Abstract
Understanding the molecular basis of natural ligand binding and activation of the glucagon-like peptide 1 (GLP1) receptor may facilitate the development of agonist drugs useful for the management of type 2 diabetes mellitus. We previously reported molecular approximations between carboxyl-terminal residues 24 and 35 within GLP1 and its receptor. In this work, we have focused on the amino-terminal region of GLP1, known to be critical for receptor activation. We developed two high-affinity, full agonist photolabile GLP1 probes having sites of covalent attachment in positions 6 and 12 of the 30-residue peptide (GLP1(7–36)). Both probes bound to the receptor specifically and covalently labeled single distinct sites. Chemical and protease cleavage of the labeled receptor identified the juxtamembrane region of its amino-terminal domain as the region of covalent attachment of the position 12 probe, whereas the region of labeling by the position 6 probe was localized to the first extracellular loop. Radiochemical sequencing identified receptor residue Tyr145, adjacent to the first transmembrane segment, as the site of labeling by the position 12 probe, and receptor residue Tyr205, within the first extracellular loop, as the site of labeling by the position 6 probe. These data provide support for a common mechanism for natural ligand binding and activation of family B G protein-coupled receptors. This region of interaction of peptide amino-terminal domains with the receptor may provide a pocket that can be targeted by small molecule agonists.
Keywords: G Protein-coupled Receptors (GPCR), Hormone Receptors, Peptide Hormones, Peptide Interactions, Peptides, Photoaffinity Labeling, Receptor Structure-function, Receptors
Introduction
Glucagon-like peptide 1 (GLP1),2 secreted by the intestinal L cells in response to food ingestion, is a glucoincretin hormone that possesses multiple physiological functions, including stimulation of glucose-dependent insulin secretion, inhibition of glucagon secretion and gastric emptying, and reduction in food intake, augmenting insulin biosynthesis, restoring β-cell sensitivity to glucose, increasing β-cell proliferation, and reducing apoptosis (1, 2). Because of these multiple antidiabetic actions and its ability to lower body weight, GLP1 receptor agonists have attracted much interest as a treatment for type 2 diabetes (3).
The GLP1 receptor is a member of the family B G protein-coupled receptors (GPCRs) that includes many important drug targets such as receptors for glucagon, glucagon-like peptide 2, gastric inhibitory polypeptide, secretin, vasoactive intestinal polypeptide, corticotropin-releasing factor, parathyroid hormone and calcitonin (4). A signature structural feature of this family is a long and structurally complex extracellular amino-terminal domain containing six conserved cysteine residues that form disulfide bonds that contribute to the development of a highly folded structure (5–9). This domain has been suggested to be the predominant domain for natural ligand binding and this is a consistent theme throughout the family (10–16). Natural ligands for family B receptors are all moderately long peptides in excess of 25 residues that have diffuse pharmacophoric domains, contributing to the complexity of their flexible interactions with the receptor amino-terminal domain. Like natural ligands for other members in the family B GPCRs, the amino-terminal portion of GLP1 is critical for the receptor selectivity and activation, whereas the carboxyl-terminal portion is critical for high affinity ligand binding (17). A tethering mechanism involving two domains of binding, with the carboxyl-terminal region of the ligand binding to the receptor amino terminus, and the amino-terminal region of the ligand possibly interacting with the receptor body, has been proposed for activation of this family of receptors (18–20).
Recently, our understanding of the molecular basis of ligand binding of the family B GPCRs has been substantially advanced with the solution of the NMR and crystal structures of the amino-terminal domains of several members (7, 9, 21–25). Although these structures suggest similarity in structural motifs and ligand binding modes, there are inconsistencies in the absolute site of ligand binding and in the positioning of the ligand in these structures (26), suggesting some variation in binding mechanisms among this family of the receptors. Of note, these structures also included that of the amino-terminal domain of the GLP1 receptor bound with an antagonist (truncated exendin-4) and the natural GLP1 ligands (24, 25). Due to the lack of the experimental data on the intact receptor structures, our current understanding of the natural ligand binding and activation of this family of GPCRs is very limited. So far, there is no consistent data for docking the amino-terminal region of the family B GPCR ligands and the orientation of the receptor amino-terminal domain relative to the core transmembrane domain (TM) is not clear.
We attempted to use the direct photoaffinity labeling approach to explore detailed spatial approximations between residues within GLP1 and those within the intact receptor. We have previously demonstrated that two carboxyl-terminal GLP1 probes incorporating a photolabile p-benzoyl-l-phenylalanine (Bpa) in positions 24 and 35 labeled receptor residues Glu133 and Glu125, respectively, both within the carboxyl-terminal region of the amino-terminal domain of the receptor (27). The amino-terminal region of family B GPCR ligands has been shown to be functionally important and to interact with the receptor body for several members (18–20). In this study, we focused on this region as possible sites of interaction with the body of the GLP1 receptor. Two photolabile probes were synthesized by incorporating a Bpa in positions 6 and 12 of the GLP1(7–36) peptide. Both probes bound specifically and saturably to the GLP1 receptor and were full agonists. They were able to covalently label single and distinct residues within the receptor. Although position 12 probe continued to label the carboxyl-terminal region of the amino-terminal domain of the GLP1 receptor, at residue Tyr145, position 6 probe indeed labeled a distinct region, at residue Tyr205 within the first extracellular loop of the receptor. These findings add important constraints to the docking of GLP1 to its receptor and provide additional insights into the molecular mechanism proposed for ligand binding and activation of family B GPCRs.
EXPERIMENTAL PROCEDURES
Materials
Human GLP1(7–36)-amide (GLP1) was purchased from Bachem (Torrance, CA). The solid-phase oxidant, N-chlorobenzenesulfonamide (IODO-BEAD), cyanogen bromide (CNBr), 2-(2-nitrophenylsulfenyl)-3-methyl-3-bromoindolenine (Skatole), and m-maleimidobenzoyl-N-hydroxysulfosuccinimide ester were purchased from Pierce Chemical Co. Phenylmethylsulfonyl fluoride, 3-isobutyl-1-methylxanthine, and N-(2-aminoethyl-1)-3-aminopropyl glass beads were from Sigma. ProSieve pre-stained protein standards were from Cambrex (Rochland, IN). Seeblue plus2 pre-stained and Multimark multicolored standards, and 10% NuPAGE gels were from Invitrogen. Endoproteinase Lys-C (Lys-C) was from Roche Applied Science. N-Glycosidase F (PNGase F) was from Proenzyme (San Leandro, CA). Soybean trypsin inhibitor (STI), Fetal Clone II, and tissue culture medium were from Invitrogen. Bovine serum albumin was from Serologicals (Norcross, GA). All other reagents were of analytical grade.
Synthetic Peptides
The photolabile GLP1 probe, [Bpa12, Arg26,34]GLP1(7–36) (Bpa12 probe), was designed to incorporate a photolabile p-benzoyl-l-phenylalanine (Bpa) to replace a phenylalanine in position 12 within the amino-terminal region of the ligand. Another photolabile probe, [Bpa6,Arg26,34]GLP1(7–36) (Bpa6 probe), was designed to incorporate a Bpa at the amino-terminal extension of the GLP1 ligand to minimize the negative functional impact on the critical residue, His7. Both probes contain a naturally occurring Tyr residue in position 19 as a site for radioiodination. Lysine residues in positions 26 and 34 were replaced by arginines to prevent Lys-C cleavage and this has previously been shown to be well tolerated (27, 28). They were synthesized by manual solid-phase techniques and purified by reversed-phase HPLC using procedures as previously described (29). Both probes and the natural GLP1 peptide were radioiodinated using a brief exposure for 15 s to the solid-phase oxidant, IODO-BEAD, as described previously (30). The radioiodinated peptides were purified by reversed-phase HPLC to yield specific radioactivities of ∼2,000 Ci/mmol.
Receptor Sources
A Chinese hamster ovary cell line stably expressing the wild-type human GLP1 receptor was utilized as a source of receptor (CHO-GLP1R) (31). It was cultured in Ham's F-12 medium supplemented with 5% Fetal Clone II on Falcon tissue culture plasticware in a 5% CO2 environment at 37 °C. Cells were passaged approximately twice a week and lifted mechanically before use.
A new GLP1 receptor mutant was generated to introduce an additional site for CNBr cleavage, replacing residue Phe143 with a methionine (F143M). Additionally, another new GLP1 receptor mutant representing alanine replacement of the site of labeling by the Bpa12 probe (Y145A) was prepared. Both mutants were prepared using the QuikChange Site-directed Mutagenesis kit from Stratagene (La Jolla, CA), with sequences verified by direct DNA sequencing. They were expressed transiently in COS-1 cells (American Type Culture Collection, Manassas, VA) after transfection using a modification of the DEAE-dextran method (32). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% Fetal Clone II in a humidified atmosphere containing 5% CO2 at 37 °C and harvested mechanically 72 h after transfection.
Plasma membranes were prepared from the above receptor-expressing cells using discontinuous sucrose gradient centrifugation (33). They were suspended in Krebs-Ringers/HEPES (KRH) medium (25 mm HEPES, pH 7.4, 104 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm KH2PO4, 1.2 mm MgSO4) containing 0.01% STI and 1 mm phenylmethylsulfonyl fluoride and stored at −80 °C until use.
Ligand Binding
The photolabile Bpa6 and Bpa12 probes were characterized to access their ability to bind receptor-bearing CHO-GLP1R cells using conditions that have been previously established (31). In brief, ∼200,000 CHO-GLP1R cells per well in 24-well plates were incubated with a constant amount of the radioligand, 125I-GLP1 (5–10 pm), in the presence of increasing concentrations of the nonradiolabeled Bpa6 or Bpa12 or control GLP1 (0 to 1 μm) in KRH medium containing 0.01% STI and 0.2% bovine serum albumin for 1 h at room temperature (reaction volume, 500 μl). After incubation, the cell-bound radioligand was separated from free radioligand by washing the cells twice with ice-cold KRH medium containing 0.1% STI and 0.2% bovine serum albumin. Cells were then lysed with 0.5 n NaOH and bound radioactivity was quantified using a γ-counter. Nonspecific binding was determined in the presence of 1 μm unlabeled GLP1 and represented less than 15% of total binding. The same assay was also utilized to characterize the binding activity of COS-1 cells transiently expressing the F143M and Y145A GLP1 receptor mutants. Data were analyzed and plotted using the nonlinear regression analysis routine for radioligand binding in the Prism program version 3.02 package (GraphPad Software, San Diego, CA) and are reported as the mean ± S.E. of duplicate determinations from a minimum of three independent experiments. Binding kinetics was determined by analysis with the LIGAND program of Munson and Rodbard (34).
Biological Activity Assay
The biological activities of the Bpa6 and Bpa12 probes to stimulate CHO-GLP1R cells were assessed by measuring cAMP responses in these cells. Approximately 8,000 cells per well were grown in 96-well plates for 48 h. On the day of assay, cells were washed twice with phosphate-buffered saline and stimulated for 30 min at 37 °C with increasing concentrations of the Bpa6 or Bpa12 probe or control GLP1 (0 to 1 μm) in KRH medium containing 0.01% STI, 0.2% bovine serum albumin, 0.1% bacitracin, and 1 mm 3-isobutyl-1-methylxanthine. Reactions were terminated by removing the medium and lysis in ice-cold 6% perchloric acid for 15 min with vigorous shaking. Lysates were adjusted to pH 6 with 30% KHCO3 and the cAMP levels were assayed in a 384-well white Optiplate using a LANCE kit from PerkinElmer per the manufacturer's instructions. The assay was performed in duplicate and repeated in at least three independent experiments. This assay was also used for characterization of the F143M and Y145A GLP1 receptor mutants expressed in COS-1 cells.
Photoaffinity Labeling
Covalent labeling of the GLP1 receptor was performed using procedures described previously (35). In brief, enriched receptor-bearing GLP1R-CHO membranes (∼50 μg) were incubated with ∼0.1 nm 125I-labeled Bpa6 or Bpa12 probe in 500 μl of KRH medium containing 0.01% STI and 1 mm phenylmethylsulfonyl fluoride in the presence of increasing amounts of competing GLP1 (0 to 1 μm) in the dark for 1 h at room temperature. The reaction was then exposed to photolysis for 30 min at 4 °C using a Rayonet photochemical reactor (Southern New England Ultraviolet Co., Bradford, CT) equipped with 3500-Å lamps. The membranes were then washed twice with ice-cold KRH medium and solubilized in SDS sample buffer before being applied to 10% SDS-polyacrylamide gels. Labeled products were visualized by autoradiography and band densitometry was performed by NIH ImageJ software. The apparent molecular masses of the radioactive bands were determined by interpolation on a plot of the mobility of the ProSieve protein markers versus the log values of their apparent masses.
Peptide Mapping
This required preparation of the affinity labeled GLP1 receptor in larger scale. For this, ∼200 μg and 0.5 nm 125I-labeled Bpa6 or Bpa12 probes were used. After gel electrophoresis, labeled bands were excised, eluted, lyophilized, and ethanol-precipitated before being used for chemical and enzymatic cleavage. Deglycosylation of labeled GLP1 receptor was performed with PNGase F following the protocol described in the manual. CNBr and Lys-C were used to cleave the labeled GLP1 receptor or mutant using procedures described previously (35). Products of cleavage were separated on 10% BisTris NuPAGE gel using MES running buffer and labeled bands were detected by autoradiography. The apparent molecular weights of the radiolabeled receptor fragments were determined by interpolation on a plot of the mobility of Seeblue plus2 pre-stained or Multimark multicolored standards versus the log values of their apparent masses.
Radiochemical Sequencing
This was used to identify the specific receptor residue that was covalently labeled by each of the photolabile GLP1 probes. For the Bpa12 probe, the receptor fragment Leu144–Met204 resulting from CNBr cleavage of the F143M mutant receptor was purified to radioactive homogeneity and covalently coupled through receptor residue Cys174 to m-maleimidobenzoyl-N-succinimide-activated N-(2-aminoethyl-1)-3-aminopropyl glass beads. Manual cycles of Edman degradation were repeated, as has been reported previously (36), and the radioactivity released in each cycle was quantified using a γ-spectrometer.
For the Bpa6 probe, due to the fact that the photolabile Bpa residue was located in the first position of this peptide, it was required to prevent cleavage of the Bpa in the first cycle of radiochemical Edman degradation sequencing of the attached receptor fragment. Therefore, acetylation of the free amino group of the photolabile probe was performed with acetic anhydride after photoaffinity labeling of the receptor, but prior to CNBr cleavage, using the procedure we have previously described (19). The radioactive pure Tyr205–Met233 fragment resulting from CNBr cleavage of the acetylated wild-type receptor labeled with the Bpa6 probe was coupled through Cys226 to the glass beads for radiochemical sequencing. As control, this receptor fragment attached with the acetylated Bpa6 probe was again acetylated with acetic anhydride and used for radiochemical sequencing.
Molecular Modeling
Illustrative molecular models were prepared by homology. Separate models of the GLP1 peptide-occupied amino terminus and the helical bundle domain were prepared and oriented relative to each other. The amino terminus of the GLP1 receptor (residues 32–132) was prepared based on the crystal structure of the exendin-4(9–39)-bound GLP receptor (Protein Data Bank code 3c5t) (24). The helical bundle (residues 145–433) was based on the structure of rhodopsin (Protein Data Bank code 2z73) (37).
RESULTS
Probe Characterization
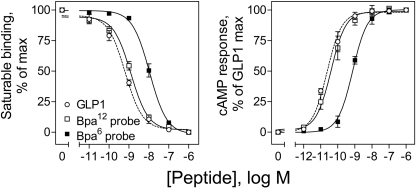
Both the Bpa6 and Bpa12 GLP1 probes were synthesized and purified to purity >99%, with their identities verified by matrix-assisted laser desorption/ionization-time of flight mass spectrometry. They bound to the GLP1 receptor specifically and saturably (GLP1, Ki = 0.7 ± 0.1 nm; Bpa6 probe, Ki = 11.2 ± 2.1 nm; Bpa12 probe, Ki = 1.2 ± 0.1 nm), and both were full agonists, stimulating maximal levels of cAMP in the CHO-GLP1R cells that were similar to that stimulated by natural GLP1, although they differed in potencies (GLP1, EC50 = 19 ± 1 pm; Bpa6 probe, EC50 = 738 ± 110 pm; Bpa12 probe, EC50 = 32 ± 7 pm). The binding affinity and potency for stimulating cAMP of the Bpa12 probe were similar to that of natural GLP1, whereas those for the Bap6 probe were lower (Fig. 1).
FIGURE 1.
Probe characterization. Left, competition-binding curves for increasing concentrations of GLP1, the Bpa6 or Bpa12 probes to displace the binding of the radioligand 125I-GLP1 to CHO-GLP1R membranes. Values represent percentages of the maximal saturable binding observed in the absence of competitor and expressed as mean ± S.E. of duplicate data from three independent experiments. Right, intracellular cAMP responses in CHO-GLP1R cells to increasing concentrations of GLP1 and the Bpa6 and Bpa12 probes. Values are expressed as mean ± S.E. of data from three independent experiments performed in duplicate, with data normalized relative to the maximal response to GLP1. Absolute basal (5.1 ± 1.3 pmol/million cells) and maximal (194 ± 38 pmol/million cells) cAMP levels were similar for all three peptides.
Photoaffinity Labeling of the GLP1 Receptor
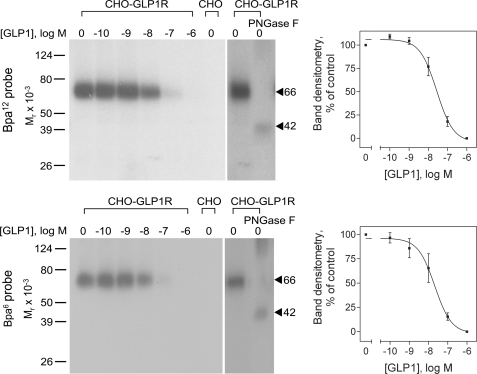
The Bpa6 and Bpa12 probes were used in photoaffinity labeling studies. As shown in Fig. 2, each of them labeled the GLP1 receptor specifically and saturably, with labeling being inhibited by GLP1 in a concentration-dependent manner (Bpa6 probe, IC50 = 24 ± 11 nm; Bpa12 probe, IC50 = 42 ± 9 nm). The receptor bands labeled with each of the probes migrated at molecular weight of approximately 66,000 and shifted to approximately 42,000 after deglycosylation with PNGase F, as expected for this receptor (27). No radioactive band was observed in affinity labeled membranes prepared from non-receptor-bearing CHO cells.
FIGURE 2.
Photoaffinity labeling of the GLP1 receptor. Left, representative autoradiographs of 10% SDS-PAGE gels used to separate the products of affinity labeling of CHO-GLP1R membranes with Bpa6 (bottom) or Bpa12 (top) probes in the presence of increasing concentrations of competing unlabeled GLP1 (from 0 to 1 μm). Shown also is the labeling of the non-receptor-bearing CHO cell membranes in the absence of competitor. The labeled GLP1 receptor with each probe migrated at molecular weight of approximately 66,000 that shifted to approximately 42,000 after deglycosylation by PNGase F as shown. Right, densitometric analysis of the competition for receptor labeling with Bpa6 (bottom) or Bpa12 (top) probes, performed in three independent experiments (mean ± S.E.).
Identification of the Bpa12 Probe-labeled Receptor Site
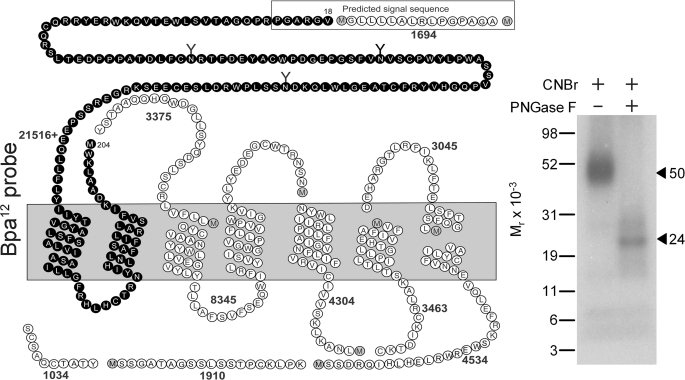
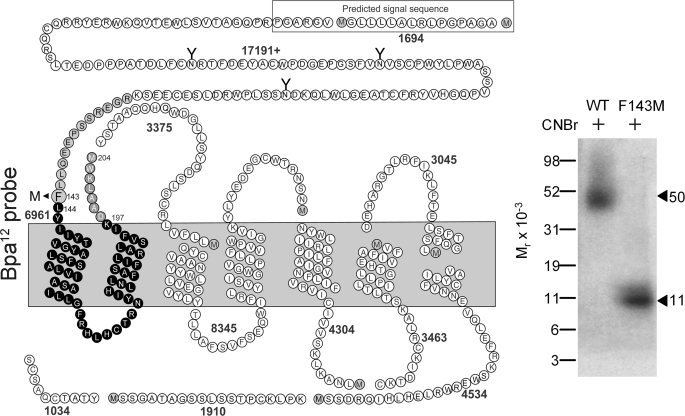
Because CNBr quantitatively cleaves proteins at the carboxyl-terminal side of methionine residues, it was chosen to gain initial insights into the regions of labeling by the Bpa12 probe. The GLP1 receptor contains 10 methionine residues and CNBr cleavage would theoretically result in 11 receptor fragments ranging in molecular masses from under 1 to more than 21 kDa, with only one of these containing sites of glycosylation (Fig. 3). Fig. 3 shows that CNBr cleavage of the labeled GLP1 receptor yielded a band that migrated at molecular weight of approximately 50,000 and shifted to approximately 24,000 after deglycosylation with PNGase F. Considering the molecular mass of the attached Bpa12 probe (3,583 Da) and the glycosylated nature of the labeled band, the labeled receptor fragment could be limited to only one candidate. This was the fragment including the extracellular amino-terminal domain, the first and second transmembrane domains, and the first intracellular loop (Val18–Met204 highlighted in black circles in Fig. 3).
FIGURE 3.
CNBr cleavage of the Bpa12 probe-labeled GLP1 receptor. Left, a diagram illustrating the theoretical fragments of the GLP1 receptor from CNBr cleavage. Right, a representative autoradiograph of a 10% NuPAGE gel used to separate the products of CNBr cleavage of the GLP1 receptor labeled with the Bpa12 probe. CNBr cleavage of the labeled GLP1 receptor yielded a fragment migrating at about Mr = 50,000 that shifted to approximately 24,000 after deglycosylation with PNGase F, indicating the site of labeling being within the large glycosylated Val18–Met204 fragment that includes the amino-terminal domain, TM1 and TM2, and the first intracellular loop of the GLP1 receptor (black circles). Data are representative of three independent experiments.
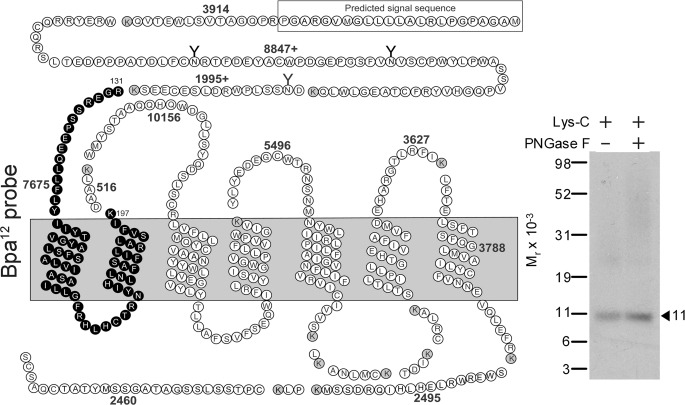
Endoproteinase Lys-C, which cleaves a protein at the carboxyl-terminal side of lysine residues, was used to further narrow down the domain of labeling by the Bpa12 probe. As shown in Fig. 4, Lys-C cleavage of the labeled GLP1 receptor yielded a band migrating at molecular weight of approximately 11,000 that did not shift further after deglycosylation. There was only one candidate to represent this, i.e. the Arg131–Lys197 fragment spanning the amino terminus, TM1, first intracellular loop, and TM2 (Fig. 4).
FIGURE 4.
Further localization of the domain of labeling with the Bpa12 probe by Lys-C cleavage. Left, a diagram illustrating the predicted sites from Lys-C cleavage of the wild-type GLP1 receptor. Right, Lys-C cleavage of the wild-type receptor yielded a band migrating at molecular weight of approximately 11,000 that did not shift further after deglycosylation with PNGase F. This indicated that the Bpa12 probe labeled the segment between Arg131 and Lys197 of the GLP1 receptor spanning the amino-terminal domain, TM1 and TM2 domains, and the first intracellular loop (bold circles). Data are representative of three independent experiments.
To further localize the site of labeling by the Bpa12 probe, a new receptor mutant, F143M, was prepared and transiently expressed in COS-1 cells. This mutant construct bound GLP1 (WT, Ki = 2.1 ± 0.5 nm; F143M, Ki = 1.4 ± 0.5 nm) and signaled similarly to the wild-type GLP1 receptor (EC50 = 32 ± 13 pm; EC50 = 47 ± 24 pm). It was efficiently and specifically labeled with the Bpa12 probe (data not shown). After CNBr cleavage, the Mr = 50,000 band from cleavage of the wild-type receptor shifted to Mr = 11,000 in the F143M mutant receptor, indicating that the site of labeling was within the segment between Leu144 and Met204 (Fig. 5). Considering the above identification by Lys-C cleavage, the site of labeling by the Bpa12 probe was between Leu144 and Lys197, within the juxtamembrane region of the amino-terminal domain of the GLP1 receptor.
FIGURE 5.
CNBr cleavage of the F143M GLP1 receptor mutant labeled with the Bpa12 probe. Left, a diagram illustrating the theoretical fragments resulting from CNBr cleavage of the F143M GLP1 receptor mutant, along with the masses of protein cores of these fragments. Right, CNBr cleavage of the labeled wild-type receptor resulted in a band migrating at molecular weight of approximately 50,000 and shifted to 11,000 in the F143M mutant receptor. This provides the definitive identification of the fragment between Leu144 and Met204 (black circles) as the domain of labeling by the Bpa12 probe. Data are representative of three independent experiments.
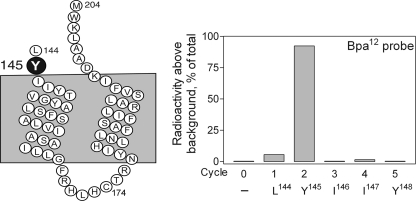
The specific site of labeling with the Bpa12 probe was identified by manual radioactive Edman degradation sequencing of the radiochemically pure CNBr fragment (Leu144–Met204) from the labeled F143M GLP1 receptor mutant. As shown in Fig. 6, a peak in eluted radioactivity appeared in cycle 2, corresponding to the covalent labeling of receptor residue Tyr145 in the juxtamembrane region of the amino-terminal domain of the GLP1 receptor.
FIGURE 6.
Identification of the receptor residue labeled with the Bpa12 probe. Left, a diagram of the CNBr fragment (Leu144–Met204) resulting from cleavage of the F143M receptor mutant. Right, representative radioactivity elution profile of Edman degradation sequencing of this fragment covalently attached with the Bpa12 probe. A peak eluted in radioactivity appeared in cycle 2, representing labeling of the receptor Tyr145 residue by the Bpa12 probe. Data are representative of three independent experiments.
Identification of the Bpa6 Probe-labeled Receptor Site
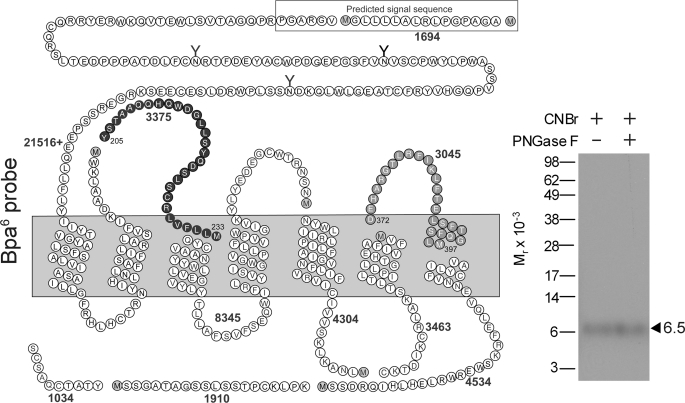
To gain an initial insight into the domain of labeling by the Bpa6 probe, we also used CNBr to cleave the labeled wild-type GLP1 receptor. As shown in Fig. 7, CNBr cleavage of the labeled GLP1 receptor yielded a band migrating at an approximate Mr = 6,000 that did not further shift after PNGase F treatment. Considering the molecular mass of the attached Bpa6 probe (3,729 Da) and the non-glycosylated nature of the labeled fragment, two fragments were felt to be candidates. These represented the Tyr205–Met233 fragment containing the ECL1 and the Asp372–Met397 fragment containing the ECL3 of the GLP1 receptor.
FIGURE 7.
CNBr cleavage of the Bpa6 probe-labeled GLP1 receptor. Left, a diagram illustrating the theoretical sites of CNBr cleavage of the GLP1 receptor. Right, CNBr cleavage of the labeled GLP1 receptor yielded a fragment migrating at molecular weight of approximately 6,500 that did not shift further after deglycosylation with PNGase F. The best candidates representing this are the fragments including the first (Tyr205–Met233, highlighted in black circles) and third (Asp372–Met397, highlighted in gray circles) extracellular loops. Data are representative of at least three independent experiments.
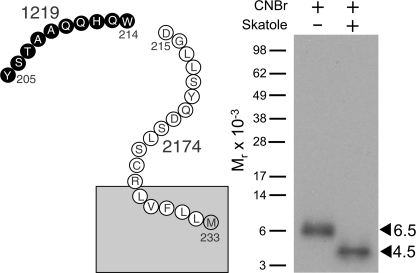
Further efforts were made to identify which of the above two candidate fragments contained the site of labeling for the Bpa6 probe. Interestingly, the Tyr205–Met233 fragment had a Trp residue at position 214, whereas the Asp372–Met397 fragment contained no Trp residues. Therefore, Skatole, which cleaves a protein at the carboxyl-terminal end of tryptophan residues, was used to further cleave the CNBr fragment labeled with the Bpa6 probe. As shown in Fig. 8, the Mr = 6,500 CNBr fragment shifted to Mr = 4,500 after Skatole cleavage. This indicated that the fragment including the ECL1 contained the site of labeling for the Bpa6 probe, likely within the Tyr205–Trp214 segment based on its migration. This was confirmed by CNBr cleavage of another M397L mutant receptor in which Met397 was mutated to a Leu (data not shown).
FIGURE 8.
Sequential CNBr and Skatole cleavage of the Bpa6 probe-labeled GLP1 receptor. Left, a diagram illustrating the predicted site resulting from Skatole cleavage of the candidate Tyr205–Met233 fragment of the wild-time receptor. Right, the non-glycosylated Mr = 6,500 band resulting from CNBr cleavage of the labeled wild-type GLP1 receptor shifted to Mr = 4,500 after further Skatole cleavage. This is most consistent with labeling of the ECL1 with the Bpa6 probe, representing the receptor region between residues Tyr205 and Met233. Based on the migration, the site of labeling with the Bpa6 probe was likely within the segment between Tyr205 and Trp214.
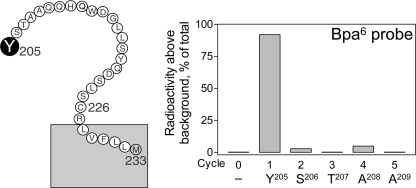
The identification of the specific site of labeling by the Bpa6 probe was achieved by sequencing the radiochemically pure Tyr205–Met233 fragment from CNBr cleavage of the acetylated wild-type GLP1 receptor. As shown in Fig. 9, a peak in eluted radioactivity appeared in the first cycle of the Edman degradation sequencing, representing covalent labeling of the receptor Tyr205 residue with the Bpa6 probe within the ECL1 of the GLP1 receptor. In the control experiment, no radioactive peak was observed when radiochemical sequencing of the same receptor fragment acetylated with acetic anhydride (data not shown).
FIGURE 9.
Identification of the receptor residue labeled with the Bpa6 probe. Left, a diagram illustrating the sequence of the CNBr fragment from the wild-type GLP1 receptor used for radiochemical sequencing. Right, a representative radioactivity elution profile of Edman degradation sequencing of the fragment resulting from CNBr cleavage of the wild-type receptor (Tyr205–Met233). A peak in radioactivity occurred in elution cycle 1, representing labeling of the receptor Tyr205 residue by the Bpa6 probe.
Functional Characterization of Receptor Site Mutants
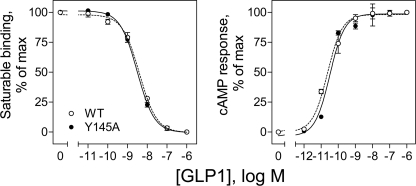
The function of GLP1 receptors incorporating an alanine replacement for each site of photoaffinity labeling was studied. The Tyr145 residue of the GLP1 receptor labeled by the Bpa12 probe was mutated to an alanine, expressed transiently in COS-1 cells, and studied for impact on the binding and biological activity of GLP1. As shown in Fig. 10, mutation of this residue had no significant effects on the binding or biological effects of GLP1. Similarly, alanine replacement of the Tyr205 residue labeled by the Bpa6 probe did not result in significant changes in GLP1 binding affinity or biological activity (38).
FIGURE 10.
Characterization of the Y145A GLP1 receptor mutant. Shown in the left panel are competition-binding curves of increasing concentrations of GLP1 to displace the binding of radioligand 125I-GLP1 to COS-1 cells expressing wild-type (WT) and Y145A GLP1 receptors. Values illustrated represent saturable binding as percentages of maximal binding observed in the absence of the competing peptide. They are expressed as the mean ± S.E. of duplicate values from a minimum of three independent experiments. Shown in the right panel are intracellular cAMP responses to increasing concentrations of GLP1 in these cells. Data points represent the mean ± S.E. of three independent experiments performed in duplicate, normalized relative to the maximal response to GLP1.
Molecular Modeling
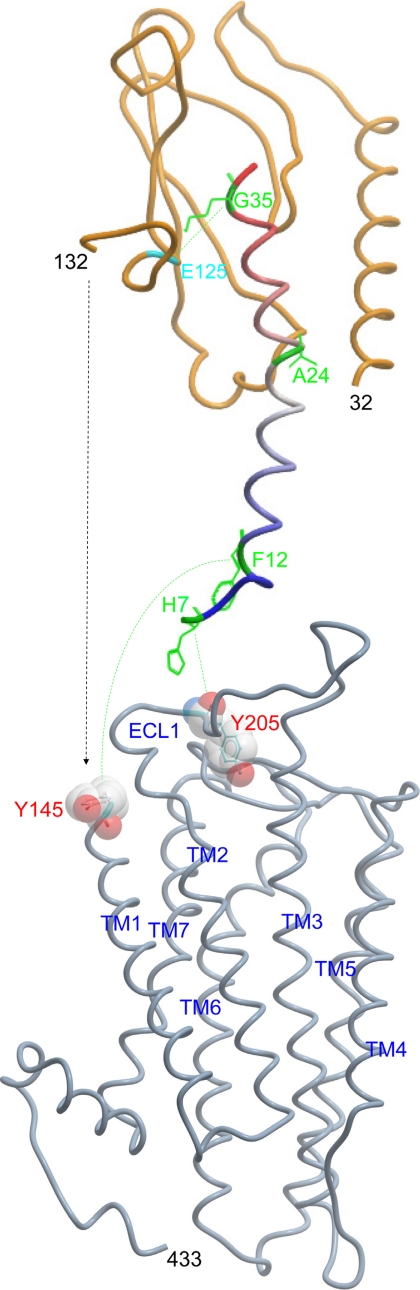
Fig. 11 illustrates the general feasibility of docking GLP1 to its receptor components, because meaningful molecular modeling of an intact receptor will require additional spatial constraints to orient the receptor amino terminus relative to its core domain and to direct the interactions between what likely represents a flexible amino terminus of the peptide and the core domain of the receptor.
FIGURE 11.
Graphic illustration of the feasibility of docking GLP1 with the two major regions of the GLP1 receptor, the amino terminus and the helical bundle core domain. Shown are homology models of docking GLP1 with the amino terminus (orange) and the helical bundle core domain (gray) of the GLP1 receptor. It should be noted that the region between residues 133 and 144 linking the amino terminus with the top of transmembrane segment was not included in this illustration because its structure was not well defined in the published crystal structures. The GLP1 peptide is colored blue to red from its amino terminus to its carboxyl terminus. Residues within GLP1 representing positions of photolabile moieties for affinity labeling (His7, Phe12, Ala24, and Gly35) are shown in green. The receptor residue, Glu125, labeled by the probe at position 35 was colored in cyan, whereas the receptor residue labeled by the probe at position 24, Glu133, is not illustrated, because it is within an unconstrained region of the receptor. The two receptor residues photoaffinity labeled by the probes at positions 12 and 6 in the current study, Tyr145 and Tyr205, respectively, are highlighted by Corey-Pauling-Koltun representation. The green-dotted lines link the sites of photoactivation with sites of covalent labeling.
DISCUSSION
Family B GPCRs are an important family of potential drug targets that regulate a wide range of endocrine and neuroendocrine functions. Our current understanding of the molecular basis of ligand binding and activation of the GLP1 receptor and other members in this family remains a major obstacle for the further development of potent receptor ligands of pharmaceutical interests. Although with recent advancement in the solution of high-resolution structures of the predominant binding domain, the amino-terminal domain, of multiple members of this family, the mechanism of activation of this group of receptors is not yet clear due to the lack of structural information of the intact receptors. In this work, we used photoaffinity labeling to explore residue-residue approximations between the functionally critical amino-terminal region of GLP1 and its intact receptor as this region has previously been shown to interact with the body of several other family members (18–20, 39).
The design of photolabile GLP1 probes to incorporate a Bpa in the amino-terminal region is challenging because this region has been shown to be critical for its function in structure-activity studies (28, 40). Replacement of His7 by alanine resulted in an 11-fold decrease in binding affinity and essentially abolished the biological activity (28). We, therefore, chose to incorporate a Bpa at the amino-terminal extension of the GLP1(7–36), in position 6. Indeed, this Bpa6 probe bound the GLP1 receptor specifically and saturably, although with affinity somewhat lower than that of natural GLP1. More importantly, this probe was a fully efficacious agonist with potency much higher than [Ala7]GLP1 (28). It is noteworthy that incorporation of the bulky, hydrophobic Bpa moiety at position 12 to replace a conserved phenylalanine was better tolerated than Ala replacement (28, 40), yielding a peptide probe that bound and signaled similarly to natural GLP1. This is important because it assures retaining the determinants of binding and activation intrinsic to the natural hormone.
The current report provided direct spatial approximation between residue 12 at the amino-terminal region of GLP1 and receptor Tyr145 residue, at the carboxyl-terminal end of the amino terminus, just above the top of the first transmembrane domain (TM1). Of note, this residue was in the same region as the receptor residues Glu125 and Glu133 that were labeled by carboxyl-terminal Bpa35 and Bpa24 GLP1 probes, respectively (27). This Tyr145 residue is adjacent to a functionally important residue, Thr149 of the GLP1 receptor, in which a T149M mutation was found in a type 2 diabetes patient that exhibited impairment of insulin secretion, insulin sensitivity, and glucose tolerance (41). This mutant has been shown to exhibit reduced binding affinity and biological activity efficacy for GLP1 in COS-7 cells (42). The site of labeling of the Bpa12 GLP1 probe within its receptor was within the analogous region of the calcitonin receptor where a mid-region position 16 probe labeled (43) and of the PTH1 receptor where a carboxyl-terminal position 33 (44) and multiple mid-region (positions 11, 13, 15, 18, and 21) PTH probes labeled (45, 46). This is also the analogous region of the VPAC1 receptor labeled by amino-terminal positions 0 and 6, and carboxyl-terminal positions 22, 24, and 28 vasoactive intestinal polypeptide probes (47–49). Interestingly, this juxtamembranous region of the amino terminus of the calcitonin receptor has been recently shown to play a critical role in small-molecule agonist action (50).
The current report also provides direct spatial approximation between the amino terminus of GLP1 and receptor residue Tyr205, a residue within the ECL1 of the GLP1 receptor. Mutagenesis has identified multiple important residues within this region for GLP1 binding that included most of the charged residues including Lys197, Asp198, Lys202, Asp215, and Arg227 (51, 52). Interestingly, double alanine scanning of this region revealed that mutation of Met204–Tyr205 to alanines resulted in a 90-fold decrease in GLP1 binding and essentially abolished the biological activity, likely due to a loss in hydrophobicity in this region (38). In that work, the authors predicted the Met204–Tyr205 residues were important for binding to GLP1 residues in the amino-terminal region. Our current work unambiguously established the spatial approximation between this Tyr205 residue and the amino terminus of the GLP1 peptide. The importance of the ECL1 in ligand binding has also been demonstrated for several other family B GPCRs by mutagenesis (32, 53, 54). Photoaffinity labeling has established this region as the domain of labeling of the PTH1 receptor for mid-region position 19 and carboxyl-terminal position 27 PTH probes (55, 56), and as that of the corticotropin-releasing factor 1 receptor for mid-region positions 17 and 22 urocortin probes (39).
Of note, alanine replacement of each of the residues labeled by the Bpa6 and Bpa12 probes (Tyr205 and Tyr145, respectively) had no impact on the binding and biological activity of the natural ligand, GLP1. This phenomenon is actually the most common in this type of study. Photoaffinity labeling as applied in this work is used to identify spatial approximation between residues within a docked peptide ligand and its receptor, rather than defining interacting residues. These distances can be used as constraints for molecular modeling, due to the direct nature of the insight, but often do not represent functionally important interactions. Indeed, if residue-residue interactions were very important to function, the replacement of the natural residue with a photolabile residue would likely not be well tolerated and might have a more profound negative impact on binding or biological activity than what was observed here.
Our current identification of the spatial approximation between the amino terminus of GLP1 and the Tyr205 residue within the ECL1 of its receptor could provide a mechanism for activation of this receptor. Generally speaking, this is consistent with the common two-domain tethering mechanism for agonist ligand binding proposed for the secretin, PTH, and calcitonin receptors (18–20). However, it should be noted that this proposed mechanism is considered a low-resolution model because multiple ligand-receptor contacts have been demonstrated within each of the two broadly defined interactions and these contacts differ from one receptor to another. For the secretin receptor, multiple residues within the mid-region and carboxyl-terminal region have been demonstrated to dock within the amino-terminal domain and so far, only amino-terminal residues 1 and 5 have been shown to interact with the top of TM6 and ECL3 (19, 57). This is the analogous region of PTH1 (18, 58) and PTH2 (59) receptors labeled by the amino-terminal PTH probes. Recently, the amino terminus of PTH has also been shown to additionally interact with a residue within TM5 of its receptor using a disulfide trapping approach (60). In addition, mid-region position 19 and carboxyl-terminal position 27 residues of PTH have also been shown to interact with the body of its receptor, with distinct residues in TM2 and ECL1, respectively (55, 56). For the corticotropin-releasing factor 1 receptor, amino-terminal positions 0 and 12 of urocortin have been shown to interact with ECL2, with midregion positions 17 and 22 of the ligand interacting with ECL1 (39). The VPAC1 receptor may use a different mechanism because as described above, both amino- and carboxyl-terminal residues of vasoactive intestinal polypeptide have been shown to interact with the juxtamembranous region of the receptor (47–49). For the GLP1 receptor in this work, the amino terminus of GLP1 was demonstrated to interact with ECL1 of its receptor, a distinct region from those of other family B GPCRs. It should be interesting to test a broader spectrum of residues in different regions of GLP1 for possible interactions with other parts of the receptor body that will ultimately help to orient the receptor amino terminus, the predominant ligand binding domain, relative to the receptor core domain.
Unfortunately, the existing crystal structures of GLP1 or the GLP analogue bound to portions of the amino terminus of the GLP1 receptor (24, 25) do not include the regions identified in the current report. These structures were most informative of the interactions between the carboxyl-terminal region of GLP1 with the receptor. The current report extends these insights to the docking of the amino terminus of the peptide. Although we established spatial approximations for two positions within this region of GLP1, we believe that these constraints are not yet adequate to propose a meaningful intact receptor model. Of note, an intact GLP1 receptor model has recently been proposed for tentative docking of both peptidyl and small molecule ligands based on very limited, indirect mutagenesis data (61); however, it does not accommodate the experimentally derived spatial approximation constraints determined in the current work. In that model (61), positions 6 and 12 of GLP1 were quite distant from their labeling sites, Tyr205 and Tyr145, 12.33 and 16.79 Å, respectively. This further indicates the need of a large amount of experimental spatial residue-residue constraints to build a credible model for flexible GLP1 docking to its receptor.
To date, we have identified four pairs of spatial approximation constraints between residues within the amino- and carboxyl-terminal regions of GLP1 and its receptor. Although these constraints provide important insights into our understanding of the molecular basis of ligand binding and activation of this important receptor and other family B GPCRs, currently, there are not adequate data to build a meaningful ligand-bound intact GLP1 receptor model.
Acknowledgments
We thank Mary L. Augustine and Alicja M. Skalecka Ball for technical assistance. We also thank Dr. Yuedong Yang, who used the SPARKS-X server at Indiana University Purdue University, Indianapolis, to prepare an illustrative molecular model.
This work was supported, in whole or in part, by National Institutes of Health Grant DK46577 and American Diabetes Association Grant 1-08-RA-42.
- GLP1
- glucagon-like peptide 1
- GPCR
- G protein-coupled receptor
- Bpa
- p-benzoyl-l-phenylalanine
- CHO
- Chinese hamster ovary
- CHO-GLP1R
- human GLP1 receptor-bearing CHO cells
- CNBr
- cyanogen bromide
- KRH
- Krebs-Ringers-HEPES
- Lys-C
- endoproteinase Lys-C
- PNGase F
- N-glycosidase F
- PTH
- parathyroid hormone
- Skatole
- 2-(2-nitrophenylsulfenyl)-3-methyl-3-bromoindolenine
- STI
- soybean trypsin inhibitor
- TM
- transmembrane domain
- HPLC
- high pressure liquid chromatography
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Baggio L. L., Drucker D. J. (2007) Gastroenterology 132, 2131–2157 [DOI] [PubMed] [Google Scholar]
- 2.Drucker D. J. (2005) Nat. Clin. Pract. Endocrinol. Metab. 1, 22–31 [DOI] [PubMed] [Google Scholar]
- 3.Estall J. L., Drucker D. J. (2006) Curr. Pharm. Des. 12, 1731–1750 [DOI] [PubMed] [Google Scholar]
- 4.Mayo K. E., Miller L. J., Bataille D., Dalle S., Göke B., Thorens B., Drucker D. J. (2003) Pharmacol. Rev. 55, 167–194 [DOI] [PubMed] [Google Scholar]
- 5.Bazarsuren A., Grauschopf U., Wozny M., Reusch D., Hoffmann E., Schaefer W., Panzner S., Rudolph R. (2002) Biophys. Chem. 96, 305–318 [DOI] [PubMed] [Google Scholar]
- 6.Dong M., Lam P. C., Gao F., Hosohata K., Pinon D. I., Sexton P. M., Abagyan R., Miller L. J. (2007) Mol. Pharmacol. 72, 280–290 [DOI] [PubMed] [Google Scholar]
- 7.Grace C. R., Perrin M. H., DiGruccio M. R., Miller C. L., Rivier J. E., Vale W. W., Riek R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12836–12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grauschopf U., Lilie H., Honold K., Wozny M., Reusch D., Esswein A., Schäfer W., Rücknagel K. P., Rudolph R. (2000) Biochemistry 39, 8878–8887 [DOI] [PubMed] [Google Scholar]
- 9.Sun C., Song D., Davis-Taber R. A., Barrett L. W., Scott V. E., Richardson P. L., Pereda-Lopez A., Uchic M. E., Solomon L. R., Lake M. R., Walter K. A., Hajduk P. J., Olejniczak E. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7875–7880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y. J., Gimpl G., Fahrenholz F. (1995) Biochem. Biophys. Res. Commun. 212, 673–680 [DOI] [PubMed] [Google Scholar]
- 11.Gourlet P., Vilardaga J. P., De Neef P., Waelbroeck M., Vandermeers A., Robberecht P. (1996) Peptides 17, 825–829 [DOI] [PubMed] [Google Scholar]
- 12.Graziano M. P., Hey P. J., Strader C. D. (1996) Receptors Channels 4, 9–17 [PubMed] [Google Scholar]
- 13.Holtmann M. H., Hadac E. M., Miller L. J. (1995) J. Biol. Chem. 270, 14394–14398 [DOI] [PubMed] [Google Scholar]
- 14.Jüppner H., Schipani E., Bringhurst F. R., McClure I., Keutmann H. T., Potts J. T., Jr., Kronenberg H. M., Abou-Samra A. B., Segre G. V., Gardella T. J. (1994) Endocrinology 134, 879–884 [DOI] [PubMed] [Google Scholar]
- 15.Stroop S. D., Nakamuta H., Kuestner R. E., Moore E. E., Epand R. M. (1996) Endocrinology 137, 4752–4756 [DOI] [PubMed] [Google Scholar]
- 16.Runge S., Wulff B. S., Madsen K., Bräuner-Osborne H., Knudsen L. B. (2003) Br. J. Pharmacol. 138, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoare S. R. (2005) Drug Discov. Today 10, 417–427 [DOI] [PubMed] [Google Scholar]
- 18.Bisello A., Adams A. E., Mierke D. F., Pellegrini M., Rosenblatt M., Suva L. J., Chorev M. (1998) J. Biol. Chem. 273, 22498–22505 [DOI] [PubMed] [Google Scholar]
- 19.Dong M., Li Z., Pinon D. I., Lybrand T. P., Miller L. J. (2004) J. Biol. Chem. 279, 2894–2903 [DOI] [PubMed] [Google Scholar]
- 20.Dong M., Pinon D. I., Cox R. F., Miller L. J. (2004) J. Biol. Chem. 279, 31177–31182 [DOI] [PubMed] [Google Scholar]
- 21.Grace C. R., Perrin M. H., Gulyas J., Digruccio M. R., Cantle J. P., Rivier J. E., Vale W. W., Riek R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pioszak A. A., Parker N. R., Suino-Powell K., Xu H. E. (2008) J. Biol. Chem. 283, 32900–32912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pioszak A. A., Xu H. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5034–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Runge S., Thøgersen H., Madsen K., Lau J., Rudolph R. (2008) J. Biol. Chem. 283, 11340–11347 [DOI] [PubMed] [Google Scholar]
- 25.Underwood C. R., Garibay P., Knudsen L. B., Hastrup S., Peters G. H., Rudolph R., Reedtz-Runge S. (2010) J. Biol. Chem. 285, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parthier C., Reedtz-Runge S., Rudolph R., Stubbs M. T. (2009) Trends Biochem. Sci. 34, 303–310 [DOI] [PubMed] [Google Scholar]
- 27.Chen Q., Pinon D. I., Miller L. J., Dong M. (2009) J. Biol. Chem. 284, 34135–34144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adelhorst K., Hedegaard B. B., Knudsen L. B., Kirk O. (1994) J. Biol. Chem. 269, 6275–6278 [PubMed] [Google Scholar]
- 29.Powers S. P., Pinon D. I., Miller L. J. (1988) Int. J. Pept. Protein Res. 31, 429–434 [DOI] [PubMed] [Google Scholar]
- 30.Ulrich C. D., 2nd, Pinon D. I., Hadac E. M., Holicky E. L., Chang-Miller A., Gates L. K., Miller L. J. (1993) Gastroenterology 105, 1534–1543 [DOI] [PubMed] [Google Scholar]
- 31.Dong M., Gao F., Pinon D. I., Miller L. J. (2008) Mol. Endocrinol. 22, 1489–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holtmann M. H., Ganguli S., Hadac E. M., Dolu V., Miller L. J. (1996) J. Biol. Chem. 271, 14944–14949 [DOI] [PubMed] [Google Scholar]
- 33.Hadac E. M., Ghanekar D. V., Holicky E. L., Pinon D. I., Dougherty R. W., Miller L. J. (1996) Pancreas 13, 130–139 [DOI] [PubMed] [Google Scholar]
- 34.Munson P. J., Rodbard D. (1980) Anal. Biochem. 107, 220–239 [DOI] [PubMed] [Google Scholar]
- 35.Dong M., Wang Y., Pinon D. I., Hadac E. M., Miller L. J. (1999) J. Biol. Chem. 274, 903–909 [DOI] [PubMed] [Google Scholar]
- 36.Ji Z., Hadac E. M., Henne R. M., Patel S. A., Lybrand T. P., Miller L. J. (1997) J. Biol. Chem. 272, 24393–24401 [DOI] [PubMed] [Google Scholar]
- 37.Murakami M., Kouyama T. (2008) Nature 453, 363–367 [DOI] [PubMed] [Google Scholar]
- 38.López de Maturana R., Treece-Birch J., Abidi F., Findlay J. B., Donnelly D. (2004) Protein Pept. Lett. 11, 15–22 [DOI] [PubMed] [Google Scholar]
- 39.Kraetke O., Holeran B., Berger H., Escher E., Bienert M., Beyermann M. (2005) Biochemistry 44, 15569–15577 [DOI] [PubMed] [Google Scholar]
- 40.Gallwitz B., Witt M., Paetzold G., Morys-Wortmann C., Zimmermann B., Eckart K., Fölsch U. R., Schmidt W. E. (1994) Eur. J. Biochem. 225, 1151–1156 [DOI] [PubMed] [Google Scholar]
- 41.Tokuyama Y., Matsui K., Egashira T., Nozaki O., Ishizuka T., Kanatsuka A. (2004) Diabetes Res. Clin. Pract. 66, 63–69 [DOI] [PubMed] [Google Scholar]
- 42.Beinborn M., Worrall C. I., McBride E. W., Kopin A. S. (2005) Regul. Pept. 130, 1–6 [DOI] [PubMed] [Google Scholar]
- 43.Dong M., Pinon D. I., Cox R. F., Miller L. J. (2004) J. Biol. Chem. 279, 1167–1175 [DOI] [PubMed] [Google Scholar]
- 44.Gensure R. C., Gardella T. J., Jüppner H. (2001) J. Biol. Chem. 276, 28650–28658 [DOI] [PubMed] [Google Scholar]
- 45.Zhou A. T., Bessalle R., Bisello A., Nakamoto C., Rosenblatt M., Suva L. J., Chorev M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3644–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wittelsberger A., Corich M., Thomas B. E., Lee B. K., Barazza A., Czodrowski P., Mierke D. F., Chorev M., Rosenblatt M. (2006) Biochemistry 45, 2027–2034 [DOI] [PubMed] [Google Scholar]
- 47.Ceraudo E., Murail S., Tan Y. V., Lacapère J. J., Neumann J. M., Couvineau A., Laburthe M. (2008) Mol. Endocrinol. 22, 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ceraudo E., Tan Y. V., Nicole P., Couvineau A., Laburthe M. (2008) J. Mol. Neurosci. 36, 245–248 [DOI] [PubMed] [Google Scholar]
- 49.Tan Y. V., Couvineau A., Murail S., Ceraudo E., Neumann J. M., Lacapère J. J., Laburthe M. (2006) J. Biol. Chem. 281, 12792–12798 [DOI] [PubMed] [Google Scholar]
- 50.Dong M., Cox R. F., Miller L. J. (2009) J. Biol. Chem. 284, 21839–21847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.López de Maturana R., Donnelly D. (2002) FEBS Lett. 530, 244–248 [DOI] [PubMed] [Google Scholar]
- 52.Xiao Q., Jeng W., Wheeler M. B. (2000) J. Mol. Endocrinol. 25, 321–335 [DOI] [PubMed] [Google Scholar]
- 53.Piserchio A., Bisello A., Rosenblatt M., Chorev M., Mierke D. F. (2000) Biochemistry 39, 8153–8160 [DOI] [PubMed] [Google Scholar]
- 54.Du K., Nicole P., Couvineau A., Laburthe M. (1997) Biochem. Biophys. Res. Commun. 230, 289–292 [DOI] [PubMed] [Google Scholar]
- 55.Gensure R. C., Shimizu N., Tsang J., Gardella T. J. (2003) Mol. Endocrinol. 17, 2647–2658 [DOI] [PubMed] [Google Scholar]
- 56.Greenberg Z., Bisello A., Mierke D. F., Rosenblatt M., Chorev M. (2000) Biochemistry 39, 8142–8152 [DOI] [PubMed] [Google Scholar]
- 57.Dong M., Lam P. C., Pinon D. I., Sexton P. M., Abagyan R., Miller L. J. (2008) Mol. Pharmacol. 74, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Behar V., Bisello A., Bitan G., Rosenblatt M., Chorev M. (2000) J. Biol. Chem. 275, 9–17 [DOI] [PubMed] [Google Scholar]
- 59.Behar V., Bisello A., Rosenblatt M., Chorev M. (1999) Endocrinology 140, 4251–4261 [DOI] [PubMed] [Google Scholar]
- 60.Monaghan P., Thomas B. E., Woznica I., Wittelsberger A., Mierke D. F., Rosenblatt M. (2008) Biochemistry 47, 5889–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin F., Wang R. (2009) J. Mol. Model. 15, 53–65 [DOI] [PubMed] [Google Scholar]