Abstract
Histone lysine methylation is a dynamic process that plays an important role in regulating chromatin structure and gene expression. Recent studies have identified Jhd2, a JmjC domain-containing protein, as an H3K4-specific demethylase in budding yeast. However, important questions regarding the regulation and functions of Jhd2 remain unanswered. In this study, we show that Jhd2 has intrinsic activity to remove all three states of H3K4 methylation in vivo and can dynamically associate with chromatin to modulate H3K4 methylation levels on both active and repressed genes and at the telomeric regions. We found that the plant homeodomain (PHD) finger of Jhd2 is important for its chromatin association in vivo. However, this association is not dependent on H3K4 methylation and the H3 N-terminal tail, suggesting the presence of an alternative mechanism by which Jhd2 binds nucleosomes. We also provide evidence that the JmjN domain and its interaction with the JmjC catalytic domain are important for Jhd2 function and that Not4 (an E3 ligase) monitors the structural integrity of this interdomain interaction to maintain the overall protein levels of Jhd2. We show that the S451R mutation in human SMCX (a homolog of Jhd2), which has been linked to mental retardation, and the homologous T359R mutation in Jhd2 affect the protein stability of both of these proteins. Therefore, our findings provide a mechanistic explanation for the observed defects in patients harboring this SMCX mutant and suggest the presence of a conserved pathway involving Not4 that modulates the protein stability of both yeast Jhd2 and human SMCX.
Keywords: Chromatin, Chromatin Histone Modification, Histone Methylation, Protein Stability, Transcription Regulation
Introduction
Covalent modifications of histones play an important role in genome maintenance and gene regulation. In particular, methylation on the side chains of lysine and arginine in histones H3 and H4 is important in regulating chromatin structure and gene transcription (1, 2). Lysine methylation is unique among the known histone modifications, as three modification states, monomethylation (me1), dimethylation (me2), and trimethylation (me3), can be generated, adding yet another level of variation to this modification “mark.” In general, methylation at H3K4, H3K36, and H3K79 has been linked to gene activation, whereas H3K9, H3K27, and H4K20 methylation is associated with repressed genes (1, 2). However, recent studies have revealed that depending on the gene context, the methylation state of a specific modified lysine can exert an opposite effect on gene expression. For example, methylation of H3K9 within the promoter or coding region can result in gene repression or activation, respectively (3). Furthermore, whereas H3K4 methylation (especially H3K4me3) is strongly associated with gene activation (1, 2, 4), H3K4me2 at certain chromatin loci plays a role in repression by preventing aberrant gene expression (5, 6).
Until recently, histone lysine methylation was thought to be stable and irreversible. However, this notion was dispelled by the identification of two different classes of lysine-specific histone demethylases, amine oxidases (e.g. LSD1) and JmjC domain-containing proteins. Both classes of enzymes catalyze lysine demethylation via an oxidation reaction that generates formaldehyde, but only JmjC class demethylases require ferrous ion (Fe2+) and α-ketoglutarate as cofactors (7, 8). Whereas LSD1 specifically targets mono- and dimethylated H3K4 or H3K9, the JmjC class consists of several subfamilies that remove methyl groups from the modified H3K4, H3K9, H3K27, or H3K36 residue (7, 8).
One member of the JmjC class is the evolutionarily conserved JARID1 family of histone demethylases, which specifically target Lys4-methylated H3 and function as transcriptional repressors (9). In addition to the JmjC catalytic domain, the JARID1 proteins also possess a conserved N-terminal motif (JmjN domain) that is strictly associated with the JmjC domain (7, 9). Metazoan JARID1 demethylases also contain several conserved functional motifs, including an ARID/BRIGHT DNA-binding domain, a C5HC2 zinc finger, and two or three plant homeodomain (PHD)2 fingers (7, 9). The ARID/BRIGHT DNA-binding domain is required for the demethylation activities of JARID1 proteins (10–13). However, it is not known whether the ARID/BRIGHT domain is essential for their association with chromatin.
Although mammalian cells encode four JARID1 H3K4 demethylases (JARID1A/RBP2, JARID1B/PLU-1, JARID1C/SMCX, and JARID1D/SMCY), only one (Jhd2) was identified in budding yeast (14–17). Human JARID1A and JARID1B have the ability to target all three states of H3K4 methylation in vivo (11, 12, 18). The yeast Jhd2 displayed activity toward all forms of Lys4-methylated H3 in vitro (14) but only toward H3K4me2 and H3K4me3 in vivo (15–17). Chromatin immunoprecipitation (ChIP) assays have revealed that in the absence of JHD2, the levels of H3K4 methylation were altered compared with the wild type (WT) during activation and attenuation of GAL1 transcription (14), suggesting a role for Jhd2 in these processes. Evidence from genetic studies indicates that Jhd2 is important for maintaining normal gene silencing at telomere and silent mating-type loci (15, 19). A recent study has shown that the protein stability and steady-state levels of Jhd2 are modulated by the proteasomal degradation following E3 ligase Not4-mediated polyubiquitination (20).
Despite these advances in our understanding of the functions of Jhd2, many fundamental questions pertaining to its substrate specificity and selectivity, domain contributions, chromatin association, and overall regulation remain unanswered. In this study, we demonstrate that Jhd2 localizes to both transcriptionally active and inactive chromatin and regulates H3K4 methylation at these loci. Upon investigating the contributions of the conserved domains in Jhd2 to its function, we found that a proper interaction between JmjN and JmjC domains is important for Jhd2 function and that Not4 controls Jhd2 protein levels by monitoring the integrity of this JmjN-JmjC interdomain interaction. Additionally, our results show that the PHD finger is important for the interaction of Jhd2 with chromatin in vivo and that this interaction is independent of H3K4 methylation.
EXPERIMENTAL PROCEDURES
Yeast Strains
Deletion mutants lacking Jhd2, Set1, Swd1, Sdc1, or Spp1 in strain YMH171 were created using PCR products containing the disrupted gene locus and the inserted KanMX4 selection module amplified from the genomic DNA template isolated from the respective BY4742-based yeast deletion strains (Open Biosystems). Jhd2 was genomically tagged with nine copies of Myc at its C terminus in YMH171 following PCR amplification using pYM6 as the template (21). MSY421, a histone H3/H4 shuffle strain (22), was used to mobilize the H3 N-terminal deletion mutant (H3(1–28Δ)). The FLAG-H2B and FLAG-H2B/set1Δ strains were derived from YZS276 (22). Detailed genotypes of the yeast strains described in this study are listed in supplemental Table S1.
DNA Constructs
All the plasmids used in this study are listed in supplemental Table S2. For overexpressing the jumonji domain-containing proteins, PCR products containing the entire open reading frame (ORF) for RPH1, GIS1, ECM5, JHD1, or JHD2 and 500 bp of DNA upstream and downstream of each ORF were mobilized into a high copy vector, YEplac112 (23). C-terminal LexA epitope-tagged Jhd2 was created by PCR amplifying the ORF of JHD2 and mobilizing it between the ADH1 promoter and a fragment of the LexA DNA-binding domain in pFBL23 (24). An XhoI-KpnI fragment containing the sequence encoding nine copies of the Myc epitope (9Myc) and the CYC1 terminator was obtained following PCR amplification and mobilized into pRS314. Subsequently, a 500-bp PCR product containing the JHD2 promoter was mobilized upstream of the region coding for 9Myc as a SacI-SpeI fragment. The entire JHD2 promoter-9Myc-CYC1 terminator module was mobilized as a SacI-KpnI fragment into pRS316. The ORF of JHD2 was then PCR-amplified and mobilized between the JHD2 promoter and the 9Myc-CYC1 terminator sequence in pRS314 or pRS316 as a SpeI-XhoI fragment. Point and truncation mutants of Jhd2 were made by PCR-based site-directed mutagenesis using pRS314-JHD2-9Myc or pRS316-JHD2-9Myc as the template. For overexpression of JHD2 or its mutant derivatives, a fragment containing the WT or mutant ORF, the promoter, and the CYC1 terminator region was excised from the pRS314-based construct and mobilized into the high copy vector pRS426. To obtain purified recombinant Jhd2 PHD finger or its variants, a sequence encoding either the WT PHD finger or its mutant derivatives was amplified by PCR and mobilized into a bacterial expression vector, pBG101 (kindly provided by the Vanderbilt Structural Biology Core). The plasmids pCS3+-6Myc and pCS3+-SMCX-6Myc were kindly provided by Ralf Janknecht (25), and the pCS3+-smcx(S451R)-6Myc construct was made by PCR-based site-directed mutagenesis. All of the constructs created using PCR amplification were verified by DNA sequencing.3
Western Blot Analysis
To determine changes in Jhd2 levels (see Fig. 2C), whole-cell extracts (WCEs) were prepared as described (26) and analyzed by Western blotting using antibodies raised against the Myc epitope (9E10; a gift from Ethan Lee) and Pgk1 (22C5, Molecular Probes) at 1:1000 and 1:5000 dilutions, respectively. Crude nuclear extracts were used to examine the levels of H3K4 methylation following the cell fractionation procedure described previously (26). The following antibodies were purchased from Millipore and used in Western blotting to detect H3K4 methylation (with dilutions indicated in parentheses): anti-H3K4me1 (1:1000), anti-H3K4me2 (1:10,000), and anti-H3K4me3 (1:2500). The histone H3 loading was monitored using anti-H3 antibody (1:7500; Active Motif). For mammalian WCEs, HeLa cells were transfected with pCS3+-6Myc, pCS3+-SMCX-6Myc, or pCS3+-smcx(S451R)-6Myc using the Lipofectamine method. Following a 2-day incubation at 37 °C, cells were washed prior to and after harvesting with ice-cold 1× phosphate-buffered saline (Sigma) and boiled in 1× Laemmli sample buffer (Bio-Rad) for 10 min. After centrifugation at 16,100 × g for 5 min, equal volumes of WCEs were subjected to Western blot analysis using anti-Myc (1:1000) and anti-β-actin (1:10,000; Sigma) antibodies.
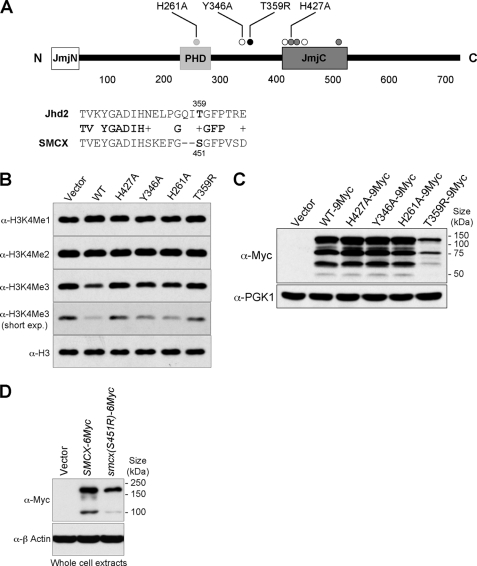
FIGURE 2.
Mutations in the conserved regions of Jhd2 can affect its activity and overall protein levels. A, shown are a schematic representation and the domain locations of Jhd2. The three conserved domains, JmjN, PHD finger, and JmjC, are shown. Dark gray circles, residues that bind to the cofactor ferrous ion (Fe2+); white circles, residues that bind to the cofactor α-ketoglutarate; light gray and black circles, a conserved residue (His261) in the PHD finger and a conserved residue (Thr359) corresponding to a human mental retardation-linked mutation site, respectively. Alignment of the Jhd2 sequence flanking Thr359 and the SMCX sequence flanking one of the sites mutated in X-linked mental retardation (Ser451) is also shown. B, shown are the results from Western blot analysis of H3K4 methylation levels in crude nuclear extracts from cells overexpressing WT JHD2 or its mutants. exp., exposure. C, shown are the results from Western blot analysis of the levels of Jhd2–9Myc or its mutant derivatives in WCEs. The level of Pgk1 served as a control for total protein loading. D, WCEs were prepared from HeLa cells expressing SMCX-6Myc or the mutant (smcx(S451R)-6Myc), and protein levels were examined using anti-Myc antibody. The level of β-actin served as a control for total protein loading.
INO1 Induction and Repression
WT and jhd2Δ strains were grown in yeast minimal synthetic complete medium without inositol (SC−Ino medium; Bio 101, Inc.) supplemented with 200 μm inositol and 2 mm choline (INO1 repression medium) (27) at 30 °C overnight. The overnight cultures were reinoculated into INO1 repression medium at 2 × 106 cells/ml and grown at 30 °C to log phase. Cells (4 × 108) were subjected to formaldehyde cross-linking for ChIP assay (INO1 repressed state). The remaining cells were harvested, washed once with SC−Ino medium, inoculated at 4 × 106 cells/ml into fresh SC−Ino medium, and grown at 30 °C for 2 h to activate INO1 expression. Again, cells (4 × 108) were set aside and subjected to formaldehyde cross-linking (INO1 induced state). To assess temporal changes in histone modification or Jhd2 occupancy following INO1 repression, cells grown in inducing medium were reinoculated into INO1 repression medium at the following initial cell densities and grown at 30 °C for the various time periods (indicated in parentheses) to obtain ∼4 × 108 cells: 7 × 106 cells/ml (20 min), 6 × 106 cells/ml (1 h), 4 × 106 cells/ml (2 h), and 2 × 106 cells/ml (4 h). Cultures grown to different incubation times following INO1 re-repression were then subjected to formaldehyde cross-linking for ChIP assay (INO1 re-repression time course).
ChIP
Double cross-linking using dimethyl adipimidate (Sigma) and formaldehyde was done essentially as described previously (28) with minor modifications. Following incubation in 10 mm dimethyl adipimidate, cells were washed and resuspended in 1× phosphate-buffered saline containing 1% formaldehyde and incubated at room temperature for 45 min with gentle agitation. The cross-linking was stopped by the addition of 130 mm glycine and incubation for 10 min at room temperature. Cells were then washed twice with 1× phosphate-buffered saline and harvested to prepare soluble chromatin for immunoprecipitating H3, H3K4me3, Jhd2-LexA, and Jhd2–9Myc using anti-H3, anti-H3K4me3, anti-LexA (Active Motif), and anti-Myc antibodies, respectively.
PCR Analysis
The data analysis of quantitative real-time PCR following ChIP was performed as described by Chandrasekharan et al. (26) with modifications. Briefly, occupancy of H3K4me3 was calculated using the 2−ΔΔCT method (Bio-Rad real-time PCR applications guide). Chromatin obtained from the set1Δ strain was used as a control to determine nonspecific immunoprecipitation, and any value obtained for this strain was subtracted from the H3K4me3 occupancy values obtained from all other strains. Soluble chromatins from yeast strains transformed with plasmid vectors were used as negative controls (“no tag”) for ChIP of Jhd2–9Myc and Jhd2-LexA. The 2−ΔCT value obtained for the negative control was subtracted from 2−ΔCT values obtained for test samples containing epitope-tagged WT or mutant Jhd2.3
Cell Fractionation
Spheroplast preparation and nuclei isolation were performed as described previously (26). WCEs were prepared by bead-beating spheroplasts in buffer A (1% SDS, 8 m urea, 10 mm MOPS (pH 6.8), 10 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, and 1 μg/ml leupeptin), followed by centrifugation at 16,100 × g for 20 min. Nuclear extracts were obtained by lysing isolated nuclei in buffer A, followed by sonication and centrifugation at 16,100 × g for 15 min. For chromatin fractions, isolated nuclei were resuspended in hypotonic solution (3 mm EDTA, 0.2 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, and 1 μg/ml leupeptin) and incubated on ice for 30 min. After centrifugation at 1700 × g for 5 min, the chromatin pellet was washed with hypotonic solution and then resuspended in buffer A. Following a brief sonication, soluble chromatin was obtained by centrifugation at 16,100 × g for 15 min. The protein concentration of the clarified lysate was measured using a Bio-Rad DC protein assay kit following the manufacturer's instructions, and an equal amount of total protein was subjected to Western blotting using anti-Myc, anti-Pgk1, or anti-H3 antibody.
Protein Synthesis Inhibition
Cycloheximide treatment of yeast was done as described previously (29) with minor modifications. Briefly, a 25-ml culture was grown to log phase, and cycloheximide was added to a final concentration of 35 μg/ml to inhibit the translation machinery. Cells (3.5 × 107) were collected at 0, 20, 40, and 60 min after cycloheximide treatment and boiled in 100 μl of 1× Laemmli sample buffer for 10 min. Following centrifugation at 16,100 × g for 5 min, equal volumes of lysates were subjected to Western blot analysis using anti-Myc or anti-Pgk1 antibody.
Proteasomal Inhibition
A log phase yeast culture (25 ml) was treated with either dimethyl sulfoxide or 0.1 mm MG132 (a proteasome inhibitor) for 30 min. The effect of proteasomal inhibition on Jhd2 or its mutant derivatives was examined in a pdr5Δ strain to allow efficient uptake of MG132 (20). WCEs were prepared in the presence of 0.1 mm MG132 and subjected to Western blotting as described above.
Tertiary Structure Prediction
The sequence of the PHD domain in Jhd2 was submitted to SWISS-MODEL structure prediction (ExPASy Proteomics Server). The software uses a homology-based search and predicts the structure utilizing solved structure(s) as a reference. The template used for the structure prediction is the solution structure of the PHD finger in JARID1D/SMCY (Protein Data Bank code 2E6R).
Immobilization of Nucleosomes
Nuclei from FLAG-H2B and FLAG-H2B/set1Δ strains were isolated as described previously (26), and the DNA content was determined by measuring the A260 of an aliquot of nuclei diluted in 1 n NaOH. Nuclei (equivalent to 660 μg of DNA) were resuspended in 200 μl of buffer B (50 mm HEPES (pH 7.6), 0.1 m KCl, 2.5 mm MgCl2, 0.25% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 5 mm N-ethylmaleimide, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, and 1 μg/ml leupeptin). Micrococcal nuclease digestion was performed twice by the addition of 5 μl of 0.1 m CaCl2 and 500 units of micrococcal nuclease (Worthington) to the nuclei suspension, followed by incubation at room temperature for 10 min, and then 4.5 μl of 0.1 m EGTA (pH 8.0) was added to stop the digestion. After centrifugation at 9300 × g for 10 min, supernatants from the two digestions were combined. The nucleosome-enriched supernatant was diluted by the addition of an equal volume of buffer B containing 10% glycerol and subjected to anti-FLAG affinity chromatography (M2, Sigma). After extensive washes with buffer B with 5% glycerol, the immobilized nucleosomes were stored at 4 °C until further use in the in vitro nucleosome binding assay. DNA was isolated from an aliquot of the immobilized nucleosomes by phenol/chloroform extraction and ethanol precipitation and resolved on a 2% agarose gel to examine the size of the nucleosomes bound to anti-FLAG antibody-conjugated agarose beads.
In Vitro Nucleosome Binding Assay
Recombinant glutathione S-transferase (GST)-tagged WT and mutant PHD fingers (amino acid 209–320) of Jhd2 were purified from Escherichia coli using glutathione-Sepharose 4B beads (GE Healthcare) following the manufacturer's instructions. The GST-tagged PHD finger (2 μg) was incubated with 5 μl of nucleosome-containing beads in 200 μl of binding buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1.5 mm MgCl2, 0.2% Triton X-100, 5% glycerol, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin A, 1 μg/ml aprotinin, and 1 μg/ml leupeptin) for 2 h at 4 °C. After three washes with binding buffer, the beads were boiled in 1× Laemmli sample buffer for 10 min. Following centrifugation at 16,100 × g for 5 min, the supernatant was subjected to Western blot analysis using anti-GST (1:20,000; GE Healthcare) and anti-FLAG (M2, 1:5000) antibodies.
RESULTS
Jhd2 Is an H3K4 Methylation-specific Demethylase Functioning at Constitutively Expressed and Inducible Genes
The JmjC domain-containing histone demethylases are evolutionarily highly conserved across many genera (7). There are five JmjC domain-containing proteins (Rph1, Gis1, Ecm5, Jhd1, and Jhd2) in budding yeast. Consistent with previous reports (15, 17), overexpression of JHD2 through a high copy, 2μ-based plasmid led to a decrease in H3K4me3 and a modest increase in H3K4me1 levels (Fig. 1A, sixth lane). Moreover, none of the JmjC domain-containing proteins exhibited any apparent demethylation of Lys79-methylated H3 under the same conditions (supplemental Fig. S1). Thus, Jhd2 is an H3K4 methylation-specific demethylase in yeast.
FIGURE 1.
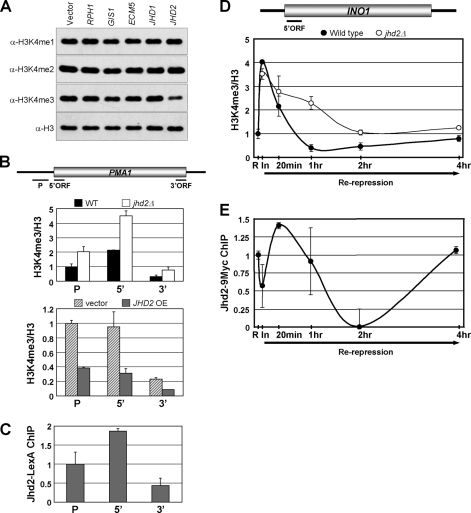
Jhd2, an H3K4-specific demethylase, functions during both active transcription and repression. A, shown are the results from Western blot analysis of H3K4 methylation levels in crude nuclear extracts obtained from yeast strains with overexpressed JmjC domain-containing proteins. B, changes in H3K4me3 levels at the PMA1 locus were analyzed by ChIP assay. The graphs depict the data obtained from the WT control and a strain lacking Jhd2 (jhd2Δ) (upper panel) and from yeast strains overexpressing JHD2 (JHD2 OE) from a high copy plasmid or containing the plasmid vector alone (lower panel). The H3K4me3 ChIP signals were first normalized to total histone H3 ChIP signals (H3K4me3/H3), and then the H3K4me3/H3 values at the promoter (P), 5′-ORF, or 3′-ORF regions in the controls and test samples were normalized to the H3K4me3/H3 value obtained for the promoter region in the WT (upper panel) or vector control (lower panel), which was set as 1. Error bars denote S.E. from two independent experiments. C, the levels of Jhd2-LexA at PMA1 were analyzed by ChIP using anti-LexA antibody. LexA immunoprecipitation/input values obtained from the no-tag control (background) were subtracted from those obtained from yeast cells with overexpressed JHD2-LexA, and the resulting differences were defined as Jhd2-LexA occupancy. The Jhd2-LexA occupancies at the 5′- and 3′-ORF regions were normalized to the promoter region, which was arbitrarily set as 1. Error bars denote S.E. from two independent experiments. D, temporal changes in H3K4me3 following the re-repression of INO1 were measured by ChIP assay. WT and jhd2Δ strains were grown in inositol/choline medium for 4 h to repress INO1 expression (R) and then grown in inositol-free medium for 2 h to induce INO1 expression (In), and INO1 was repressed again by growing cells in inositol/choline medium for the indicated time periods (Re-repression). Cells at different time points were subjected to ChIP analysis using anti-H3K4me3 and anti-H3 antibodies. The levels of H3K4me3/H3 at the 5′-ORF of INO1 in the WT or jhd2Δ mutant at different time points are shown as -fold changes relative to the H3K4me3/H3 value obtained at the repressed state for the WT, which was set as 1. Error bars denote S.E. from two independent experiments. E, temporal changes in Jhd2–9Myc occupancy following the re-repression of INO1 were measured by ChIP assay performed essentially as described for D. Chromatin-bound Jhd2–9Myc was immunoprecipitated using anti-Myc antibody. Myc immunoprecipitation/input values obtained from the no-tag control (background) were subtracted from those obtained for yeast expressing JHD2-9Myc, and the resulting difference was defined as Jhd2–9Myc occupancy. The Jhd2–9Myc occupancies at the 5′-ORF of INO1 at different time points are shown as fold changes relative to the Jhd2–9Myc occupancy obtained at the initial repressed state (R), which was set as 1. Error bars denote S.E. obtained from two independent experiments.
Given the genome-wide distribution of H3K4 methylation (30, 31) and because H3K4me3 is closely associated with gene transcription (1, 2), we tested whether Jhd2 plays a role in controlling this H3 modification during transcription. Toward this end, ChIP assays were undertaken to assess the occurrence and changes, if any, in Jhd2 occupancy and in H3K4me3 levels at the highly expressed PMA1 gene. Compared with the WT, deletion of JHD2 (jhd2Δ) or overexpression of JHD2-LexA led to a 2-fold increase or decrease in H3K4me3 at the promoter or ORF regions of PMA1, respectively (Fig. 1B). This result suggests that Jhd2 might be present on constitutively and highly expressed genes to maintain their normal H3K4 methylation levels. Indeed, ChIP data showed that Jhd2-LexA was present on the promoter and ORF regions of the PMA1 gene (Fig. 1C). Interestingly, the distribution pattern of Jhd2-LexA across the PMA1 gene was similar to that seen for H3K4me3 (Fig. 1, compare B and C). Similar results were obtained from ChIP assays of HSP104 (see Fig. 8B), which is expressed at very low levels under normal conditions (32). Given that H3K4me3 is the substrate for Jhd2, these data suggest that Jhd2 might be actively recruited during gene transcription to dynamically regulate the H3K4 methylation levels.
FIGURE 8.
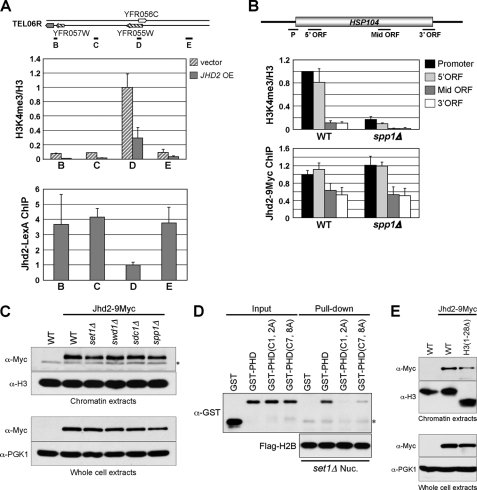
Normal distribution of Jhd2 on chromatin is independent of H3K4 methylation but requires an intact PHD finger. A, upper panel, the levels of H3K4me3 at four loci (black lines labeled B–E) in the subtelomeric region close to the right arm of chromosome 6 (TEL06R) were analyzed by ChIP assay. The graph depicts data obtained from the vector-alone control or overexpressing JHD2-LexA (JHD2 OE). The H3K4me3 levels at different loci are shown as -fold change relative to the value obtained for the vector-alone control at the D locus, which is present within an ORF. Error bars denote S.E. from two independent experiments. Lower panel, the levels of Jhd2-LexA at TEL06R were analyzed by ChIP using anti-LexA antibody. LexA immunoprecipitation/input values obtained from the no-tag control were subtracted from those obtained from strains overexpressing JHD2-LexA, and the resulting differences were defined as Jhd2-LexA occupancy. Occupancies of Jhd2-LexA at different loci were normalized to the D region. Error bars denote S.E. from two independent experiments. B, the levels of H3K4me3 and Jhd2 occupancy in HSP104 were determined by ChIP assay as described for A. Fold change in H3K4me3 levels or Jhd2 occupancy at various regions of HSP104 are shown relative to their respective levels at the promoter (P) region in the WT, which was set as 1. Error bars denote S.E. from three independent experiments. Mid, middle. C, shown are the results from Western blot analysis of the Jhd2 levels in chromatin extracts (upper panel) and WCEs (lower panel) obtained from the WT or strains lacking Set1-COMPASS complex components (Set1, Swd1, Sdc1, and Spp1). The asterisk denotes an anti-Myc cross-reacting protein. D, shown are the results from in vitro mononucleosome binding assay. The immobilized mononucleosomes isolated from set1Δ were incubated with the recombinant GST-tagged WT or mutant PHD finger of Jhd2 (2 μg). After extensive washing, the mononucleosome-bound recombinant proteins were eluted in sample buffer. Recombinant proteins (40 ng; Input) and eluates (40%) were subjected to Western blot analysis using anti-GST antibody. An aliquot of the eluate (10%) was also used as a control to show the equal loading of immobilized mononucleosomes, as detected using anti-FLAG antibody. The asterisk denotes the light chain of mouse IgG. E, shown are the results from Western blot analysis of the Jhd2 levels in chromatin extracts (upper panel) and WCEs (lower panel) prepared from the WT and H3(1–28Δ) strains. set1Δ Nuc., set1Δ nucleosome.
Next, to test whether Jhd2 also has a role in the regulation of H3K4 methylation in an inducible gene, we measured the kinetics of H3K4me3 levels on the INO1 gene in WT and jhd2Δ cells during the activated and repressed states. Additionally, we also examined Jhd2 occupancy at the 5′-ORF region of INO1 under the same conditions. Transcription of INO1, encoding inositol-1-phosphate synthase, is activated or repressed by the absence or presence of inositol/choline in the medium, respectively (33). As shown in Fig. 1D, WT and jhd2Δ cells displayed similar levels of H3K4me3 in the INO1 5′-ORF region both during the initial repressed state and following induction. Upon re-repression, the levels of H3K4me3 decreased rapidly in WT cells. However, in the absence of JHD2, the reduction of H3K4me3 levels was delayed, and it persisted for 2 h following re-repression compared with the WT, where it persisted for <1 h. Importantly, ChIP data confirmed that the Jhd2 occupancy decreased at the 5′-ORF of INO1 upon induction but increased immediately upon re-repression (Fig. 1E). Taken together, our results show that Jhd2 is required for the rapid removal of the H3K4me3 mark upon transcriptional repression of INO1.
The Cofactor-binding Residues and the PHD Finger Are Important for Jhd2 Activity
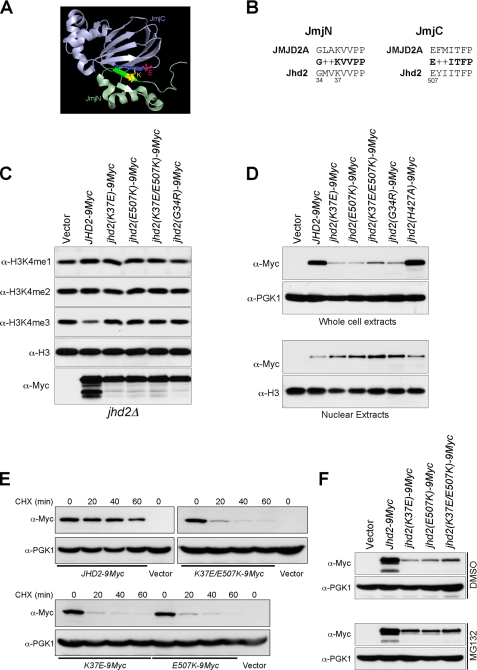
The JmjC domain-containing enzymes require iron (Fe2+) and α-ketoglutarate as cofactors for their activities (34). All the highly conserved cofactor-binding residues, except Tyr346 (one of the α-ketoglutarate-binding residues), reside in the JmjC catalytic domain of Jhd2 (Fig. 2A). Besides the JmjC domain, Jhd2 also contains a JmjN domain and a PHD finger (Fig. 2A). To investigate the importance of these different domains for Jhd2 function, we created point mutations in the conserved residues that bind α-ketoglutarate (Y346A) or Fe2+ (H427A) or that reside in the PHD finger (H261A). Compared with the WT, overexpression of these jhd2 mutant alleles showed that whereas jhd2(H261A) and jhd2(Y346A) caused only a modest decrease in H3K4me3 levels, jhd2(H427A) had no apparent reduction (Fig. 2B). Thus, binding to cofactors and maintaining an intact PHD finger are important for the function of Jhd2 in vivo.
T359R, a Substitution Mutation Corresponding to a Human SMCX Mutation Linked to Mental Retardation, Affects both the Demethylase Function and Global Protein Level of Jhd2
Mutations in SMCX (JARID1C), a human homolog of Jhd2, have been associated with X-linked mental retardation (35–37). Patients carrying one of the well characterized X-linked mental retardation mutations in SMCX (S451R) developed mental retardation and also showed mild deformities in the tongue and fingers (36). However, the molecular consequence(s) of this mutation was not known. Ser451 resides in a region that is highly conserved from yeast to human (Fig. 2A, lower panel) (7). To investigate the effect of S451R on SMCX function in vivo, we generated a corresponding mutation, T359R, in Jhd2. Unlike overexpression of WT JHD2, overexpression of jhd2(T359R) did not reduce H3K4me3 levels (Fig. 2B, sixth lane), suggesting that this mutation might abrogate the enzymatic activity and create a null allele of Jhd2. Interestingly, the T359R mutation also resulted in a severe reduction in the steady-state protein levels compared with WT JHD2 and the other jhd2 alleles (jhd2(H427A), jhd2(H261A), and jhd2(Y346A)) (Fig. 2C). This result suggests that Thr359 might be important for regulating the protein stability of Jhd2. Next, we examined whether Ser451 in SMCX is important for maintaining protein stability in human cells. To this end, we ectopically expressed 6Myc-SMCX and 6Myc-smcx(S451R) in HeLa cells. Western analyses revealed that similar to Jhd2(T359R), the total protein levels of 6Myc-SMCX(S451R) were lower than those of WT 6Myc-SMCX (Fig. 2D), suggesting a conserved role for Thr359 and Ser451 in maintaining the protein stabilities of Jhd2 and SMCX, respectively. Importantly, this decrease in the protein levels of SMCX(S451R) likely accounts for the observed defects in patients harboring this mutation (36).
The Structural Integrity of Jhd2 Is Important for Its Protein Stability
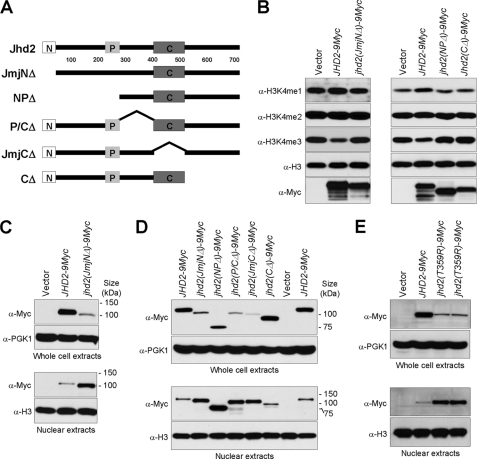
Although the JmjN domain is highly conserved, it does not exist in all JmjC domain-containing histone demethylases (7). For example, the JmjN domain is absent in the H3K36 demethylases such as human JHDM1 (JHDM1A and JHDM1B) and yeast Jhd1. To determine the contribution of this domain to Jhd2 function, we created a deletion derivative lacking the JmjN domain (Jhd2(JmjNΔ)) and examined the effect of this truncation on the ability of Jhd2 to remove H3K4 methylation (Fig. 3A). Unlike the WT, overexpression of jhd2(JmjNΔ) did not reduce H3K4 methylation (Fig. 3B), indicating that the JmjN domain is essential for Jhd2 function. Intriguingly, the protein levels of overexpressed jhd2(JmjNΔ) appeared reduced compared with those of the WT (Fig. 3B). Indeed, a drastic reduction in the steady-state levels of Jhd2(JmjNΔ) was evident when the proteins were expressed from the endogenous JHD2 promoter using a low copy (CEN-based) plasmid (Fig. 3C). This result suggests that the structural integrity of Jhd2 is important for maintaining its whole-cell protein levels. In contrast, the levels of Jhd2(JmjNΔ) in the nuclear extracts were higher than those of the WT (Fig. 3C). Given the increase in nuclear Jhd2(JmjNΔ) levels, it is conceivable that the lack of H3K4 demethylation in this mutant (Fig. 3B) is likely due to the truncation and not due to reduced global protein levels (Fig. 3C, upper panel). Taken together, these novel findings put forth a possibility that the overall protein stability and nuclear localization of Jhd2 might be regulated by its protein structure.
FIGURE 3.
The global protein level and subcellular localization of Jhd2 are sensitive to its structural integrity. A, shown is a schematic representation of Jhd2 and its truncation mutants. The JmjN, PHD, and JmjC domains are represented as shown in Fig. 2A. B, crude nuclear extracts were prepared from the jhd2Δ strain containing vector alone or overexpressing WT or mutant JHD2. The levels of H3K4 methylation were examined by Western blotting. C, the levels of Jhd2 and Jhd2(JmjNΔ) in the WCEs (upper panel) and nuclear extracts (lower panel) were examined by Western blotting. Pgk1 and H3 were used as loading controls in the WCEs and nuclear extracts, respectively. D, shown are the results from Western blot analysis of the levels of WT Jhd2 and its truncation mutants in the WCEs (upper panel) and nuclear extracts (lower panel). E, shown are the results from Western blot analysis of the levels of Jhd2 and Jhd2(T359R) in the WCEs (upper panel) and nuclear extracts (lower panel).
Previous studies have revealed the presence of two SMCX splice variants in human cells, and both proteins exhibit in vivo H3K4 demethylation. Whereas the longer form of SMCX is predominantly a nuclear protein (38–40), its shorter splice variant (lacking the C-terminal 120 amino acids) is present at very low levels in the nucleus (25). Taken together, these studies and our findings suggest that controlling the nuclear localization of SMCX and Jhd2 might be a mode to regulate their H3K4 demethylation on chromatin. Therefore, we tested the levels of Jhd2 and its mutant derivatives in addition to Jhd2(JmjNΔ) within the nucleus (Fig. 3A). As expected, all deletions of Jhd2 abolished its function as a demethylase (Fig. 3B) (data not shown). Moreover, except for the C-terminal deletion mutant (Jhd2(CΔ)), all of the mutants showed decreased steady-state protein levels and increased nuclear protein levels similar to Jhd2(JmjNΔ) (Fig. 3D). These results show that any change in the structural integrity of the region encompassing the JmjN and JmjC domains of Jhd2 that abrogates its enzymatic function will lead to a decrease in its global protein levels but increase its nuclear protein levels. Next, we tested whether these changes in Jhd2 levels in the whole-cell and nuclear extracts are due to the loss of its enzymatic activity. To this end, we examined the whole-cell and nuclear levels of the catalytically dead allele Jhd2(H427A) (Fig. 2B). As shown in Fig. 4C, both the global and nuclear levels of Jhd2(H427A) are similar to those of the WT. This result shows that the loss of enzymatic activity alone does not alter the protein levels of Jhd2 and strongly implicates the protein structural perturbations induced by the deletions as the primary cause for the observed changes in its global and nuclear protein levels. Given that the human X-linked mental retardation mutation-mimetic change in Jhd2 (T359R) also showed a drastic reduction in steady-state protein levels (Figs. 2C and 3E) and loss of demethylase function (Fig. 2B) similar to that seen for the deletion mutants (Fig. 3, B–D), we tested the protein levels of this point mutant in the nucleus. Indeed, similar to the deletion derivatives, the nuclear protein levels of Jhd2(T359R) were increased compared with the WT (Fig. 3E), suggesting that this point mutation alone is sufficient to perturb the structural integrity of Jhd2 and alter its global and nuclear protein levels. Collectively, our findings suggest that the protein stability of Jhd2 might be regulated by its structure and not by its enzymatic activity.
FIGURE 4.
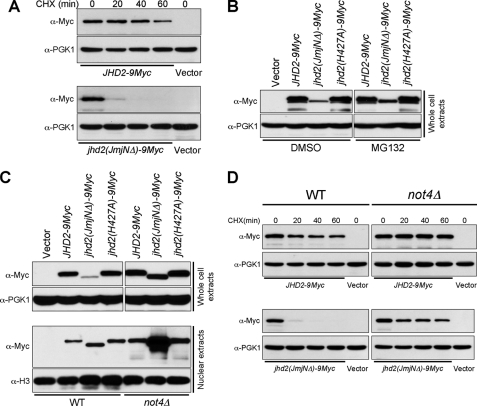
Loss of the JmjN domain in Jhd2 causes protein instability mediated by the E3 ligase Not4 and the proteasome. A, cells containing WT Jhd2 or the Jhd2(JmjNΔ) mutant were treated with 35 μg/ml cycloheximide (CHX) for the indicated periods of time to terminate protein synthesis. Equal amount of WCEs were subjected to Western blotting using anti-Myc and anti-Pgk1 antibodies. B, shown are the results from Western blot analysis of the levels of WT Jhd2 and its mutants in WCEs obtained from cells treated with dimethyl sulfoxide (DMSO; vehicle) or with MG132. C, the levels of WT Jhd2 and its mutants in WCEs and nuclear extracts obtained from control (WT) and not4Δ strains were detected using anti-Myc antibody. D, shown are the results from Western blot analysis of the levels of WT Jhd2 and the Jhd2(JmjNΔ) mutant in WCEs obtained from control (WT) and not4Δ strains treated with cycloheximide for the indicated times.
To test whether the protein stability of Jhd2 is compromised by mutations, we examined the global levels of WT Jhd2 and a mutant (Jhd2(JmjNΔ)) following termination of protein synthesis using cycloheximide. As shown in Fig. 4A, deletion of the JmjN domain caused rapid degradation of Jhd2. Compared with WT Jhd2 levels, Jhd2(JmjNΔ) was barely detectable at 20 min after cycloheximide treatment, and it was completely absent at 40 min following the translational arrest. Recently, E3 ligase Not4-mediated polyubiquitination followed by proteasomal degradation was shown to modulate the steady-state levels of Jhd2 (20). Therefore, we tested whether the compromised protein stability of Jhd2(JmjNΔ) is due to a high protein turnover mediated by the proteasome. Supporting this possibility, proteasomal inhibition using MG132 resulted in increased steady-state levels of Jhd2(JmjNΔ) (Fig. 4B). We further tested whether Not4 is involved in this proteasome-mediated degradation of Jhd2(JmjNΔ). Indeed, the steady-state levels of Jhd2(JmjNΔ) were restored nearly to WT levels in the not4Δ strain (Fig. 4C), and this mutant protein was highly stabilized in cells lacking Not4 (Fig. 4D). This finding suggests a protein structure-monitoring role for Not4 in maintaining Jhd2 levels. Additionally, the nuclear protein levels of Jhd2(JmjNΔ) were increased in not4Δ compared with both WT Jhd2 and Jhd2(H427A) (Fig. 4C). Taken together, our findings reveal that the structural integrity of Jhd2 is a crucial determinant for maintaining its protein stability mediated by Not4.
The Interaction between JmjN and JmjC Domains Is Important for Jhd2 Function
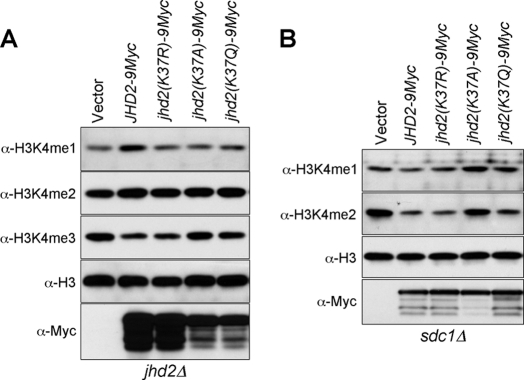
The crystal structure of the catalytic core domain of human JMJD2A, an H3K9me3- and H3K36me3-specific demethylase, has revealed a physical interaction between the two antiparallel β-strands in its JmjN and JmjC domains (Fig. 5A) (41). The residues constituting each β-strand of the JmjN and JmjC domains are highly conserved among the demethylases containing both these domains (7), implicating an important role for the interdomain interactions in regulating demethylation. Indeed, it has been proposed that this JmjN-JmjC domain interaction might cause a conformational change in the JMJD2A catalytic core to activate its enzymatic function (41). Although differing in their substrate specificity, the strong sequence similarity between the two β-strands in the JmjN and JmjC domains of JMJD2A and Jhd2 (Fig. 5B) suggests that an interdomain interaction might also occur in Jhd2 to regulate its function. To test this possibility, we mutated the conserved residues in the β-strands in the JmjN and JmjC domains of Jhd2 and assessed their effects on its enzymatic function. Unlike the WT, overexpression of jhd2(G34R), jhd2(K37E), and jhd2(E507K) did not reduce H3K4me3 levels (Fig. 5C), indicating that these conserved residues are important for Jhd2 function. Moreover, similar to the jhd2(T359R) null mutant, the global protein levels were reduced, and nuclear protein levels were increased in the JmjN and JmjC domain mutants (Fig. 5D), suggesting that these mutations might also perturb the structural integrity and elicit the proteasome-mediated protein degradation response (20). Indeed, the protein stabilities of Jhd2(K37E) and Jhd2(E507K) were severely compromised (Fig. 5E), and MG132-mediated proteasomal inhibition resulted in increased steady-state levels of these two Jhd2 mutants (Fig. 5F). The close proximity between Lys37 in the JmjN domain and Glu507 in the JmjC domain suggests that a charge-based interaction might occur between these two residues (Fig. 5A). Therefore, we created a charge-switching double mutation (K37E/E507K) and tested whether it can restore Jhd2 function compared with the single-site mutations (K37E and E507K). Overexpression of jhd2(K37E/E507K) did not reduce H3K4 methylation (Fig. 5C), indicating a disruption of the enzymatic function due to the introduced mutations. However, its whole-cell protein levels were modestly increased compared with Jhd2(K37E) or Jhd2(E507K) (Fig. 5, D–F). This result suggests that a charge-based interaction might occur between Lys37 and Glu507. However, the charged side chains in K37E and E507K might not be present in a correct orientation for efficient interdomain interaction, leading to the loss of enzymatic function. Alternatively, maintaining a positively charged residue at position 37 in the JmjN domain and a negative one at position 507 in the JmjC domain might be important for the structural integrity and for the interdomain interaction. Supporting this possibility, only overexpression of a Jhd2 mutant containing the charge-conserved substitution mutation (K37R), but not jhd2(K37A) and jhd2(K37Q), retained demethylation function and reduced H3K4me3 levels similar to overexpression of the WT (Fig. 6A).
FIGURE 5.
Residues that might mediate the JmjN-JmjC domain interaction are critical for the global protein levels and subcellular localization of Jhd2. A, shown is the crystal structure of the JmjN (light green) and JmjC (light blue) domains of JMJD2A (Protein Data Bank code 2GP5). The two domains interact with each other through one β-strand in the JmjN domain (green) and another one in the JmjC domain (blue). The two residues involved in ionic interaction, lysine in the JmjN domain (yellow) and glutamate in the JmjC domain (pink), are represented by the ball-and-stick model. B, shown is the protein sequence alignment of residues involved in the JmjN-JmjC domain interaction of JMJD2A and Jhd2. + denotes similar residues. C, shown are the results from Western blot analysis of H3K4 methylation levels in the crude nuclear extracts prepared from jhd2Δ containing a high copy plasmid alone or overexpressing WT JHD2 or its mutants. D, WCEs (upper panel) and nuclear extracts (lower panel) were obtained from the jhd2Δ strain containing low copy plasmids containing WT JHD2 or its mutants and subjected to Western blotting using anti-Myc, anti-Pgk1, and anti-H3 antibodies. E, shown are the results from Western blot analysis of the levels of WT Jhd2 and its mutants in WCEs obtained from cells treated with cycloheximide (CHX) for the indicated periods of time. F, shown are the results from Western blot analysis of the levels of WT Jhd2 and its mutants in WCEs obtained from cells treated with dimethyl sulfoxide (DMSO; vehicle) or with MG132.
FIGURE 6.
The positive charge at position 37 in the JmjN domain is important for the demethylase function of Jhd2 toward all three states of H3K4 methylation. Shown are the results from Western blot analysis of H3K4 methylation levels in crude nuclear extracts prepared from jhd2Δ (A) and sdc1Δ (B) cells overexpressing WT JHD2 or its mutant derivatives.
In in vitro assays, Jhd2 has been shown to demethylate H3K4me1-, H3K4me2-, or H3K4me3-containing peptides (14). However, only H3K4me3 levels were reduced when Jhd2 was overexpressed in yeast cells (Figs. 1–3, 5, and 6) (15, 17). Thus, the ability of Jhd2 to demethylate H3K4me2 or H3K4me1 in vivo is probably masked by the conversion of H3K4me3 into H3K4me2 or of H3K4me2 into H3K4me1, respectively. To test this possibility, we overexpressed WT JHD2 and its mutant alleles (jhd2(K37R), jhd2(K37A), and jhd2(K37Q)) in a yeast strain lacking H3K4me3 due to the deletion of SDC1, a Set1-COMPASS subunit (42). Consistent with its in vitro activity and supporting our “masking” hypothesis, overexpression of WT JHD2 (Figs. 6B and 7C, second lane) and jhd2(K37R), but not jhd2(K37A) (Fig. 6B, third and fourth lanes), reduced H3K4me2 and H3K4me1 in sdc1Δ compared with the control (vector alone). Therefore, in addition to H3K4me3, Jhd2 also demethylates H3K4me2 and H3K4me1 in vivo. Interestingly, overexpression of the acetylation-mimetic jhd2(K37Q) allele, which could not demethylate H3K4me3 (Fig. 6A, fifth lane), reduced only H3K4me2 levels (Fig. 6B, fifth lane). This result suggests that the interdomain interaction between the JmjN and JmjC domains through their two β-strands might play a role in modulating the demethylation function of Jhd2 toward different methylation states of H3K4 and that this interaction might be regulated by the acetylation of Lys37 within the JmjN domain.
FIGURE 7.
The two zinc fingers in the PHD domain are important for the function of Jhd2 as an H3K4 demethylase. A, shown is a speculative structure for the PHD finger of Jhd2 as predicted by SWISS-MODEL, a knowledge-based protein-modeling algorithm (ExPASy Proteomics Server). The two zinc-coordinating sites, each composed of four residues (cysteine or histidine), are shown. B and C, shown are the results from Western blot analysis of H3K4 methylation levels in crude nuclear extracts prepared from jhd2Δ (B) and sdc1Δ (C) strains overexpressing WT JHD2 or PHD finger mutant variants. D, nuclei isolated from the WT and set1Δ strains containing FLAG-H2B were digested using micrococcal nuclease to solubilize chromatin. The released soluble chromatin was immobilized onto anti-FLAG antibody M2-conjugated agarose beads. The DNA fragments isolated from immobilized chromatin resolved on 2% agarose gel show the presence of predominantly mononucleosomes (∼146 bp). Marker, 100-bp DNA ladder. E, shown are the results from in vitro mononucleosome binding assay. The immobilized mononucleosomes obtained from the WT were incubated with recombinant GST-tagged WT or mutant PHD fingers of Jhd2 (2 μg). After extensive washing, the mononucleosome-bound recombinant proteins were eluted in sample buffer. Recombinant proteins (40 ng; Input) and eluates (40%) were subjected to Western blot analysis using anti-GST antibody. An aliquot of the eluate (10%) was also used as a control to show the equal loading of immobilized mononucleosomes, as detected using anti-FLAG antibody. The asterisk denotes the light chain of mouse IgG. WT Nuc., WT nucleosome. F, shown are the results from ChIP analysis of the levels of WT Jhd2 or PHD finger mutant variants at HSP104. The levels of Jhd2 or its mutant variants at the promoter (P), 5′-ORF, middle (Mid) ORF, and 3′-ORF regions were normalized to the level of WT Jhd2 at the promoter region, which was set as 1. Error bars denote S.E. from three independent experiments.
The PHD Finger Is Required for the Demethylation Activity and Chromatin Association of Jhd2
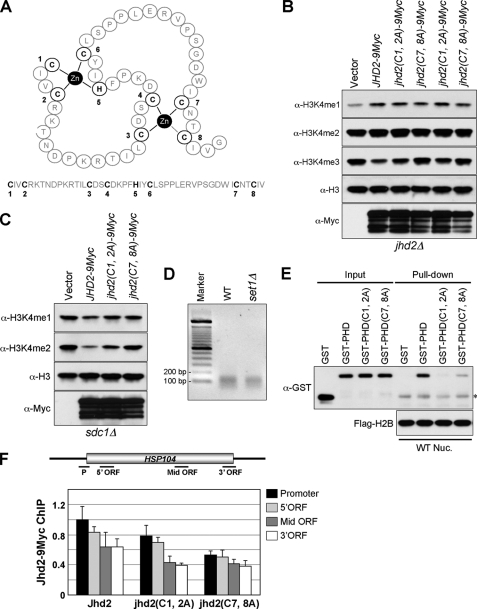
The PHD finger between the JmjN and JmjC domains of the JARID1 family of H3K4 demethylases contains a conserved C4HC3 zinc-binding motif (Fig. 2A) (7). Based on the solution structure of the PHD finger in human SMCY (JARID1D, an H3K4 demethylase; Protein Data Bank code 2E6R), the three cysteine residues (Cys1, Cys2, and Cys6) together with the histidine residue (His5) present in the Jhd2 PHD finger can be predicted to form the first zinc finger, whereas the remaining cysteine residues (Cys3, Cys4, Cys7, and Cys8) form the second one (Fig. 7A). To determine the role for these two zinc-coordinating sites in Jhd2 function, we disrupted the PHD finger by creating two double mutants, jhd2(C1A,C2A) and jhd2(C7A,C8A). Whereas overexpression of jhd2(C1A,C2A) resulted in a reduction of H3K4me3, overexpression of jhd2(C7A,C8A) had no apparent effect (Fig. 7B). Moreover, only overexpression of jhd2(C1A,C2A) in sdc1Δ caused a reduction in H3K4me2, but not jhd2(C7A,C8A) (Fig. 7C). Collectively, these results show that disruption of the second zinc-binding site within the PHD finger abrogates the demethylase function, and therefore, it is more critical for Jhd2 activity toward H3K4me2 and H3K4me3 than the first one.
Many, if not all, PHD-containing proteins associate with chromatin through the direct interaction between their PHD fingers and the histones (43, 44). Therefore, it is conceivable that the loss of demethylation in the PHD finger mutants may be due to a poor association of Jhd2 with chromatin. To test this possibility, we performed in vitro nucleosome binding assays. To obtain mononucleosomes, nuclei isolated from the WT yeast strain containing FLAG-H2B were digested extensively with micrococcal nuclease, followed by immunoprecipitation using anti-FLAG antibody-agarose beads. Agarose gel electrophoresis showed that the DNA isolated from the immobilized nucleosomes was ∼150 bp in length, confirming the presence of mainly mononucleosomes (Fig. 7D). Subsequently, equal amounts of immobilized mononucleosomes were used to pull down the purified recombinant GST-tagged PHD finger of Jhd2 (GST-PHD) or its mutants (GST-PHD(C1A,C2A) and GST-PHD(C7A,C8A)). As shown in Fig. 7E, GST-PHD, but not GST alone, was able to interact with the immobilized nucleosome. However, compared with GST-PHD, both PHD finger mutants (GST-PHD(C1A,C2A) and GST-PHD(C7A,C8A)) showed reduced binding to the nucleosome (Fig. 7E, seventh and eighth lanes). This reduced chromatin binding of the PHD finger mutants agrees well with their debilitated demethylation compared with the WT (Fig. 7, B and C). However, GST-PHD(C7A,C8A) showed better nucleosome binding than GST-PHD(C1A,C2A) (Fig. 7E, seventh and eighth lanes), even though only the jhd2(C7A,C8A) PHD finger mutation abrogated the in vivo demethylation activity of Jhd2 (Fig. 7, B and C). Therefore, to further confirm whether the PHD finger in Jhd2 is important for its in vivo chromatin association, we performed ChIP assays to assess the occupancy of C-terminally 9Myc epitope-tagged Jhd2 and its PHD finger mutants (Jhd2(C1A,C2A) and Jhd2(C7A,C8A)) on the HSP104 gene. Consistent with their effects on Jhd2 demethylation function, the occupancy of Jhd2(C7A,C8A) was more profoundly reduced on the promoter and ORF regions of HSP104 than Jhd2(C1A,C2A) compared with the WT (Fig. 7F). Collectively, our findings show that the structural integrity of the PHD finger, especially the second zinc-coordinating site, is important for the association of Jhd2 with chromatin in addition to its contributions to the H3K4 demethylation activity.
Chromatin Association of Jhd2 Is Independent of H3K4 Methylation and the H3 N-terminal Tail Region
Recently, PHD fingers have emerged as a class of specialized modules that bind to the trimethylated lysine residues on histones, such as H3K4, H3K9, and H3K36 (44, 45). Intriguingly, upon investigating the role and occupancy of Jhd2 in the subtelomeric region using ChIP assays, we found an inverse correlation between H3K4me3 and Jhd2 occupancy (Fig. 8A). Overexpression of JHD2-LexA led to a decrease in H3K4me3 at both the proximal region of the YFR055W ORF (5′-ORF) and the ORF-free regions close to the telomere at the right end of chromosome 6 (Fig. 8A, upper panel). Unexpectedly, the levels of Jhd2-LexA at the ORF-free regions were higher compared with those of the actively transcribed YFR055W, even though the H3K4me3 levels at the proximal region of YFR055W were ∼10-fold greater compared with the ORF-free regions (Fig. 8A). This result suggests that the chromatin association of Jhd2 mediated by its PHD finger is not dependent on H3K4me3. To test this possibility, we performed ChIP assays to determine the Jhd2 occupancy on the HSP104 gene in an spp1Δ strain that has reduced levels of H3K4me3 (Fig. 8B). No change in Jhd2 occupancy was evident in spp1Δ compared with the WT, suggesting that the association of Jhd2 with chromatin is independent of H3K4me3.
To further confirm that H3K4me3 is not a prerequisite for the chromatin binding of Jhd2, we performed chromatin fractionation analysis to assess the global levels of Jhd2 on chromatin in the WT or mutants lacking subunits of the Set1-COMPASS complex. To this end, nuclei isolated from the WT and mutants were gently lysed using hypotonic solution to obtain chromatin as described (26, 46). To detect chromatin-bound Jhd2–9Myc expressed from its endogenous promoter, equal amounts of chromatin from each strain were subjected to Western analyses using anti-Myc antibody. Initially, the levels of Jhd2–9Myc on chromatin were assessed in spp1Δ and sdc1Δ mutants, wherein H3K4me3 levels were either reduced or completely abolished, respectively. As shown in Fig. 8C, the chromatin-bound Jhd2–9Myc levels in spp1Δ and sdc1Δ were similar to those in the WT. This result confirms that the chromatin association of Jhd2 is independent of H3K4me3. Additionally, compared with the WT, no change in chromatin-bound Jhd2–9Myc levels was seen in set1Δ and swd1Δ mutants that completely lack H3K4 methylation (Fig. 8C). This finding suggests that the chromatin binding of Jhd2 is independent of not only H3K4me3 but also all forms of H3K4 methylation.
To determine whether the chromatin association of Jhd2 mediated by its PHD finger is indeed independent of H3K4 methylation, we performed in vitro nucleosome binding assays as described above (Fig. 7, D and E). To this end, we used mononucleosomes isolated from the set1Δ strain that lacks H3K4 methylation (Fig. 7D and supplemental Fig. S2) and assessed the ability of the Jhd2 PHD finger or its mutant derivatives to bind to the immobilized nucleosomes. As shown in Fig. 8D, the binding of GST-PHD and its mutant derivatives to mononucleosomes from the set1Δ strain was similar to their binding to mononucleosomes isolated from the WT strain containing H3K4 methylation (Fig. 7E and supplemental Fig. S2). This finding confirms the need for an intact PHD finger to interact with chromatin and, importantly, suggests that the in vitro interaction between the Jhd2 PHD finger and mononucleosomes is not dependent on H3K4 methylation.
In addition to their binding to trimethyllysine-containing histones, several PHD fingers, including the N-terminal PHD finger of human JARID1A H3K4 demethylase, have been shown recently to bind to non-methylated H3K4-containing peptides composed of amino acids 1–20 (47–50). Interestingly, our chromatin fractionation analysis showed that Jhd2 associated with chromatin in vivo even in the absence of the H3 N-terminal tail region lacking amino acids 1–28 (H3(1–28Δ)) (Fig. 8E). Therefore, our finding suggests that Jhd2 might be binding to some other region(s) of H3 either unmodified or modified by methylation. It is conceivable that Jhd2 might associate with methylated H3K36 or H3K79 modified by methyltransferase Set2 or Dot1, respectively (51). However, demethylation of H3K4me3 still occurred in set2Δ and dot1Δ upon overexpression of JHD2 (supplemental Fig. S3). Thus, the in vivo association of Jhd2 with chromatin is not dependent on H3K4, H3K36, or H3K79 methylation.
DISCUSSION
Function of Jhd2 in Transcriptionally Active and Inactive Chromatin Regions
Several studies have identified Jhd2 as an H3K4-specific demethylase in yeast (14–17). However, evidence for its occupancy and distribution on chromatin has been lacking. Our ChIP analyses demonstrated for the first time that Jhd2 binds to the promoter and coding regions of genes with either high (PMA1) or low (HSP104) expression (Figs. 1C, 7F, and 8B). Moreover, deletion or overexpression of JHD2 resulted in a 2-fold increase and decrease in H3K4me3 levels at the promoter and ORF regions of PMA1, respectively (Fig. 1B). Thus, our results suggest that Jhd2 might be actively recruited during gene transcription to dynamically regulate H3K4 methylation levels.
Besides active genes, we also showed that Jhd2 functions during the activated state, the repressed (uninduced) state, and the attenuation phase following repression of the inducible INO1 gene. Similar to its association with the constitutively expressed genes, Jhd2 also associated with the 5′-ORF region of INO1 during transcriptional induction (Fig. 1E). However, the levels of Jhd2 at the induced state were 2-fold less compared with the initial repressed state (Fig. 1E). Although this decrease in Jhd2 levels might account for the 4-fold increase in the H3K4me3 levels following induction, H3K4me3 levels were not increased in the activated state of INO1 in the absence of Jhd2 (Fig. 1D). Therefore, taken together, these data suggest that even though Jhd2 is present at INO1 during the activated state, it might play only a minor role in regulating the H3K4me3 levels, and the robust increase in H3K4me3 levels during INO1 induction is a result of the dominant Set1-COMPASS complex-mediated methylation. We found that during re-repression, the levels of H3K4me3 at the 5′-ORF of INO1 were decreased rapidly in the WT, but not in jhd2Δ (Fig. 1D). Furthermore, our ChIP data also revealed that Jhd2 dissociates from the INO1 5′-ORF region upon induction and quickly reassociates with this region upon re-repression (Fig. 1E). Thus, Jhd2 might play a role in the rapid repression of these induced genes by actively removing this active H3 modification mark upon re-repression. To the best of our knowledge, this is the first demonstration that shows the dynamics of histone demethylation and chromatin occupancy of a JmjC domain-containing protein during activation and repression of an inducible gene.
Similar to INO1, H3K4me3 levels are increased during GAL1 gene re-repression in the absence of Jhd2 (14). However, it is not known whether H3K4 demethylation by Jhd2 is sufficient to reestablish the repressed state of GAL1. Our ChIP analysis did not detect any prolonged occupancy of Rpb1 (the largest subunit of RNA polymerase II) on INO1 in jhd2Δ upon reestablishment of its repression (data not shown), suggesting the involvement of other mechanism(s) in the re-repression of INO1 and perhaps GAL1. Indeed, deletion of both JHD2 and JHD1, an H3K36-specific demethylase, led to higher GAL1 RNA levels during induction compared with those in the WT or single deletion mutants (14). These findings suggest that Jhd2 and Jhd1 (i.e. H3K4 and H3K36 demethylation) might play a redundant role in the re-repression of activated genes in yeast. Altogether, our results demonstrate that Jhd2 can dynamically associate with chromatin to modulate H3K4 methylation levels on both active (PMA1 and HSP104) and repressed (INO1) genes in yeast. These observations are in contrast to the JARID1 family H3K4 demethylases from various species, which have been shown to exert their effects only on repressed genes (9).
Jhd2 has been shown to be important for maintaining normal telomeric silencing (15). In addition, demethylation of H3K4 has been proposed to be a rate-limiting step in the formation of silent chromatin at the mating-type loci, as loss of JHD2 delays the establishment of silencing in these regions (19). However, there is no evidence that Jhd2 localizes to these regions to exert its effect on the formation of silent chromatin. Our ChIP data showed that when overexpressed, Jhd2 localized and reduced H3K4me3 levels across the telomeric regions of the right arm of chromosome 6 (TEL06R) (Fig. 8A). In keeping with the fact that H3K4 is hypomethylated at telomeres and silent mating-type loci (52), our results suggest that Jhd2 plays an active role in maintaining low levels of H3K4 methylation required for the formation of silent chromatin in these transcriptionally inactive regions.
Role of the JmjN Domain in Mediating Jhd2 Demethylase Function
The crystal structure of the catalytic core domain of human JMJD2A (an H3K9me3- and H3K36me3-specific demethylase) shows extensive contacts between the two antiparallel β-strands in its JmjN and JmjC domains (Fig. 5A) (41). Given the strong sequence similarity between the β-strands in the JmjN and JmjC domains of JMJD2A and Jhd2 (Fig. 5B), our results suggest that this JmjN-JmjC domain interaction might also occur in Jhd2 and that, importantly, this interdomain interaction is required for Jhd2 activity in vivo (Figs. 5C and 6A). Comparison of the substrate preferences of all known JmjC domain-containing histone demethylases has revealed that enzymes with only a JmjC domain prefer mono- and dimethylated substrates, whereas those containing both JmjN and JmjC domains (JmjN/JmjC) demethylate either di- and trimethylated lysines or all forms of substrates (9). Therefore, the difference in substrate specificity, especially trimethylated lysines targeted by the JmjN/JmjC domain-containing demethylases, is likely to be imparted by the JmjN domain through its interaction with the JmjC domain. Indeed, we found that different amino acid substitutions at the conserved Lys37 in the JmjN domain exerted differential affects on Jhd2 function. Whereas the charge-conserved mutation (jhd2(K37R)) maintained normal Jhd2 activity in vivo, jhd2(K37A) totally abolished demethylation (Fig. 6, A and B). In contrast, jhd2(K37Q) demethylated only H3K4me2, but not H3K4me3 and H3K4me1 (Fig. 6, A and B). These results suggest that the interdomain interaction between the two β-strands of the JmjN and JmjC domains might play a role in modulating the substrate specificity of Jhd2. Finally, given that the Lys-to-Gln substitution in jhd2(K37Q) is an acetylation-mimetic mutation, it will be interesting to determine whether the JmjN-JmjC domain interaction is modulated by acetylation of Lys37 within the JmjN domain.
Regulation of Not4-mediated Protein Degradation of Jhd2
Our results demonstrate that except for the C-terminal deletion mutant (Jhd2(CΔ)), all of the Jhd2 truncation mutants showed decreased steady-state protein levels (Figs. 3, C and D). In contrast, it has been shown that deletion of the PHD finger alone has no apparent effect on Jhd2 protein levels (20). These findings suggest that any change in the structural integrity of the region encompassing the JmjN and JmjC domains (but not the PHD finger) of Jhd2 can lead to a decrease in its global protein levels. In keeping with this hypothesis, we found that point mutations in the conserved antiparallel β-strands involved in JmjN-JmjC interdomain interaction (Fig. 5D), but not those in the conserved C4HC3 zinc-binding motif of the PHD finger (Fig. 7B), also caused a decrease in Jhd2 protein levels. Collectively, these results indicate that any perturbation of the proper interdomain interaction between the JmjN and JmjC domains can adversely affect the steady-state levels of Jhd2.
The mechanism by which Not4 recognizes and targets Jhd2 for degradation is not fully understood. Importantly, we showed that the Jhd2(JmjNΔ) protein is stabilized and that its global levels were restored nearly to WT levels in the not4Δ strain (Fig. 4, C and D), suggesting a protein structure-monitoring role for Not4 in controlling Jhd2 levels. However, the Jhd2(JmjNΔ) mutant is apparently less stable than WT Jhd2 even in the absence of Not4 (Fig. 4D), indicating that other parallel pathway(s) may also play a role in the structural quality control of Jhd2. Collectively, on the basis of our findings, we propose that Not4 might modulate the protein levels of Jhd2 by monitoring its structural integrity, especially the proper interaction between the JmjN and JmjC domains.
Jhd2 localizes to both the cytoplasm and nucleus (data not shown) (53). The differential subcellular localization of the two SMCX (human homolog of Jhd2) splice variants suggests that controlling the nuclear localization of SMCX and Jhd2 might be a mode to regulate their H3K4 demethylation on chromatin (25, 38–40). Intriguingly, whereas the deletion and point mutations of Jhd2 resulted in a decrease in steady-state levels, their levels in the nucleus were all higher compared with WT Jhd2 (Figs. 3, D and E, and 5D). Furthermore, in the not4Δ mutant, the global Jhd2(JmjNΔ) levels were increased and were equivalent to those of the WT, but the nuclear levels were dramatically increased and exceeded the WT levels (Fig. 4C). These results reveal a link between the structural integrity and subcellular localization of Jhd2. Our results are reminiscent of those seen for the yeast transcription factor Msn2, which, upon activation by certain stress conditions (glucose exhaustion, chronic stress, or low protein kinase A activity), accumulates in the nucleus (54), and an increase in its protein degradation under these conditions also results in a reduction in its overall whole-cell protein levels. Therefore, we propose that through an unknown mechanism(s) (e.g. increased nuclear import or decreased nuclear export), the Jhd2 mutants accumulate in the nucleus. Subsequently, due to the improper JmjN-JmjC interdomain interaction, the altered forms of Jhd2 are then targeted for rapid degradation by Not4, leading to lower whole-cell protein levels compared with WT Jhd2.
Finally, like Jhd2(P/CΔ) (Fig. 3D), Jhd2(T359R) also displayed lower steady-state but higher nuclear levels than Jhd2 (Figs. 2C and 3E). These results indicate that residues, including Thr359, located between the PHD finger and JmjC domain are important for the structural integrity of Jhd2. Interestingly, the global levels of human SMCX(S451R), a mutation corresponding to Jhd2(T359R), were lower than those of WT SMCX (Fig. 2D), suggesting a conserved role for Thr359 and Ser451 in regulating the protein stabilities of Jhd2 and SMCX, respectively. Because human Not4 can also polyubiquitinate SMCX in vitro (20), our findings suggest the presence of a conserved pathway involving Not4 that modulates the protein stability of both yeast Jhd2 and human SMCX.
Controlling the Chromatin Association of Jhd2
Exactly how the H3K4 demethylases belonging to the JARID1 family interact with chromatin is not fully understood. Although not present in Jhd2, all of the JARID1 H3K4 demethylases from higher eukaryotes harbor an ARID/BRIGHT DNA-binding domain, which is required for their demethylation activities (10–13). However, it is not known whether the ARID/BRIGHT domain is essential for their association with chromatin. In this study, we have shown that the PHD finger is important for Jhd2 to associate with chromatin and for its in vivo activity (Fig. 7). The PHD finger in most JmjC domain-containing demethylases and transcription factors binds to either H3 containing methylated lysine (H3K4, H3K9, H3K27, and H3K36) or the unmodified H3 N-terminal tail region (43, 44). However, whereas the PHD finger is important for the chromatin association of Jhd2 in vivo (Fig. 7F), it binds to mononucleosomes independent of H3K4 methylation in in vitro binding assays (Figs. 7E and 8D). Moreover, the in vivo association of Jhd2 with chromatin is not dependent on H3K4, H3K36, or H3K79 methylation (Fig. 8C and supplemental Fig. S3) and is independent of the first 28 amino acids in H3 (Fig. 8E). Collectively, our findings put forth a novel possibility that through its PHD finger, Jhd2 might associate with chromatin via binding to H3 outside of its N-terminal tail region or, alternatively, via binding to some other histones.
Acknowledgments
We thank Dr. Ethan Lee for generously providing the anti-Myc antibody and Drs. Mitch Smith, Brian Strahl, and Tony Weil for the yeast strains. We also thank Dr. Ralf Janknecht for providing the SMCX plasmids and the Vanderbilt University Center for Structural Biology for the bacterial expression vector.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA109355. This work was also supported by the Vanderbilt-Ingram Cancer Center and the Kleberg Foundation.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3, Tables S1 and S2, and additional references.
DNA primers used in this study are available upon request.
- PHD
- plant homeodomain
- ChIP
- chromatin immunoprecipitation
- WT
- wild type
- ORF
- open reading frame
- WCE
- whole-cell extract
- MOPS
- 4-morpholinepropanesulfonic acid
- GST
- glutathione S-transferase.
REFERENCES
- 1.Berger S. L. (2007) Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 3.Vakoc C. R., Mandat S. A., Olenchock B. A., Blobel G. A. (2005) Mol. Cell 19, 381–391 [DOI] [PubMed] [Google Scholar]
- 4.Eissenberg J. C., Shilatifard A. (2010) Dev. Biol. 339, 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berretta J., Pinskaya M., Morillon A. (2008) Genes Dev. 22, 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinskaya M., Gourvennec S., Morillon A. (2009) EMBO J. 28, 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klose R. J., Kallin E. M., Zhang Y. (2006) Nat. Rev. Genet. 7, 715–727 [DOI] [PubMed] [Google Scholar]
- 8.Shi Y., Whetstine J. R. (2007) Mol. Cell 25, 1–14 [DOI] [PubMed] [Google Scholar]
- 9.Secombe J., Eisenman R. N. (2007) Cell Cycle 6, 1324–1328 [DOI] [PubMed] [Google Scholar]
- 10.Lee M. G., Norman J., Shilatifard A., Shiekhattar R. (2007) Cell 128, 877–887 [DOI] [PubMed] [Google Scholar]
- 11.Xiang Y., Zhu Z., Han G., Ye X., Xu B., Peng Z., Ma Y., Yu Y., Lin H., Chen A. P., Chen C. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19226–19231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamane K., Tateishi K., Klose R. J., Fang J., Fabrizio L. A., Erdjument-Bromage H., Taylor-Papadimitriou J., Tempst P., Zhang Y. (2007) Mol. Cell 25, 801–812 [DOI] [PubMed] [Google Scholar]
- 13.Tu S., Teng Y. C., Yuan C., Wu Y. T., Chan M. Y., Cheng A. N., Lin P. H., Juan L. J., Tsai M. D. (2008) Nat. Struct. Mol. Biol. 15, 419–421 [DOI] [PubMed] [Google Scholar]
- 14.Ingvarsdottir K., Edwards C., Lee M. G., Lee J. S., Schultz D. C., Shilatifard A., Shiekhattar R., Berger S. L. (2007) Mol. Cell. Biol. 27, 7856–7864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang G., Klose R. J., Gardner K. E., Zhang Y. (2007) Nat. Struct. Mol. Biol. 14, 243–245 [DOI] [PubMed] [Google Scholar]
- 16.Seward D. J., Cubberley G., Kim S., Schonewald M., Zhang L., Tripet B., Bentley D. L. (2007) Nat. Struct. Mol. Biol. 14, 240–242 [DOI] [PubMed] [Google Scholar]
- 17.Tu S., Bulloch E. M., Yang L., Ren C., Huang W. C., Hsu P. H., Chen C. H., Liao C. L., Yu H. M., Lo W. S., Freitas M. A., Tsai M. D. (2007) J. Biol. Chem. 282, 14262–14271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klose R. J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., Tempst P., Gilliland D. G., Zhang Y., Kaelin W. G., Jr. (2007) Cell 128, 889–900 [DOI] [PubMed] [Google Scholar]
- 19.Osborne E. A., Dudoit S., Rine J. (2009) Nat. Genet. 41, 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mersman D. P., Du H. N., Fingerman I. M., South P. F., Briggs S. D. (2009) Genes Dev. 23, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. (1999) Yeast 15, 963–972 [DOI] [PubMed] [Google Scholar]
- 22.Sun Z. W., Allis C. D. (2002) Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 23.Gietz R. D., Sugino A. (1988) Gene 74, 527–534 [DOI] [PubMed] [Google Scholar]
- 24.Béranger F., Aresta S., de Gunzburg J., Camonis J. (1997) Nucleic Acids Res. 25, 2035–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim T. D., Shin S., Janknecht R. (2008) Biochem. Biophys. Res. Commun. 366, 563–567 [DOI] [PubMed] [Google Scholar]
- 26.Chandrasekharan M. B., Huang F., Sun Z. W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16686–16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schüller H. J., Schorr R., Hoffmann B., Schweizer E. (1992) Nucleic Acids Res. 20, 5955–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner R. G., Nelson Z. W., Gottschling D. E. (2005) Mol. Cell. Biol. 25, 6123–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belle A., Tanay A., Bitincka L., Shamir R., O'Shea E. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., Young R. A. (2005) Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 31.Millar C. B., Grunstein M. (2006) Nat. Rev. Mol. Cell Biol. 7, 657–666 [DOI] [PubMed] [Google Scholar]
- 32.Sanchez Y., Lindquist S. L. (1990) Science 248, 1112–1115 [DOI] [PubMed] [Google Scholar]
- 33.Chen M., Hancock L. C., Lopes J. M. (2007) Biochim. Biophys. Acta 1771, 310–321 [DOI] [PubMed] [Google Scholar]
- 34.Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. (2006) Nature 439, 811–816 [DOI] [PubMed] [Google Scholar]
- 35.Jensen L. R., Amende M., Gurok U., Moser B., Gimmel V., Tzschach A., Janecke A. R., Tariverdian G., Chelly J., Fryns J. P., Van Esch H., Kleefstra T., Hamel B., Moraine C., Gecz J., Turner G., Reinhardt R., Kalscheuer V. M., Ropers H. H., Lenzner S. (2005) Am. J. Hum. Genet. 76, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos C., Rodriguez-Revenga L., Madrigal I., Badenas C., Pineda M., Milà M. (2006) Eur. J. Hum. Genet. 14, 583–586 [DOI] [PubMed] [Google Scholar]
- 37.Tzschach A., Lenzner S., Moser B., Reinhardt R., Chelly J., Fryns J. P., Kleefstra T., Raynaud M., Turner G., Ropers H. H., Kuss A., Jensen L. R. (2006) Hum. Mutat. 27, 389. [DOI] [PubMed] [Google Scholar]
- 38.Christensen J., Agger K., Cloos P. A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K. H., Salcini A. E., Helin K. (2007) Cell 128, 1063–1076 [DOI] [PubMed] [Google Scholar]
- 39.Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H. H., Whetstine J. R., Bonni A., Roberts T. M., Shi Y. (2007) Cell 128, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 40.Tahiliani M., Mei P., Fang R., Leonor T., Rutenberg M., Shimizu F., Li J., Rao A., Shi Y. (2007) Nature 447, 601–605 [DOI] [PubMed] [Google Scholar]
- 41.Chen Z., Zang J., Whetstine J., Hong X., Davrazou F., Kutateladze T. G., Simpson M., Mao Q., Pan C. H., Dai S., Hagman J., Hansen K., Shi Y., Zhang G. (2006) Cell 125, 691–702 [DOI] [PubMed] [Google Scholar]
- 42.Dehé P. M., Géli V. (2006) Biochem. Cell Biol. 84, 536–548 [DOI] [PubMed] [Google Scholar]
- 43.Bienz M. (2006) Trends Biochem. Sci. 31, 35–40 [DOI] [PubMed] [Google Scholar]
- 44.Baker L. A., Allis C. D., Wang G. G. (2008) Mutat. Res. 647, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., Allis C. D. (2005) Cell 121, 859–872 [DOI] [PubMed] [Google Scholar]
- 47.Ooi S. K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S. P., Allis C. D., Cheng X., Bestor T. H. (2007) Nature 448, 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan F., Collins R. E., De Cegli R., Alpatov R., Horton J. R., Shi X., Gozani O., Cheng X., Shi Y. (2007) Nature 448, 718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Org T., Chignola F., Hetényi C., Gaetani M., Rebane A., Liiv I., Maran U., Mollica L., Bottomley M. J., Musco G., Peterson P. (2008) EMBO Rep. 9, 370–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang G. G., Song J., Wang Z., Dormann H. L., Casadio F., Li H., Luo J. L., Patel D. J., Allis C. D. (2009) Nature 459, 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shilatifard A. (2006) Annu. Rev. Biochem. 75, 243–269 [DOI] [PubMed] [Google Scholar]
- 52.Bernstein B. E., Humphrey E. L., Erlich R. L., Schneider R., Bouman P., Liu J. S., Kouzarides T., Schreiber S. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8695–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 54.Durchschlag E., Reiter W., Ammerer G., Schüller C. (2004) J. Biol. Chem. 279, 55425–55432 [DOI] [PubMed] [Google Scholar]