Abstract
Classical arabinogalactan proteins partially defined by type II O-Hyp-linked arabinogalactans (Hyp-AGs) are structural components of the plant extracellular matrix. Recently we described the structure of a small Hyp-AG putatively based on repetitive trigalactosyl subunits and suggested that AGs are less complex and varied than generally supposed. Here we describe three additional AGs with similar subunits. The Hyp-AGs were isolated from two different arabinogalactan protein fusion glycoproteins expressed in tobacco cells; that is, a 22-residue Hyp-AG and a 20-residue Hyp-AG, both isolated from interferon α2b-(Ser-Hyp)20, and a 14-residue Hyp-AG isolated from (Ala-Hyp)51-green fluorescent protein. We used NMR spectroscopy to establish the molecular structure of these Hyp-AGs, which share common features: (i) a galactan main chain composed of two 1→3 β-linked trigalactosyl blocks linked by a β-1→6 bond; (ii) bifurcated side chains with Ara, Rha, GlcUA, and a Gal 6-linked to Gal-1 and Gal-2 of the main-chain trigalactosyl repeats; (iii) a common side chain structure composed of up to six residues, the largest consisting of an α-l-Araf-(1→5)-α-l-Araf-(1→3)-α-l-Araf-(1→3- unit and an α-l-Rhap-(1→4)-β-d-GlcUAp-(1→6)-unit, both linked to Gal. The conformational ensemble obtained by using nuclear Overhauser effect data in structure calculations revealed a galactan main chain with a reverse turn involving the β-1→6 link between the trigalactosyl blocks, yielding a moderately compact structure stabilized by H-bonds.
Keywords: Carbohydrate Structure, Glycoconjugate, Glycoprotein, Glycoprotein Structure, Glycosylation, Arabinogalactan Proteins, Hydroxyproline-rich Glycoproteins, Plant Cell Walls, Type II Arabinogalactan
Introduction
Hydroxyproline-rich glycoproteins of the cell surface comprise groups of related structural proteins, including the extensins that form cell wall scaffolding networks essential for cytokinesis (1) and the classical arabinogalactan proteins (2) that are largely at the membrane wall interface (3) and have diverse functions (4). O-Hyp6 glycosylation characterizes the hydroxyproline-rich glycoproteins and is of much interest as it defines molecular properties and, hence, biological function. Arabinogalactan proteins are highly glycosylated mainly with O-Hyp-arabinogalactan polysaccharides (5, 6). Extensins are less highly glycosylated mainly with small O-Hyp arabinooligosaccharides (7, 8), whereas the related proline-rich proteins are minimally glycosylated also with arabinooligosaccharides (9). Peptide sequence directs O-Hyp glycosylation by the addition of small oligosaccharides to contiguous Hyp residues and larger acidic arabinogalactan polysaccharides to clustered noncontiguous Hyp (10, 11). For example, clustered Ala-Hyp and Ser-Hyp are typical AGP glycosylation motifs (12), whereas the Hyp residues in repetitive blocks of Ser-Hyp2 orSer-Hyp4 are arabinosylated. The “hyperglycosylation” of closely related AGPs complicates their purification, a problem that can be overcome by expressing single individual AGPs as GFP fusion glycoproteins, the hydrophobic GFP tag enabling chromatographic purification (13, 14). This approach also allows purification of neo-AGPs containing single repeating AGP glycosylation motifs, for example (Ala-Hyp)n or (Ser-Hyp)n, for base-catalyzed peptide bond hydrolysis. Base hydrolysis releases alkali-stable Hyp-arabinogalactan glycoamino acids, designated Hyp-AGs (5, 6), that can be further purified by size-exclusion chromatography. Here we report the complete structural elucidation of three such Hyp-AGs ranging in size from 14 to 22 sugar residues. The structures were determined using multidimensional homonuclear and heteronuclear NMR spectroscopy in conjunction with molecular simulations in the presence of water.
Ala-Hyp-polysaccharide-1 isolated earlier from (Ala-Hyp)51-GFP expressed in tobacco cells represented the first complete structure of a small Hyp-AG (15) characteristic of type II arabinogalactans, which are defined by β-1,3-linked galactose residues. Ala-Hyp-polysaccharide-1 consisted of a 5-residue 1,3-β-d-Gal backbone interrupted by a single β-1,6 linkage connecting Gal-3 and Gal-4 (numbered from the reducing end). Galactose residues 1 and 2 had small-bifurcated side-chain substituents at C-6. The side chains possessed a single β-d-Gal di-substituted at C-3 with α-l-arabinose di- or trisaccharides and at C-6 with β-d-glucuronic acid or α-l-rhamnosylglucuronic acid (12, 15). We proposed that ∼15-residue repetitive blocks of decorated β-(1–3) trigalactosyl subunits connected by β-1,6 linkages create the larger arabinogalactan polysaccharides of AGPs, consistent with the small blocks separated by periodate-sensitive residues suggested earlier (16, 17).
As type II arabinogalactans are often considered intractably complex, and because Ala-Hyp-polysaccharide-1 was only a single example of a Hyp-AG subunit, we determined the structure of three additional Hyp-AGs derived from two different AGP motifs, repetitive Ala-Hyp and Ser-Hyp. Two of the Hyp-AGs designated interferon-polysaccharide-1 (interferon Hyp-polysaccharide 1) and interferon-polysaccharide-2 were isolated from a fusion glycoprotein of human interferon α2b fused to a (Ser-Hyp)20 AGP glycomodule (18). The third Hyp-AG, designated Ala-Hyp polysaccharide-2, was isolated from (Ala-Hyp)51-GFP similar to Ala-Hyp-polysaccharide-1 described earlier (15). Here we identified the fundamental similarities between these Hyp-arabinogalactans and determined if the non-glycosylated domains (interferon α2 versus GFP) or AGP motifs (Ser-Hyp versus Ala-Hyp repeats) influenced the glycan structure. Significantly, the six-residue galactan backbone of these new Hyp-AGs consisted of two β-1,3-linked galactosyl trisaccharides connected by a β-1,6 linkage. Such “decorated” ∼15-residue trisaccharide subunits likely constitute the fundamental building blocks of type II arabinogalactan polysaccharides; hence, they are far less complex than commonly supposed (4, 19). Finally, the NMR analyses and molecular modeling of the glycans revealed major conformers that include a moderately compact folded structure.
EXPERIMENTAL PROCEDURES
Gene Construction and Expression in Tobacco Cells
Genes encoding Interferon α2-(Ser-Hyp)20 and (Ala-Hyp)51-GFP were constructed and expressed as described in detail earlier (12, 18). Briefly, proteins were targeted for secretion using a tobacco extensin signal sequence, and gene expression was under control of the 35 S cauliflower mosaic virus promoter. The genes were subcloned into the plant transformation vector pBI121 and expressed in tobacco Bright Yellow-2 cells selected and maintained as described earlier (12, 18).
Isolation of the Interferon α2-(Ser-Hyp)20 and (Ala-Hyp)51-GFP Fusion Glycoproteins
Interferon α2-(Ser-Hyp)20 and (Ala-Hyp)51-GFP were isolated from the medium of suspension-cultured tobacco Bright Yellow-2 cells by hydrophobic interaction chromatography on a phenyl-Sepharose column as described earlier (18).
Isolation of Hyp-arabinogalactans
Two hundred mg of Interferon α2-(Ser-Hyp)20 were hydrolyzed in 20 ml of 0.44 n NaOH solution at 108 °C for 18 h. The cooled solution was titrated to pH 7.8 with cold 1 n HCl and then freeze-dried. Hydrolysates were fractionated on an analytical Superdex-Peptide column (HR 10/30, GE Healthcare) equilibrated in 20% acetonitrile (aqueous) and eluted at a flow rate of 0.3 ml/min (15).
Fractions (0.6 ml total volume each) were freeze-dried and analyzed for Hyp and monosaccharides colorimetrically or by gas chromatography using methods described earlier (18). The fraction containing the most Hyp (fraction 16 described in Ref. 18, interferon Hyp-polysaccharide-1) and a fraction containing later-eluting Hyp-glycans (fraction 18, Ref 18, interferon Hyp-polysaccharide-2) were rerun on the Superdex column, freeze-dried, and then used for NMR analyses. The Hyp-glycan Ala-Hyp-polysaccharide-2 from (Ala-Hyp)51-GFP was isolated by a combination of cation exchange and gel filtration chromatography as described earlier (15, 20).
NMR Spectroscopy
A 1-mg sample of each Hyp-AG was dissolved in 0.5 ml of 99.996% D2O (Cambridge Isotope Laboratories, Andover, MA). NMR experiments were carried out either at 55 °C on a Bruker DMX-800 equipped with a cryoprobe or at 25 °C on a Bruker DMX-600 spectrometer equipped with a triple-resonance probe and three-axis gradient coils. The parallel data sets include one-dimensional 1H, two-dimensional 1H-homonuclear correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY) (mixing time 60 and 90 ms), rotating frame NOE spectroscopy (ROESY) (200 ms), and nuclear Overhauser effect spectroscopy (NOESY) (mixing time 150, 300 and 500 ms), and two-dimensional 13C,1H heteronuclear single quantum coherence (HSQC) and heteronuclear multiple bond coherence (HMBC) NMR spectra. In addition, several more interferon Hyp-polysaccharide-1 experiments were recorded in an effort to resolve assignment ambiguities such as magnitude COSY with one-, two-, and three-step relay transfer and two-dimensional 13C,1H heteronuclear HSQC-TOCSY and HSQC-NOESY together with diffusion-ordered spectroscopy for measuring the diffusion constant. Water suppression was achieved by either presaturation or WATERGATE techniques. Data were processed with NMRPipe (21) and visualized using NMRView (22) Chemical shifts were referenced to an external standard: 4,4-dimethyl-4-silapentane-1-sulfonic acid.
NMR Structure Calculations
Interferon Hyp-polysaccharide-1 was constructed in an arbitrary extended conformation using the LEaP module of Amber 10 (23). The starting model was subjected to a restrained simulated annealing conformational search protocol to obtain an ensemble of structures consistent with the NMR data. All assigned NOESY cross-peaks were classified as strong (1.8–2.7 Å), medium (1.8–3.7 Å), weak (1.8–5.0 Å), and very weak (1.8–6.0 Å) interproton distance restraints according to their intensities. Beyond these bounds, a quadratic penalty potential was applied with a force constant of 20 kcal mol−1 Å−2. A total of 49 distance restraints were used for interferon Hyp-polysaccharide-1 of which 34 were assigned non-ambiguously to protons in sequential residues. Cross-peaks that correspond to non-sequential assignments gave rise to ambiguous restraints where either more than one proton pair contributes to the NOESY volume or unambiguous assignment was not possible. Ambiguous peaks were interpreted as an < r−6 >−1/6 averaged value of the contributing interproton distances.
For a restrained molecular dynamics conformational search, initial models were energy-minimized without restraints and subjected to 200 simulated annealing cycles. A cycle started from the structure obtained in the previous cycle and included 10-ps heating from 300 to 1000 K followed by an equilibration of 10 ps at 1000 K without any restraints applied. The force constants for all restraints were then scaled gradually from 0 to the final values during 20 ps at 1000 K followed by cooling the system to 300 K over 30 ps. Atomic interactions within the system were calculated using the Glycam06 parameter set for sugars (24) and Generalized Born solvation (igb = 2) with monovalent salt concentration corresponding to 0.1 m. The last structures in each cycle were energy-minimized using the same parameters and restraints as described above. Ten best models for an average structure were selected based on NMR restraint violations and the potential energy of the molecule to undergo further refinement in explicit water and counterion environment. Each of these model structures was placed in a truncated octahedral box of about 5000 TIP3P water molecules and two K+ counterions to neutralize the total charge. In one case we used Ca2+ to neutralize the charge of the uronic acids. Parameters related to water and counterions were taken from the standard Amber libraries. The system was energy-minimized and then heated to 300 K at constant volume during 50 ps, whereas the solute was kept under positional restraints with a force constant of 25 kcal mol−1 Å−2. The positional restraints were gradually removed over 300 ps at constant pressure (1 atm) and temperature (300 K), and a production phase was initiated for 2 ns with the full set of restraints applied. The final structures were energy-minimized and used for subsequent analysis. The hydrodynamic radius was calculated for the model structures using HYDROPRO Version 7c2 (25).
RESULTS
The Structures of Interferon Hyp-polysaccharide-1, interferon Hyp-polysaccharide-2, and Ala-Hyp-polysaccharide-2 were determined based on earlier composition analyses and on the chemical shifts (15, 26) observed in one-dimensional 1H NMR spectra and two-dimensional COSY, TOCSY, HSQC, and HMBC NMR spectra as follows:
Primary Structure of Interferon Hyp-polysaccharide-1
Size-fractionated base hydrolysates of Interferonα2-(Ser-Hyp)20 yielded a single peak containing sugar and Hyp residues, described earlier (26). Two subfractions were chosen for structural analyses; one contained the major Hyp-AG species, designated Interferon Hyp-polysaccharide-1, with 22 glycosyl residues estimated by the Hyp to monosaccharide molar ratios, and a second fraction contained a smaller, less abundant Hyp-arabinogalactan of 20 glycosyl residues, designated interferon Hyp-polysaccharide-2.
Sugar Composition and Configuration
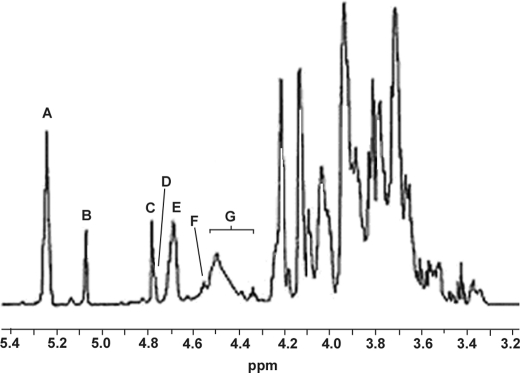
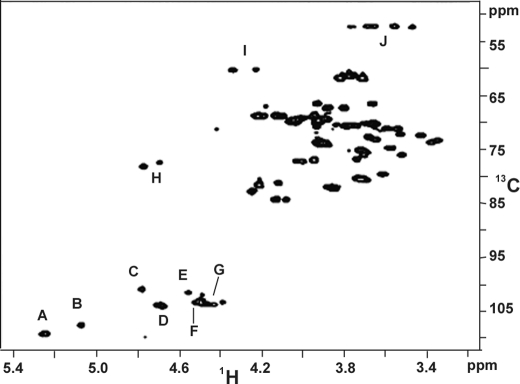
The integrated areas in the anomeric region in a one-dimensional 1H NMR spectrum (Fig. 1) corroborated the molar ratios Hyp Gal10 Ara8 GlcU2 Rha2 of interferon Hyp-polysaccharide-1 (26), confirming interferon Hyp-polysaccharide-1 as a 22-residue arabinogalactan attached to Hyp. The anomeric configurations were confirmed by the corresponding anomeric carbon signals in the HSQC spectrum (Fig. 2, supplemental Table I). Interferon Hyp-polysaccharide-1 contained a total of eight α-l-Araf residues; 6 of the 8 α-Araf residues were either terminal, 1,5-linked, or 1,3-linked (∼5.25 ppm), and 2 gave signals that were consistent with being both terminal and 1→ 5-linked to l-Araf residues (5.087 ppm). Interferon Hyp-polysaccharide-1 also contained 2 α-l-Rhap residues (4.797 ppm), 2 β-d-GlcUAp residues (∼4.5 ppm), and a total of 10 β-d-Galp residues; 1 was O-linked to Hyp (4.57 ppm), 5 were consistent with a galactan backbone (∼ 4.70 ppm) (15), and 4 others that produced signals typical of side-chain β-d-Galp residues (4.40–4.54 ppm). The chemical shift at 4.786 ppm identified 4-H of Hyp.
FIGURE 1.
1H NMR spectrum of Hyp-AG interferon Hyp-polysaccharide-1 at 55 °C. Signals A and B were assigned respectively to H-1 of six and two α-l-Ara residues, signal C to H-1 of two α-l-Rha residues, signal D (shoulder peak of C) to H-4 of Hyp, and signal E to the five β-d-Gal residues of the galactan backbone. Signal F was assigned to H-1 of β-d-Gal linked to Hyp and signal G to H-1 of the four side chain β-d-Gal residues and two β-d-GlcUA residues.
FIGURE 2.
HSQC spectrum of Hyp-AG Interferon Hyp-polysaccharide-1 at 55 °C. Cross-peaks A and B were assigned to H-1/C-1 of α-l-Ara, C to H-1/C-1 of α-l-Rha, D to the H-1/C-1 of β-d-Gal residues of the galactan backbone, and E to H-1/C-1 of β-d-Gal linked to Hyp. Cross-peak F was assigned to H-1/C-1 of β-d-GlcUA residues, G to H-1/C-1 of side-chain β-d-Gal, H to H-4/C-4 of Hyp, I to H-2/C-2 of Hyp, and J to H-5/C-5 of Hyp.
The Interferon Hyp-polysaccharide-1 Hyp-galactose Linkage
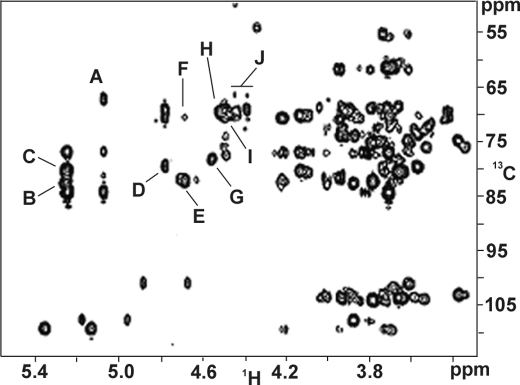
Signals arising from the Hyp residue were also identified in the TOCSY (supplemental Fig. 1) and HSQC spectra (Fig. 2). The chemical shifts are shown in supplemental Table I. The H-4 and C-4 resonances characteristic of non-glycosylated Hyp shifted downfield from 4.62 and 70.6 to 4.786 and 78.02 ppm, respectively, judging from the HSQC spectrum. This indicated that the hydroxyl group of Hyp was galactosylated (15, 18). The HMBC spectrum (Fig. 3) confirmed this in cross-peak G, which arose from C-4 of Hyp (78.02 ppm) and H-1 of a β-d-Galp residue (4.57) ppm, designated G1 in Fig. 4 and supplemental Table I.
FIGURE 3.
HMBC spectrum of Hyp-AG interferon Hyp-polysaccharide-1 at 55 °C. This helped identify the interferon Hyp-polysaccharide-1 monosaccharide sequence. Cross-peak A correlated Ara H-1 with Ara C-5, cross-peak B correlated Ara H-1 with Ara C-3, cross-peak C correlated Ara H-1 with side-chain Gal C-3, cross-peak D correlated Rha H-1 with GlcUA C-4, cross-peak E correlated backbone Gal H-1 with backbone Gal C-3, cross-peak F correlated backbone Gal H-1 with backbone Gal C-6, cross-peak G correlated G1 H-1 with Hyp C-4, cross-peak H correlated GlcUA H-1 with side-chain Gal C-6, and cross-peaks I and J correlated side-chain Gal H-1 to backbone Gal C-6.
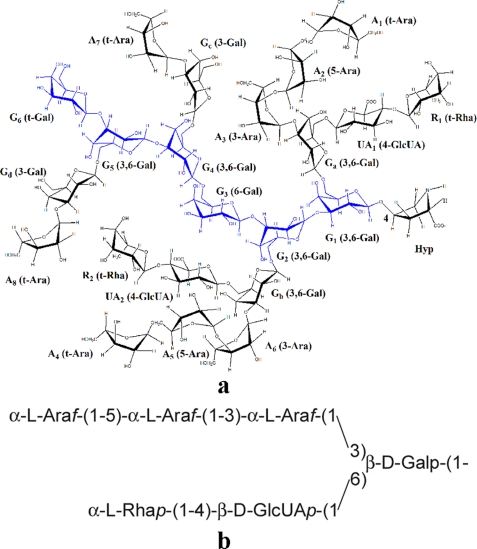
FIGURE 4.
Interferon Hyp-polysaccharide-1 and its canonical side chain. a, the primary structure of the 22-residue Hyp-arabinogalactan Interferon Hyp-polysaccharide-1 is shown. Residue labeling includes the linkage of each residue and corresponds to that featured in supplemental Table I. The Hyp residue is positioned to the right, beneath rhamnose residue R1. b, the canonical bifurcated AG side chain is shown.
Galactan Backbone and Side Chain Gal Residues
In addition to G1 linked to Hyp, there were another five β-d-Galp residues in the interferon Hyp-polysaccharide-1 galactan backbone (designated G1–6 in supplemental Table I and Figs. 4 and 5) judging by H-1 resonances at ∼4.70 ppm in the one-dimensional 1H spectrum. Four of the five backbone Gal residues were 3-linked to each other and to G1, as deduced from cross-peaks E in the HMBC spectrum (Fig. 3), which correlated backbone Gal H-1 signals with backbone Gal 3-C signals. A fifth backbone Gal participated in a 1→ 6 linkage to another backbone Gal residue deduced from cross-peak F in the HMBC spectrum. Thus, based on our earlier work (15) and the NMR spectra presented here, the interferon Hyp-polysaccharide-1 galactan backbone had the structure β-d-Galp-(1→3)-β-d-Galp-(1→3)-β-d-Galp-(1→6)-β-d-Galp-(1→3)-β-d-Galp-(1→3)-β-d-Galp-(1→4)-O-Hyp.
FIGURE 5.
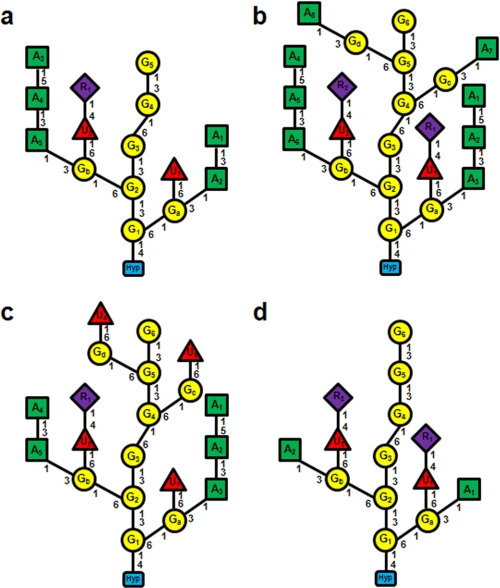
Comparison of the primary structures of Hyp-AGs. a, Ala-Hyp-polysaccharide-1 (15). b, interferon Hyp-polysaccharide-1. c, interferon Hyp-polysaccharide-2. d, Ala-Hyp-polysaccharide-2. The yellow circles indicate β-d-Galp residues, the green squares indicate α-l-Araf, the purple diamonds indicate α-l-Rhap, the red triangles indicate β-d-GlcUAp, and the blue blocks indicate Hyp. The residue labels inside the shapes (A3, G1, etc.) correspond to the residues described under “Results,” Fig. 4, and the supplemental Tables I–V.
The final 4 of the 10 Gal residues of interferon Hyp-polysaccharide-1 occurred in side chains (designated Ga, Gb, Gc, Gd in supplemental Table I and Figs. 4 and 5) linked 1→6 to the backbone Gal residues. This was deduced from one-dimensional 1H and two-dimensional 13C,1H HMBC spectra. Resonances at 4.40–4.54 ppm in the one-dimensional 1H NMR spectrum were consistent with four Gal side chains attached to the galactan backbone (15), and cross-peaks I and J in the HMBC spectrum indicated that the four side-chain Gal residues were 1→6 linked to backbone Gal residues.
Interferon Hyp-polysaccharide-1 Side-chain Composition and Linkages
The two α-l-Rha residues, designated R1 and R2, were terminal, deduced by a comparison of their assigned chemical shifts (supplemental Table I) obtained from TOCSY (supplemental Fig. 1) and HSQC spectra (Fig. 2) with those of earlier characterized Ala-Hyp-polysaccharide-1 (15). Rhamnose residues R1 and R2 were linked to O-4 of β-d-GlcUAp (UA1 and UA2 of supplemental Table I), deduced from cross-peak D in the HMBC spectrum (Fig. 3). Cross-peak H in the HMBC spectrum indicated side-chain Gal residues were substituted at O-6 with glucuronic acid residues UA1 and UA2. The same chemical shifts arising from α-l-Rhap, β-d-GlcUAp, and side-chain β-d-Galp residues were identified earlier on side-chain Gal residues in Ala-Hyp-polysaccharide-1 (15). This indicated the side chains were attached to backbone Gal residues nearest Hyp; therefore, we assigned the two Rha-(1→4)-GlcUA subunits to side-chain Gal residues closest to Hyp, Ga, and Gb (supplemental Table I and Figs. 4 and 5) to give two side chains: α-l-Rhap-(1→4)-β-d-GlcUAp-(1→6)-β-d-Ga and -Gb.
The Ara residues occurred in small side chains that were 3-linked to the side-chain Gal residues. The HMBC spectrum cross-peak A identified Ara 5-linked to another Ara (Fig. 3). It arose from H-1 of α-l-Araf residues (5.087 ppm, A1 and A4 in Fig. 4a) and 5-C of other Ara residues (67.1 ppm, A2 and A5 in Fig. 4a) This was consistent with the one-dimensional 1H NMR spectrum that indicated there were two Ara-(1→5)-Ara linkages in interferon Hyp-polysaccharide-1. HMBC signals arising from the ring carbon atoms of A1 and A4 (C-2, C-3, and C-4) showed that they were terminal residues (15); hence, two diarabinosyl structures occurred having the structure α-l-Araf-(1→5)-α-l-Araf-(1→).
The HSQC C-1/H-1 signals at 109.2 ppm and ∼5.25 ppm arose from six Ara residues (A2, A3, A5, A6, A7, A8 in supplemental Table I, Figs. 4 and 5) and indicated two other types of linkages in the arabinose side chains. Cross-peak B in the HMBC spectrum indicated linkage of α-l-Araf to O-3 of another α-l-Araf residue (3-C signal at 83.88 ppm) as in the structure α-l-Araf-(1→3)-α-l-Araf (i.e. A2 to A3 and A5 to A6), whereas cross-peak C indicated α-l-Araf-(1→3)-β-d-Gal linkages (side-chain Gal residues Ga-d in supplemental Table I). The side-chain Gal H-3 chemical shifts (∼3.73 ppm) (15) present in the TOCSY spectrum (supplemental Fig. 1) indicated each of the four side-chain Gal residues was O-3 substituted. As four Ara residues were attached to Gal, and the other four occurred in two α-l-Araf-(1→5)-α-l-Araf-(1→ units, we concluded that two of the arabinosyl side chains had the structure α-l-Araf-(1→5)-α-l-Araf-(1→3)-α-l-Araf-(1→3)-Galsc, and two had the structure α-l-Araf-(1→3)-Galsc.
A comparison of the interferon Hyp-polysaccharide-1 HSQC and HMBC spectra with those of Ala-Hyp-polysaccharide-1 (15) showed that the interferon Hyp-polysaccharide-1 anomeric proton/carbon chemical shifts arising from Gc and Gd (∼103.6/4.447 and 103.2/4.41 ppm), the side-chain Gal residues furthest from Hyp in Fig. 4, differed from the other side-chain Gal residues, Ga and Gb (both ∼103.4/4.50). Furthermore, the signals from Ga and Gb in Interferon Hyp-polysaccharide-1 were identical to those of Ala-Hyp-polysaccharide-1 characterized earlier (15). Therefore, we designated the specific side-chain Gal residues in the two α-l-Araf-(1→5)-α-l-Araf-(1→3)-α-l-Araf-(1→3)-Gal units as Ga and Gb. They were part of the two bifurcated six-residue side chains of Interferon Hyp-polysaccharide-1. The Gal residues in the two α-l-Araf-(1→3)-side-chain Gal units were designated Gc and Gd (supplemental Table I, Figs. 4 and 5).
Interferon Hyp-polysaccharide-1 Long-range Interactions
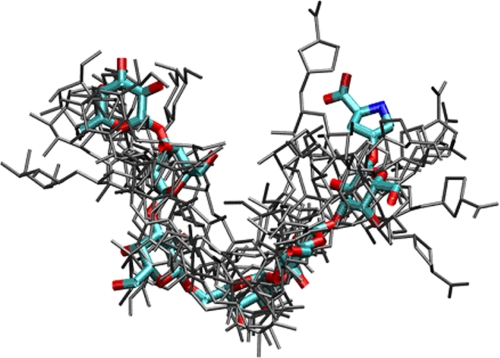
Lowering the temperature of NMR analyses from 55 to 25 °C (see the chemical shift assignments in supplemental Table II) provided the following lines of evidence for a folded Interferon Hyp-polysaccharide-1 conformer (Fig. 6).
FIGURE 6.
Molecular model of Interferon Hyp-polysaccharide-1. A range of galactan backbone conformations (in gray) is consistent with the experimental NOE data. A representative conformation is shown in color with the terminal Hyp residue upper right.
A diffusion-ordered spectroscopy spectrum (supplemental Fig. 2) gave a diffusion coefficient of (1.58 ± 0.1) ×10−10 m2/s. Using the Stokes-Einstein equation, this value corresponds to a hydrodynamic radius of 15.6 ± 1.0 Å, which is consistent with a globular folded structure and close to the calculated value for model structures of 13.9 ± 0.3 Å (see below).
The intensity of NOEs arising from protons between sequential O-linked sugar residues, for example H-1 of R1 and H-4 of glucuronic acid residue UA1 (supplemental Table III and Fig. 3) suggested a somewhat restricted conformation around glycosidic links rather than free rotation. Furthermore, the single set of chemical shifts rules out the existence of several stable conformations.
Although the major spectral region was not amenable to unequivocal analysis due to considerable resonance overlap, a region possessing unique chemical shifts showed two NOE clusters at ∼5.25/4.50 and ∼5.25/4.70 ppm. The first cluster included three NOEs indicating long-range interactions; (i) δ 5.241/δ 4.526 attributed to H-1 of Ara residue A7 or A8 and H-1 of Gal residue Ga; (ii) δ 5.233/δ 4.500 attributed to H-1 of A7 or A8 and H-1 of Gb, UA1, or UA2; (iii) δ 5.262/δ 4.511 attributed to H-1 of Ara A3 or A6 and H-1 of UA1 or UA2. NOEs (i) and (ii) indicate a side chain Ara of the second trigalactosyl unit is close to the side-chain Gal/UA residues of the first trigalactosyl unit; this suggests that the β-1–6 linkage between two trigalactosyl units allows the main chain to fold. The second cluster also included diagnostic NOEs; (i) 5.237/4.694 ppm attributed to H-1 of Ara A7 or A8 and H-1of Gal G2; (ii) 5.265/4.700 ppm attributed to H-1 of A3 or A6 and H-1 of G4. The intensity of these NOEs indicates a distance of 6 Å between the respective protons that suggests the molecule is folded. The possible effect of spin diffusion can be excluded because these NOEs could be observed with a short mixing time of 150 ms.
Primary Structure of Interferon Hyp-polysaccharide-2
Neutral sugar, uronic acid, and Hyp analyses of interferon Hyp-polysaccharide-2 gave a molar ratio of Hyp Gal10 Ara5 GlcUA4 Rha. The interferon Hyp-polysaccharide-2 1H NMR spectrum (supplemental Fig. 4) gave a very similar signal pattern in the anomeric proton region as interferon Hyp-polysaccharide-1, except the peak area ratios differed, as interferon Hyp-polysaccharide-2 contained two more GlcUA and one less Rha and only five Ara residues. Of the five α-Araf residues, only one (anomeric proton signal at ∼5.08 ppm) was 5-linked to another Ara; the other four (anomeric proton signal at ∼5.24 ppm) were terminal, 1,3-linked, or 1,5-linked. The α-Rhap residue was terminal, and of the 10 β-Galp residues, 6 were backbone Gal residues. Five of the backbone Gal residues gave H-1 signals at 4.68–4.71 ppm, and the sixth, G1 in supplemental Table IV, was linked to Hyp. The other four Gal residues were part of the side chains (4.39–4.49 ppm). The H-1 signals of β-GlcUAp were not resolved from those of the side-chain Gal; however, the signal peak integral combined with chemical analyses of interferon Hyp-polysaccharide-2 indicated it had four GlcUA residues. Together, the 1H NMR spectrum and the sugar analyses indicated interferon Hyp-polysaccharide-2 was a 20-sugar residue Hyp-arabinogalactan (Fig. 5c). We assigned the chemical shifts of interferon Hyp-polysaccharide-2 (supplemental Table IV) using two-dimensional TOCSY (supplemental Fig. 5), HSQC (supplemental Fig. 6), and HMBC (supplemental Fig. 7) spectra discussed below.
Hyp-Gal Linkage and Galactan Backbone
Interferon Hyp-polysaccharide-2 had the same galactan backbone structure as interferon Hyp-polysaccharide-1. The Hyp-Gal linkage was established by cross-peak G in the HMBC spectrum (supplemental Fig. 7). Like interferon Hyp-polysaccharide-1, the galactan backbone of interferon Hyp-polysaccharide-2 was composed of five β-d-Galp residues (G2-G6 in supplemental Table IV) and six Gal linked to Hyp (G1). Cross-peak E indicated the backbone Gal residues were mainly 1→3-linked, although a 1→6 link occurred between G3 and G4 (cross-peak F in supplemental Fig. 7 and Fig. 5c).
Interferon Hyp-polysaccharide-2 Side Chains
The side-chain structures were similar to those of interferon Hyp-polysaccharide-1, but generally smaller. The four side-chain Gal residues evident in the one-dimensional 1H NMR spectrum were attached to backbone Gal residues through the O-6 position, deduced from cross-peaks I and J in the HMBC spectrum (supplemental Fig. 7) (side-chain Gal H-1 at 4.39–4.49 ppm to backbone Gal 6-C at ∼70.0 ppm). Interferon Hyp-polysaccharide-2 had only one terminal α-l-Rhap but four β-d-GlcUAp residues. Cross-peak D in the HMBC spectrum indicated an α-l-Rhap-(1→4)-β-d-GlcUAp unit, and the chemical shifts of the other GlcUA residues (supplemental Table IV) indicated they were unsubstituted and, therefore, terminal. Cross-peak H showed that all four GlcUA residues were β-1→ 6-linked to side-chain Gal residues. In Fig. 5c, we assigned the Rha-(1→4)-GlcUA unit to one of the two side-chain Gal residues closest to Hyp, Gb, based on earlier work with Ala-Hyp-polysaccharide-1 (15); however, we had no direct evidence for this assignment, and any one of the four side-chain Gal residues were candidates.
HMBC cross-peaks A, B, and C (supplemental Fig. 7) corresponded to the following side chain arabinosyl units: α-l-Ara-(1→5)-α-l-Ara, α-l-Araf-(1→3)-α-l-Araf, and α-l-Araf-(1→3)-Gal (15). These units were part of two arabinosyl side chains, α-l-Ara-(1→5)-α-l-Ara-(1→3)-α-l-Araf-(1→3 and α-l-Araf-(1→3)-α-l-Araf-(1→3, that we assigned to side-chain Gal residues Ga and Gb rather than Gc and Gd because the H-3 resonances (supplemental Table IV, Fig. 5c) in the TOCSY spectrum (supplemental Fig. 5) indicated residues Gc and Gd were unsubstituted at O-3. However, we could not discern the precise distribution of the diarabinosyl and triarabinosyl units between Ga and Gb.
Primary Structure of Ala-Hyp-polysaccharide-2
Ala-Hyp-polysaccharide-2 derived from base-catalyzed hydrolysates of (Ala-Hyp)51-enhanced green fluorescence protein was a 14-sugar residue Hyp-AG determined from the peak areas of its one-dimensional 1H NMR spectrum (not shown) and sugar/Hyp analyses corresponding to Hyp1 Gal8 Ara2 GlcUA2 Rha2, one residue less than Hyp1 Gal7 Ara5 GlcUA2 Rha of Ala-Hyp-polysaccharide-1 (15). The chemical shifts of Ala-Hyp-polysaccharide-2 (supplemental Table V) were assigned based on HSQC (supplemental Fig. 8) and HMBC (supplemental Fig. 9) spectra compared with those of Ala-Hyp-polysaccharide-1.
Ala-Hyp-polysaccharide-2 Hyp-Gal Linkage and Galactan Backbone
Cross-peak G of Ala-Hyp-polysaccharide-2 in the HMBC spectrum established the linkage of β-d-Galp (G1) to position 4 of Hyp (Fig. 5c). G1 with five other backbone β-d-Galp residues comprised the galactan backbone, confirmed by the four β-d-Galp-(1→3)-Gal linkages evident in cross-peak E and by a single β-d-Galp-(1→6)-Gal linkage (cross-peak F). Thus, Ala-Hyp-polysaccharide-2 had one more backbone Gal residue than Ala-Hyp-polysaccharide-1.
Ala-Hyp-polysaccharide-2 Side Chains
Like Ala-Hyp-polysaccharide-1, interferon Hyp-polysaccharide-1, and interferon Hyp-polysaccharide-2 side chain Gal residues (Ga and Gb) of Ala-Hyp-polysaccharide-2 were attached to the galactan backbone through 1–6 linkages, deduced from cross-peak I. These two Gal side chains were assigned to G1 and G2 (Fig. 5c, supplemental Table V), as the chemical shifts were like those already characterized in Ala-Hyp-polysaccharide-1 (15).
Like the other Hyp-AGs, the β-d-GlcUAp residues of Ala-Hyp-polysaccharide-2 O-4 substituted with terminal α-l-Rhap residues (cross-peak D in supplemental Fig. 9 and Table V). Furthermore, the GlcUA residues were O-6-linked to Galsc deduced from cross-peak H. Therefore, Ala-Hyp-polysaccharide-2 contained two side-chain units of α-l-Rhap-(1→4)-β-d-GlcUAp-(1→6)-β-d-Gal.
Judging by the C/H chemical shifts at δ 82.3/4.21 ppm in the HSQC spectrum (supplemental Fig. 8 and Table V), the two α-l-Araf residues were terminal residues. Cross-peak C in the HMBC spectrum indicated two side-chain units of α-l-Araf-(1→3)-β-d-Gal.
NMR Structure Calculations of the Hyp-AGs
Model structures of interferon Hyp-polysaccharide-1 were generated by simulated annealing, and the 10 best models were further refined in explicit water and ion environment. These models, consistent with NOE distance information obtained at 25 °C (supplemental Table III), depict an overall folded structure with backbone residues Gal-6 and Gal-1 in proximity forming a sharp bend or “reverse turn” formed by the β-1,6-link between repetitive β-1,3-linked trigalactosyl subunits. We note that some conformational flexibility was seen in the NMR ensemble without violating the experimental distance information (Fig. 6). Significantly, ∼10 intramolecular H-bonds stabilized interactions in which uronic acid carboxyls appeared close enough to chelate Ca2+, a possibility supported by NOE-restrained molecular dynamics simulation of interferon Hyp-polysaccharide-1 in explicit water and Ca2+ (supplemental Fig. 10). The chelated ion remained strongly bound to the uronic acids without violating the available NOE distance data throughout the simulation. As an additional test of the global properties of the structural models, we calculated the hydrodynamic radius of the structures as 13.9 ± 0.3 Å.
DISCUSSION
Complete elucidation of type II arabinogalactan structure has been a major goal since the isolation of Hyp-AGs more than 30 years ago (5, 6). Churms et al. (16, 27, 28) notably suggested a repetitive structure with “AG substituents showing blocks of 1,3–1inked galactan backbone interrupted by periodate susceptible residues (kinked region).” The structure of Ala-Hyp-polysaccharide-1 supported the repetitive subunit hypothesis and also suggests that the arabinogalactan structure is highly conserved in both classical AGPs and the less abundant chimeric glycoproteins of the cell surface (29). As Ala-Hyp-polysaccharide-1 was only the first Hyp-AG (15), we characterized additional Hyp-AGs to test the possibility that AGs are highly varied structures dictated by regional peptide sequence and too complex for structural elucidation. We expressed the two most frequently occurring AGP motifs Ser-Hyp and Ala-Hyp in tobacco Bright Yellow-2 cells as chimeric fusion glycoproteins with non-glycosylated partners interferon α2b and green fluorescence protein, respectively. This allowed comparison of Hyp-AGs isolated from Ser-Hyp and Ala-Hyp repeats in the fusion proteins interferon α2-(Ser-Hyp)20 (15, 30) and (Ala-Hyp)51-GFP.
Interferon α2-(Ser-Hyp)20 yielded Hyp-AGs that ranged from 18 to 26 residues. Interferon Hyp-polysaccharide-1 was the largest characterized and contained 22 residues corresponding to the composition Gal10 Ara8 GlcUA2 Rha2. Hyp-AGs isolated from (Ala-Hyp)51-GFP were marginally smaller and somewhat more disperse (14–25 residues). Significantly, all the newly characterized Hyp-AGs (Fig. 5) had a six-residue galactan main chain consisting of two β-1,3-linked trigalactosyl blocks linked by a β-1,6 linkage. Ala-Hyp-polysaccharide-1 also shared this main-chain structure except that it lacked the sixth (terminal) Gal residue (15).
The side-chain substituents linked to the galactan main chain were similar in position and composition. The small Hyp-AGs, Ala-Hyp-polysaccharide-1, and Ala-Hyp-polysaccharide-2 contained only two side chains attached to main-chain Gal residues G1 and G2 (15) and differed from each other only in their Rha and Ara content (Fig. 5). In contrast, the larger Hyp-AGs, interferon Hyp-polysaccharide-1 and interferon Hyp-polysaccharide-2 each had four side chains ranging from two to six glycosyl residues linked to G1, G2, G4, and G5 (Figs. 4 and 5). These three new Hyp-AGs and Ala-Hyp-polysaccharide-1 shared an ∼15-residue subunit consisting of a repetitive trisaccharide with two bifurcated acidic side chains (Figs. 4–6), each with a maximum of six residues; these side chains are attached to C-6 of G1 and G2, i.e. the first and second Gal residues, numbered from the reducing end of the galactan backbone.
This relatively invariant Hyp-AGs structure with bifurcated side chains is apparently widespread; it is consistent with compositional data from diverse species (16, 27, 28) and with the fact that AGPs from diverse species selectively co-precipitate with the β-Yariv reagent. The six-residue side chain of interferon Hyp-polysaccharide-1 is identical with the gum arabic side chain in the legume Acacia senegal (31) except for the addition of terminal 5-linked Araf in interferon Hyp-polysaccharide-1. Furthermore, neither the type of Hyp-AGs peptide motif (Ser-Hyp versus Ala-Hyp) nor the attached non-glycosylated domain (interferon versus GFP) affected the composition or structure of this 15-residue glycan subunit, again consistent with a highly conserved structure. The “type II” (i.e. β-1,3-linked) Hyp-AGs represented here by interferon Hyp-polysaccharide-1, interferon Hyp-polysaccharide-2, Ala-Hyp-polysaccharide-2 (Figs. 4 and 5), and Ala-Hyp-polysaccharide-1 (15) confirm that tobacco Bright Yellow-2 cell Hyp-AGs consist of small ∼15-residue subunit repeats with a common composition and linkage pattern differing mainly in the number of trigalactosyl subunits decorated with two side chains each composed of up to 6 residues. Thus, larger Hyp-AGs of up to 150 sugar residues (5, 6) may consist of ∼10 repetitive subunits. There are variations on the major 15-residue theme. For example, some Arabidopsis Hyp-AGs lack rhamnose (32) and likely contain fucose; furthermore, some monosaccharide residues may be modified by acetylation (33) or 4-O-methylation (34).
Although interferon Hyp-polysaccharide-1, interferon Hyp-polysaccharide-2, and Ala-Hyp-polysaccharide-2 each had a complete 15-residue subunit, i.e. a galactose trisaccharide with 6-residue bifurcated side chains, these Hyp-AGs also contained incomplete subunits with truncated side chains, for example (α-l-Ara-(1→3)-Gal) in interferon Hyp-polysaccharide-1 and (β-d-GlcUA-(1→6)-Gal) in interferon Hyp-polysaccharide-2. These variations, particularly the incomplete Ala-Hyp-polysaccharide-1 Gal backbone, argue for biosynthesis of Hyp-AGs via stepwise saccharide addition to an AGP polypeptide or alternatively en bloc transfer of incomplete lipid-linked arabinogalactan intermediates (15) or even sugar trimming after transfer, although compelling evidence for degradative turnover of AGPs has not yet been observed. The existence of AGP microheterogeneity like that shown here may account for notions of Hyp-AGs as intractably complex. However, a small Hyp-arabinogalactan containing only 4 different sugars and 7 different glycosidic linkages is a relatively simple structure compared, for example, with the complex rhamnogalacturonan-II pectic polysaccharide with 12 different sugar residues linked by more than 20 different glycosidic linkages (35).
Signaling functions and roles as determinants of cell fate dominate current AGP discussion. However, the location of classical AGPs at the cell surface and their sheer physical abundance (3) predict primarily structural functions. Structural conservation implies that Hyp-AGs play similar conserved roles in both classical AGPs and AGP chimeras (3, 29). Nevertheless, despite considerable speculation (4, 36), specific biological roles of classical AGPs and their Hyp-AGs remain to be elucidated. The ubiquity of AGPs at the cell surface and involvement of classical AGPs in numerous fundamental developmental processes is clear (3, 37–41), yet AGP function at molecular levels is relatively unexplored. The current structural elucidation including computer simulations support structural roles for classical AGPs as follows.
Exclusively β-1–3-linked galactans belong to the compact hollow helix polysaccharide family (42). However, interspersed β-1–6-linkages form “kinks” (17) that may be analogous to the classical reverse β-turns of polypeptides. A molecular model (Fig. 6) depicts interferon Hyp-polysaccharide-1 as a folded polysaccharide that forms a moderately compact spheroid consistent with both the NOE data and the hydrodynamic radius determined experimentally.
Modeling also revealed other conserved features of interferon Hyp-polysaccharide-1 that may contribute to its stability and molecular function. They include the general stabilizing role of ∼10 intramolecular H-bonds and shrinkage of conformational space (43) by the bulky bifurcated AG side chains; these limit the conformational AG landscape particularly when restrained even further in vivo by their attachment to the polypeptide backbone.
A conserved core structure for type II arabinogalactans suggests a conserved function and some speculations about the precise roles played by the Hyp-arabinogalactans that decorate naturally occurring AGPs and the numerous AGP chimeras that populate the plasma membrane-cell wall interface. Indeed, in tobacco cells complete coverage of the plasma membrane by classical AGPs is likely (3).
Although the sheer abundance of classical AGPs at the cell surface suggests they (and their glycans) are structural molecules, roles for AGPs in signal transduction might arise from the composition and presentation of residues at the periphery of the glycans, the regions that also exhibit microheterogeneity. The compact folded structure deduced here for interferon Hyp-polysaccharide-1 indicates that the side chains are readily available for homophilic and heterophilic interactions and the galactan backbone somewhat less so, especially near the reducing end of the polysaccharide where the larger side chains shield the galactan backbone.
For example, although all Hyp-arabinogalactans possess a galactan backbone with arabinosyl side chains, some lack the abundant uronic acid residues prevalent in the Hyp-arabinogalactans of tobacco (44, 45) where charge repulsions of the uronic acid residues may form a compression buffer at the cell surface analogous to animal proteoglycan compression buffers (15). On the other hand, the close proximity of glucuronic acid residues may favor Ca2+ chelation (supplemental Fig. 10), which may be relevant to processes involving calcium signaling (45). Finally, wall AGPs may play a role in regulating cell extension, perhaps by acting as pectic plasticizers (3). The structures described here may help test these hypotheses.
Supplementary Material
Acknowledgments
We gratefully acknowledge the School of Life Sciences and the University of Sussex. We are also grateful to Charles Cottrell of the Ohio State University Campus Chemical Instrument Center for contributions to the NMR analyses.
This work was supported, in whole or in part, by National Institutes of Health Grant 2-P41-RR05351-06 (to the Complex Carbohydrate Research Center). This work was also funded by United States Department of Agriculture Grants 2004-34490-14579), Herman Frasch Foundation Grant 526-HF02, The Ohio University Biomimetic, Nanoscience, and Nanotechnology Program Grant GC0013845), and by the Ohio University Molecular and Cellular Biology Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables I–V and Figs. 1–10.
- Hyp
- hydroxyproline
- AGP
- arabinogalactan-protein
- AG
- arabinogalactan
- GFP
- green fluorescent protein
- COSY
- homonuclear correlation spectroscopy
- TOCSY
- total correlation spectroscopy
- NOE
- nuclear Overhauser effect
- NOESY
- NOE spectroscopy
- HSQC
- heteronuclear single quantum coherence
- HMBC
- heteronuclear multiple bond coherence.
REFERENCES
- 1.Cannon M. C., Terneus K., Hall Q., Tan L., Wang Y., Wegenhart B. L., Chen L., Lamport D. T., Chen Y., Kieliszewski M. J. (2008) Proc. Natl. Acad Sci. 105, 2226–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamport D. T. (1970) Annu. Rev. Plant Physiol. 21, 235–270 [Google Scholar]
- 3.Lamport D. T., Kieliszewski M. J., Showalter A. M. (2006) New Phytol. 169, 479–492 [DOI] [PubMed] [Google Scholar]
- 4.Seifert G. J., Roberts K. (2007) Annu. Rev. Plant Biol. 58, 137–161 [DOI] [PubMed] [Google Scholar]
- 5.Pope D. G. (1977) Plant Physiol. 59, 894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamport D. T. A. (1977) in Recent Advances in Phytochemistry (Loewus F. A., Runeckles V. C. eds) pp. 79–115, Plenum Publishing Corp., New York [Google Scholar]
- 7.Lamport D. T. (1967) Nature 216, 1322–1324 [Google Scholar]
- 8.Lamport D. T., Miller D. H. (1971) Plant Physiol. 48, 454–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kieliszewski M., de Zacks R., Leykam J. F., Lamport D. T. A. (1992) Plant Physiol. 98, 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieliszewski M. J., Lamport D. T. A. (1994) Plant J. 5, 157–172 [DOI] [PubMed] [Google Scholar]
- 11.Kieliszewski M. J. (2001) Phytochemistry 57, 319–323 [DOI] [PubMed] [Google Scholar]
- 12.Tan L., Leykam J. F., Kieliszewski M. J. (2003) Plant Physiol. 132, 1362–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shpak E., Leykam J. F., Kieliszewski M. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14736–14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Z. D., Tan L., Showalter A. M., Lamport D. T., Kieliszewski M. J. (2002) Plant J. 31, 431–444 [DOI] [PubMed] [Google Scholar]
- 15.Tan L., Qiu F., Lamport D. T., Kieliszewski M. J. (2004) J. Biol. Chem. 279, 13156–13165 [DOI] [PubMed] [Google Scholar]
- 16.Churms S. C., Stephen A. M., Siddiqui I. R. (1981) Carbohydr. Res. 94, 119–122 [Google Scholar]
- 17.Fincher G. B., Stone B. A., Clarke A. E. (1983) Annu. Rev. Plant Physiol. 34, 47–70 [Google Scholar]
- 18.Xu J., Tan L., Goodrum K. J., Kieliszewski M. J. (2007) Biotechnol. Bioeng. 97, 997–1008 [DOI] [PubMed] [Google Scholar]
- 19.Serpe M. D., Nothnagel E. A. (1999) Adv. Bot. Res. 30, 207–289 [Google Scholar]
- 20.Tan L. (2003) O-Glycosylation Motifs in Arabinogalactan-Proteins, Ph.D. dissertation, Ohio University; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 22.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 23.Case D. A., Darden T. A., Cheatham T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Crowley M., Walker R. C., Zhang W., Merz K. M., Wang B., Hayik S., Roitberg A., Seabra G., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Mathews D. H., Seetin M. G., Sagui C., Babin V., Kollman P. A. (2008) AMBER 10, University of California, San Francisco., University of California, San Francisco [Google Scholar]
- 24.Kirschner K. N., Yongye A. B., Tschampel S. M., González-Outeiriño J., Daniels C. R., Foley B. L., Woods R. J. (2008) J. Comput. Chem. 29, 622–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García De La Torre J., Huertas M. L., Carrasco B. (2000) Biophys. J. 78, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J., Shpak E., Gu T., Moo-Young M., Kieliszewski M. J. (2005) Biotechnol. Bioeng. 90, 578–588 [DOI] [PubMed] [Google Scholar]
- 27.Churms S. C., Merrifield E. H., Stephen A. M. (1983) Carbohydr. Res. 123, 267–279 [DOI] [PubMed] [Google Scholar]
- 28.Churms S. C., Stephen A. M. (1984) Carbohydr. Res. 133, 105–123 [Google Scholar]
- 29.Borner G. H., Lilley K. S., Stevens T. J., Dupree P. (2003) Plant Physiol. 132, 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill J. M., Alewood P. F., Craik D. J. (2000) Eur. J. Biochem. 267, 4649–4657 [DOI] [PubMed] [Google Scholar]
- 31.Defaye J., Wong E. (1986) Carbohydr. Res. 150, 221–231 [Google Scholar]
- 32.Xu J., Tan L., Lamport D. T., Showalter A. M., Kieliszewski M. J. (2008) Phytochemistry 69, 1631–1640 [DOI] [PubMed] [Google Scholar]
- 33.Serpe M. D., Nothnagel E. A. (1995) Plant Physiol. 109, 1007–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsumuraya Y., Ogura K., Hashimoto Y., Mukoyama H., Yamamoto S. (1988) Plant Physiol. 86, 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Neill M. A., Ishii T., Albersheim P., Darvill A. G. (2004) Annu. Rev. Plant Biol 55, 109–139 [DOI] [PubMed] [Google Scholar]
- 36.Gaspar Y., Johnson K. L., McKenna J. A., Bacic A., Schultz C. J. (2001) Plant Mol. Biol. 47, 161–176 [PubMed] [Google Scholar]
- 37.Chaves I., Regalado A. P., Chen M., Ricardo C. P., Showalter A. M. (2002) Physiol. Plant. 116, 548–553 [Google Scholar]
- 38.Guan Y., Nothnagel E. A. (2004) Plant Physiol. 135, 1346–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahata K., Takeuchi M., Fujita M., Azuma J., Kamada H., Sato F. (2004) Plant Cell Physiol. 45, 1658–1668 [DOI] [PubMed] [Google Scholar]
- 40.Gens J. S., Fujiki M., Pickard B. G. (2000) Protoplasma 212, 115–134 [DOI] [PubMed] [Google Scholar]
- 41.Pennell R. I., Knox J. P., Scofield G. N., Selvendran R. R., Roberts K. (1989) J. Cell Biol. 108, 1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rees D. A. (1977) Polysaccharide Shapes, pp. 1–80, Chapman & Hall, London [Google Scholar]
- 43.Xia J., Daly R. P., Chuang F. C., Parker L., Jensen J. H., Margulis C. J. (2007) J. Chem. Theory Comput. 3, 1629–1643 [DOI] [PubMed] [Google Scholar]
- 44.Bacic A., Churms S. C., Stephen A. M., Cohen P. B., Fincher G. B. (1987) Carbohydr. Res. 162, 85–93 [Google Scholar]
- 45.Gane A. M., Craik D., Munro S. L., Howlett G. J., Clarke A. E., Bacic A. (1995) Carbohydr. Res. 277, 67–85 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.