, i.e. those transforming toluene/xylenes into benzoate/methylbenzoates. The benzoate generated from toluene can be degraded through the pWW0-encoded meta pathway genes or through the chromosomally encoded benzoate ortho pathway because this “chromosomal” pathway is also induced in the presence of upper pathway substrates (15, 47). However, Crc also exerts a strong repressing effect on expression of the chromosomal ortho pathway for benzoate (6, 31).
One of the genes that showed a target for Crc was xylN, which encodes an outer membrane protein involved in xylene uptake (48). Although aromatic compounds can diffuse across bacterial membranes, particularly when present in high (millimolar) concentrations, high specificity transport systems increase the efficiency and rate of substrate uptake. These become particularly important when the aromatic compound in question is present in low (micromolar) concentrations, as commonly occurs in natural environments (49, 50). Reducing the levels of the XylN porin would therefore diminish the uptake and the internal concentration of the aromatic compounds that act as effectors of the XylR transcriptional regulator, thereby generating a multiplicative effect on the repression of the pathway. The overall effect of Crc reducing the uptake of the pathway substrates, the transcriptional activation of the pathway genes, and the translation of some of the pathway enzymes probably assures a very fast repressive response if cells metabolizing an aromatic substrate suddenly find that a preferred compound has become available. In fact, translational regulation is generally believed to provide a rapid response to changes in environmental or physiological conditions and is therefore an important step in the regulation of gene expression (51).
The regulatory processes that control the expression of the TOL pathway have been studied in depth. Earlier efforts were directed to elucidate the mechanisms responsible for the transcriptional regulation of the pathway genes. The results presented in this work show the presence of an additional and complex layer of regulation that occurs at the post-transcriptional level in response to the presence of a preferred carbon source and that depends on the Crc protein. Interestingly, the two layers of regulation are connected because Crc reduces the levels of the XylR and XylS transcriptional regulators and, presumably, the entry of the pathway substrates that act as effectors of these regulators. The final effect is a reduced, but not abolished, expression of the TOL pathway genes when preferred compounds are present. This emphasizes the complexity of the regulation of the TOL pathway and its finely tuned integration into cell physiology.
Supplementary Material
Acknowledgments
We are grateful to J. C. Oliveros for providing the bioinformatic tools for analyzing the DNA sequences, to V. de Lorenzo for providing the recombinant antibody against XylR, to J. L. Martínez for critical reading of the manuscript, and to L. Yuste for excellent technical assistance.
This work was supported by grants from the Spanish Ministry of Science and Innovation (Grants BFU2009-07009/BMC and CSD2007-00005) and from the Instituto de Salud Carlos III (Spanish Network for Research into Infectious Diseases; Grant REIPI RD06/0008), co-financed by the European Development Regional Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- CCR
- carbon catabolite repression
- 3MBA
- 3-methylbenzyl alcohol
- BADH
- benzyl alcohol dehydrogenase
- BZDH
- benzaldehyde dehydrogenase
- C23O
- catechol-2,3-dioxygenase
- TOL pathway
- toluene/xylene assimilation pathway
- nt
- nucleotide(s)
- PTSNtr
- nitrogen phosphotransferase system.
REFERENCES
- 1.Dos Santos V. A., Heim S., Moore E. R., Strätz M., Timmis K. N. (2004) Environ. Microbiol. 6, 1264–1286 [DOI] [PubMed] [Google Scholar]
- 2.Görke B., Stülke J. (2008) Nat. Rev. Microbiol. 6, 613–624 [DOI] [PubMed] [Google Scholar]
- 3.Deutscher J. (2008) Curr. Opin. Microbiol. 11, 87–93 [DOI] [PubMed] [Google Scholar]
- 4.Rojo F. (March10, 2010) FEMS Microbiol. Rev. DOI: 10.1111/j.1574–6976.2010.00218.x [DOI] [PubMed] [Google Scholar]
- 5.Hester K. L., Madhusudhan K. T., Sokatch J. R. (2000) J. Bacteriol. 182, 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morales G., Linares J. F., Beloso A., Albar J. P., Martínez J. L., Rojo F. (2004) J. Bacteriol. 186, 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreno R., Martínez-Gomariz M., Yuste L., Gil C., Rojo F. (2009) Proteomics 9, 2910–2928 [DOI] [PubMed] [Google Scholar]
- 8.Williams P. A., Murray K. (1974) J. Bacteriol. 120, 416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greated A., Lambertsen L., Williams P. A., Thomas C. M. (2002) Environ. Microbiol. 4, 856–871 [DOI] [PubMed] [Google Scholar]
- 10.Ramos J. L., Marqués S., Timmis K. N. (1997) Annu. Rev. Microbiol. 51, 341–373 [DOI] [PubMed] [Google Scholar]
- 11.Inouye S., Nakazawa A., Nakazawa T. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5182–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramos J. L., Mermod N., Timmis K. N. (1987) Mol. Microbiol. 1, 293–300 [DOI] [PubMed] [Google Scholar]
- 13.Gallegos M. T., Marqués S., Ramos J. L. (1996) J. Bacteriol. 178, 2356–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González-Pérez M. M., Ramos J. L., Marqués S. (2004) J. Bacteriol. 186, 1898–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domínguez-Cuevas P., González-Pastor J. E., Marqués S., Ramos J. L., de Lorenzo V. (2006) J. Biol. Chem. 281, 11981–11991 [DOI] [PubMed] [Google Scholar]
- 16.Inouye S., Nakazawa A., Nakazawa T. (1985) J. Bacteriol. 163, 863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugouvieux-Cotte-Pattat N., Köhler T., Rekik M., Harayama S. (1990) J. Bacteriol. 172, 6651–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo V., Cases I., Herrero M., Timmis K. N. (1993) J. Bacteriol. 175, 6902–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duetz W. A., Marqués S., de Jong C., Ramos J. L., van Andel J. G. (1994) J. Bacteriol. 176, 2354–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holtel A., Marqués S., Möhler I., Jakubzik U., Timmis K. N. (1994) J. Bacteriol. 176, 1773–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marqués S., Holtel A., Timmis K. N., Ramos J. L. (1994) J. Bacteriol. 176, 2517–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cases I., de Lorenzo V., Pérez-Martín J. (1996) Mol. Microbiol. 19, 7–17 [DOI] [PubMed] [Google Scholar]
- 23.Duetz W. A., Marqués S., Wind B., Ramos J. L., van Andel J. G. (1996) Appl. Environ. Microbiol. 62, 601–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duetz W. A., Wind B., Kamp M., van Andel J. G. (1997) Microbiology 143, 2331–2338 [DOI] [PubMed] [Google Scholar]
- 25.Aranda-Olmedo I., Marín P., Ramos J. L., Marqués S. (2006) Appl. Environ. Microbiol. 72, 7418–7421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aranda-Olmedo I., Ramos J. L., Marqués S. (2005) Appl. Environ. Microbiol. 71, 4191–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacGregor C. H., Wolff J. A., Arora S. K., Phibbs P. V., Jr. (1991) J. Bacteriol. 173, 7204–7212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuste L., Rojo F. (2001) J. Bacteriol. 183, 6197–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hester K. L., Lehman J., Najar F., Song L., Roe B. A., MacGregor C. H., Hager P. W., Phibbs P. V., Jr., Sokatch J. R. (2000) J. Bacteriol. 182, 1144–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno R., Ruiz-Manzano A., Yuste L., Rojo F. (2007) Mol. Microbiol. 64, 665–675 [DOI] [PubMed] [Google Scholar]
- 31.Moreno R., Rojo F. (2008) J. Bacteriol. 190, 1539–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno R., Marzi S., Romby P., Rojo F. (2009) Nucleic Acids Res. 37, 7678–7690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnleitner E., Abdou L., Haas D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21866–21871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 7458–7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Vol. 3, Appendix A2.2, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 36.Morales G., Ugidos A., Rojo F. (2006) Environ. Microbiol. 8, 1764–1774 [DOI] [PubMed] [Google Scholar]
- 37.Yuste L., Hervás A. B., Canosa I., Tobes R., Jiménez J. I., Nogales J., Pérez-Pérez M. M., Santero E., Díaz E., Ramos J. L., de Lorenzo V., Rojo F. (2006) Environ. Microbiol. 8, 165–177 [DOI] [PubMed] [Google Scholar]
- 38.Fraile S., Roncal F., Fernández L. A., de Lorenzo V. (2001) J. Bacteriol. 183, 5571–5579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. (1987) Gene 56, 125–135 [DOI] [PubMed] [Google Scholar]
- 40.Worsey M. J., Williams P. A. (1975) J. Bacteriol. 124, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sala-Trepat J. M., Evans W. C. (1971) Eur. J. Biochem. 20, 400–413 [DOI] [PubMed] [Google Scholar]
- 42.Abril M. A., Michan C., Timmis K. N., Ramos J. L. (1989) J. Bacteriol. 171, 6782–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dean D., Nomura M. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 3590–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerretti D. P., Mattheakis L. C., Kearney K. R., Vu L., Nomura M. (1988) J. Mol. Biol. 204, 309–329 [DOI] [PubMed] [Google Scholar]
- 45.Møller T., Franch T., Udesen C., Gerdes K., Valentin-Hansen P. (2002) Genes Dev. 16, 1696–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urban J. H., Vogel J. (2007) Nucleic Acids Res. 35, 1018–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowles C. E., Nichols N. N., Harwood C. S. (2000) J. Bacteriol. 182, 6339–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasai Y., Inoue J., Harayama S. (2001) J. Bacteriol. 183, 6662–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nichols N. N., Harwood C. S. (1997) J. Bacteriol. 179, 5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Díaz E., Ferrández A., Prieto M. A., García J. L. (2001) Microbiol. Mol. Biol. Rev. 65, 523–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romby P., Springer M. (2003) Trends Genet. 19, 155–161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.